Abstract
The Toxoplasma gondii phosphoinositide-specific phospholipase C gene (TgPI-PLC) was cloned, sequenced and expressed in Escherichia coli and its enzymatic characteristics were investigated. TgPI-PLC is present in the genome as a single-copy gene consisting of 22 exons interrupted by 21 introns, and encodes a polypeptide of 1097 amino acids with a predicted molecular mass of 121 kDa. In addition to the conserved catalytic X and Y domains, TgPI-PLC contains an apparent N-terminal PH domain, an EF hand motif and a C-terminal C2 domain. When compared with mammalian δ-type PI-PLC, TgPI-PLC has an additional extended N-terminus and two insertions in the region between the X and Y domains, with a 31–35% identity over the whole sequence. Recombinant TgPI-PLC, as well as the native enzyme obtained from crude membrane extracts of the parasite, was more active with phosphatidylinositol than with phosphatidylinositol 4,5-bisphosphate as substrate. Indirect immunofluorescence analysis using an affinity-purified antibody against TgPI-PLC revealed that this enzyme localizes in the plasma membrane of the parasites.
Keywords: calcium; inositol 1,4,5-trisphosphate; phosphatidyl-inositol 4,5-bisphosphate; phosphoinositide; phospholipase C (PLC); Toxoplasma gondii
Abbreviations: DAG, diacylglycerol; GPI, glycosylphosphatidylinositol; HFF, human foreskin fibroblast; IP3, D-myo-inositol 1,4,5-trisphosphate; IPTG, isopropyl β-D-thiogalactoside; NES, nuclear export signal; ORF, open reading frame; PBS-T, PBS containing 0.1% Tween 20; PH, pleckstrin homology; PI, phosphatidylinositol; PIP2, phosphatidylinositol 4,5-bisphosphate; PI-PLC, phosphoinositide-specific phospholipase C; TgPI-PLC, Toxoplasma gondii PI-PLC; rTgPI-PLC, recombinant TgPI-PLC; PLC, phospholipase C; RIPA, radioimmunoprecipitation analysis; SAG1, surface antigen 1; sulpho-NHS-biotin, sulpho-N-hydroxysuccinimidobiotin
INTRODUCTION
PI-PLCs (phosphoinositide-specific phospholipases C) catalyse the hydrolysis of PIP2 (phosphatidylinositol 4,5-bisphosphate) to IP3 (D-myo-inositol 1,4,5-trisphosphate) and DAG (sn-1,2-diacylglycerol) [1,2]. The products of this reaction function as second messengers in eukaryotic cells. The soluble IP3 stimulates release of Ca2+ from intracellular stores [1]. The membrane-resident DAG stimulates protein phosphorylation by activating various protein kinase C isoenzymes [2]. So far, 13 mammalian PI-PLC isoenzymes have been cloned, and they are divided into six classes (β-, γ-, δ-, ϵ-, ζ- and η-type) on the basis of their structure and activation mechanism [3]. It seems very likely that PI-PLC-δ evolved first, because every PI-PLC cloned so far from a non-mammalian species (for example Dictyostelium, yeast, higher plants, Chlamydomonas) is a δ-isoform [4]. Results from several laboratory groups [5–8] have suggested that, at least in yeasts, PI-PLC-δ is required for a number of nutritional and stress-related responses. It has also been postulated that PI-PLC-δ could have a role in differentiation of Dictyostelium discoideum [9]. Transcription of this PI-PLC-δ appears to be enhanced during cell aggregation, decreases during slug formation and increases in the culminating fruit body [9]. D. discoideum PI-PLC-δ is G-protein-coupled [10].
The knowledge of the functions of PI-PLCs in parasitic protozoa is very limited. In Trypanosoma cruzi, a novel lipid-modified PI-PLC [11] is involved in the differentiation of trypomastigote to amastigote forms [12] and has also been postulated to be involved in the removal of GPI (glycosylphosphatidylinositol)-anchored proteins [13]. Roles for a PI-PLC in both Toxoplasma gondii invasion and egress from the host cells have been proposed [14–16]. Ethanol, which is a potent trigger of Ca2+-stimulated microneme secretion [14], stimulated an increase in T. gondii IP3, implying that this second messenger may mediate intracellular Ca2+ release [15]. Consistent with this observation, xestospongin C, an IP3 receptor antagonist, inhibited microneme secretion and blocked parasite attachment and invasion of host cells [15]. It was proposed that T. gondii possesses an intracellular Ca2+ release channel with properties of the IP3/ryanodine receptor super-family [15]. A role for a T. gondii PI-PLC in parasite egress from the dying host has also been postulated on the basis of studies with the PI-PLC inhibitor U-73122 [16]. It was shown that permeabilized Toxoplasma-infected cells pre-incubated with U-73122, but not with the inactive analogue U-73343, prevented parasite egress in the presence of extracellular buffer, and that parasite egress depended on the intracellular Ca2+ increase stimulated by the decrease in the external K+ concentration [16].
In the present paper, we report the cloning, sequencing, and expression of a gene (TgPI-PLC) encoding a PI-PLC from T. gondii. The gene product (TgPI-PLC) was found to be associated with the plasma membrane. The enzyme is similar to δ-type PI-PLCs of mammals and plants, but, in contrast with other PI-PLCs, when tested under some specific conditions of Ca2+ and Mg2+ concentrations, it is more active against PI (phosphatidylinositol) than against PIP2.
MATERIALS AND METHODS
Cultures
T. gondii tachyzoites (RH and 2F1 strains) were cultivated in primary HFFs (human foreskin fibroblasts) maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% foetal calf serum (HyClone) and an antibiotic mixture of penicillin and streptomycin, at 37 °C and 5% CO2, and purified as described by Moreno and Zhong [17]. The transgenic 2F1 strain stably expressing the β-galactosidase gene was a gift from Dr L. David Sibley (Washington University School of Medicine, St. Louis, MO, U.S.A.).
Isolation of cDNA and genomic DNA clones of TgPI-PLC and sequencing
Using Trypanosoma cruzi PI-PLC (GenBank® accession number AF093565) to search the T. gondii genome database (http://ToxoDB.org/) [18], DNA contig sequences ranging from 100 to 300 bp were found to be highly similar to Trypanosoma cruzi PI-PLC. Specific primers against these shorter fragments were used to amplify T. gondii cDNA by PCR in a PTC-100 Programmable Thermal Controller (MJ Research), and resulted in an unique 1.3 kb PCR product, which subsequently was cloned into pCR 2.1-TOPO vector (Invitrogen Life Technologies) and sequenced. A BLAST search of the GenBank® database revealed that this 1.3 kb cDNA clone (Ta1.3) encoded a protein fragment with high identity with other known PLCs. Subsequently the Ta1.3 clone was used as a DNA probe to screen T. gondii tachyzoite cDNA and genomic libraries (both kindly provided by the AIDS Reference and Reagent Repository, U.S. National Institutes of Health, Bethesda, MD, U.S.A.). Approx. 2×106 plaques of the cDNA library were screened using [α-32P]dCTP-labelled Ta1.3. Three positive cDNA clones were obtained, and subsequent DNA sequencing confirmed that all three cDNA clones were identical. There was overlap of all isolated cDNA clones with the 3′ end of the Ta1.3 clone. To obtain the full-length cDNA sequence, 5′-RACE (rapid amplification of cDNA 5′ ends) was performed using a kit from Invitrogen Life Technologies, according to the manufacturer's instructions. Approx. 2×105 plaques of the genomic library were screened using the [α-32P]dCTP-labelled Ta1.3 clone at high stringency according to the manufacturer's instructions (Stratagene). Three positive plaques were isolated and confirmed to be identical by DNA sequencing. The whole genomic DNA was sequenced by primer walking. DNA sequencing was performed using a BigDye Terminator Cycle sequencing kit and a 373A DNA Automatic Sequencer (PerkinElmer Applied Biosystems) at the Biotechnology Center, University of Illinois at Urbana-Champaign. The whole cDNA sequence of TgPI-PLC was spliced with program EditView 1.0.1. The sequence alignment was performed using the ClustalW alignment program available at the Biology WorkBench 3.2 (http://workbench.sdsc.edu/).
Southern blot analysis
Total genomic DNA from tachyzoites was isolated by phenol extraction, digested with different restriction enzymes that cut at sites not contained within the coding region, separated on a 0.8% agarose gel and transferred on to nylon membranes. The blot was probed with a [α-32P]dCTP-labelled Ta1.3 probe using standard methods [19].
Expression and purification of recombinant TgPI-PLC (rTgPI-PLC)
The whole ORF (open reading frame) of TgPI-PLC was amplified using T. gondii cDNA with forward primer, 5′-GGCTAGCATGGAGAGACAGACGTCTTCG-3′, and reverse primer, 5′-GGCTAGCTCACACCAAGGCCCCCGGTGG-3′, in which the underlined nucleotides are the introduced NheI sites to allow TgPI-PLC to be cloned into the expression vector pET28a (Novagen). PCR was performed using FastStart Taq DNA polymerase (Roche Applied Science), and PCR products were inserted into the pCR 2.1-TOPO TA vector and subcloned into pET28a vector, which was linearized with NheI and dephosphorylated with bacterial alkaline phosphatase (Invitrogen Life Technologies). The recombinant construct TgPI-PLC/pET28a was an inframe fusion of TgPI-PLC with an N-terminal His6 tag encoded in the vector and confirmed by DNA sequencing. The recombinant construct TcPI-PLC/pET28a was transformed into E. coli BL21-CodonPlus(DE3)-RIPL strain (Stratagene) and the transformants were inoculated into 1 litre of Luria–Bertani broth medium supplemented with 30 μg/ml kanamycin, 50 μg/ml chloramphenicol and 75 μg/ml streptomycin at 37 °C. When the culture density reached a D600 of 0.5, the expression of the TgPI-PLC gene was induced by addition of 0.25 mM IPTG (isopropyl β-D-thiogalactoside) and incubated at 16 °C for 72 h.
Unless indicated otherwise, the rTgPI-PLC protein was purified according to the method of Ghosh et al. [20] using the resin pre-charged with Ni2+. All procedures were performed at 4 °C. Briefly, the induced cultures were harvested and washed twice with cold PBS. The cell pellets were resuspended in Buffer A (0.5 M NaCl, 20 mM Hepes, 5 mM imidazole/HCl and 25% glycerol, pH 7.9) and incubated on ice for 20 min after the addition of 10 mg/ml lysozyme. After sonication on ice (three times for 20 s each using a Branson Sonifier 450 at 15% amplitude with 30 s intervals), the lysate was incubated with DNase and RNase for 20 min with gentle shaking to reduce the lysate viscosity. After centrifugation at 20000 g for 30 min, the supernatant was loaded into the pre-wet His•Bind Quick 900 Cartridges (Novagen) with Buffer A and was sequentially washed with Buffer A and Buffer B (0.5 M NaCl, 20 mM Hepes, 60 mM imidazole and 25% glycerol, pH 7.9). The rTgPI-PLC was finally eluted with a linear gradient of imidazole (200–1000 mM) in 0.5 M NaCl, 20 mM Hepes and 25% glycerol, pH 7.9. The eluted fractions containing PI-PLC activity were pooled, dialysed, divided into aliquots and stored at −80 °C. The purity of rTgPI-PLC was determined by SDS/10% PAGE, and the protein concentration was determined using a protein assay agent (Bio-Rad Laboratories) using BSA as a standard.
Assay of enzyme activity of rTgPI-PLC
The purified rTgPI-PLC activity was measured by the release of water-soluble radioactivity from [3H]inositol-labelled PIP2 or PI. Briefly, the substrate solutions of cold PIP2 (Avanti Polar Lipids) and [inositol-2-3H(n)]PIP2 (PerkinElmer Life Sciences) or unlabelled PI (Sigma) and [myo-inositol-2-3H(n)]PI (PerkinElmer Life Sciences) in organic solvent were mixed, and the solvent was thoroughly evaporated to dryness under a stream of nitrogen. The standard assay employed a reaction mixture containing 15000–20000 c.p.m. of [3H]PIP2 or [3H]PI, 40 μM unlabelled PIP2 or PI, 0.1% sodium deoxycholate, 50 mM Hepes/HCl (pH 7.0), 0.2 mM dithiothreitol, 2.5 mM EGTA, 3 mM MgCl2, 0–10 mM CaCl2 and an appropriate amount of purified rTgPI-PLC protein to give a final volume of 50 μl. The free Ca2+ concentrations was obtained using a CaCl2/EGTA buffer and were calculated with the MaxChelator programme Webmaxc standard (http://www.stanford.edu/~cpatton/webmaxc/webmaxcS.htm). The reaction was allowed to proceed at 30 °C for 20 min and was stopped by the addition of 0.5 ml of chloroform/methanol/concentrated HCl (200:100:0.6, by vol.), followed by 0.15 ml of 5 mM EGTA in 1 M HCl. The samples were mixed well through vigorous vortex-mixing for 30 s, and subjected to centrifugation at 20000 g for 5 min at room temperature (25 °C). The upper phase (aqueous) was taken and was dissolved in 5 ml of a liquid-scintillation fluid Ecolume (ICN Biomedical). After mixing thoroughly, the samples were counted in a liquid-scintillation analyser TRI-CARB 2100TR (Packard Instrument Company).
Neomycin and compound 48/80 (Sigma) were freshly prepared in sterile distilled water. U-73122 (Calbiochem) was reconstituted in chloroform, divided into small aliquots, evaporated to dryness under a stream of nitrogen and frozen at −20 °C. The aliquots were reconstituted in ethanol before use.
SDS/PAGE and Western blot analyses
The protein samples were subjected to SDS/10% PAGE, and the electrophoresed proteins were transferred on to nitrocellulose membranes using a Bio-Rad transblot apparatus. Following transfer, the membrane blots were blocked with 3% fish gelatin (Sigma) in PBS-T (PBS containing 0.1% Tween 20) at 4 °C overnight. The blots were incubated with the 1:2000 dilution of purified guinea-pig anti-TgPI-PLC serum at room temperature for 1 h. The blots were washed with PBS-T four times for 15 min each. After incubation with a 1:10000 dilution of peroxidase-conjugated AffiniPure goat anti-guinea-pig IgG (H+L) antibody (Jackson ImmunoResearch Laboratories), they were washed four times for 15 min. The immunoblots were visualized on blue-sensitive X-ray films (Midwest Scientific) using an ECL® (enhanced chemiluminescence) detection kit (Amersham Biosciences) according to the manufacturer's instructions.
Generation and affinity-purification of antibodies
The epitope analysis of the whole TgPI-PLC protein sequence with Mac Vector program (Immunological Resource Center, University of Illinois at Urbana-Champaign) predicted that the 285-amino-acid polypeptide near the C-terminus corresponding to amino acids 613–907 (see Figure 1B) presented the highest antigenicity. To obtain the recombinant polypeptide antigen, the forward primer, 5′-GGAATTCCCGGGTCTGAGCATTTCTC-3′ (the underlined nucleotides are an EcoRI site) and reverse primer, 5′-GCTCGAGACGCGCGGGATTTCGCAT-3′ (the underlined nucleotides are an XhoI site) were used to amplify TgPI-PLC cDNA encoding amino acids 613–907 (see Figure 1C). The resulting PCR product was cloned into pET28a expression vector to create a partial TgPI-PLC–His6 fusion construct allowing purification of the recombinant protein on nickel–agarose columns. The recombinant plasmid was transformed into host E. coli BL21(DE3) (Novagen), and overexpression of recombinant protein was obtained through induction with 1 mM IPTG at 37 °C for 4 h. The expressed protein present in inclusion bodies was purified using His•Bind Quick 900 Cartridges under denaturing conditions (6 M urea) according to the manufacturer's instructions. The eluted recombinant protein was dialysed against 0.5 M NaCl and 0.1 M NaHCO3 (pH 8.3), and concentrated by Microcon centrifugal filter devices (Millipore). The purity of antigen protein was determined by SDS/10% PAGE.
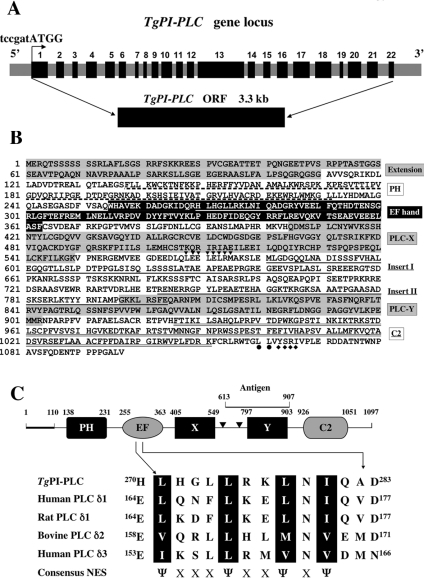
Figure 1. Organization and predicted protein sequence of TgPI-PLC.
(A) Schematic map of the TgPI-PLC gene locus. The relative position of exons in the genomic DNA is indicated by black boxes with numbers. The grey boxes show the 5′ end flanking region, the introns and the 3′ end flanking region. A Kozak translation initiation site (tccgatATGG, where the lower-case letters indicate untranslated sequence) is indicated above with a bent arrow. The isolated TgPI-PLC cDNA clone (5.5 kb) contains a 3.3 kb ORF. (B) Deduced amino acid sequence of the TgPI-PLC cDNA clone. The unique N-terminal extension and the catalytic X and Y domains are highlighted in grey. The additional two inserts between the X and Y domains are underlined. The consensus NES of TgPI-PLC (H270LHGLLRKLNIQAD283) within the EF hand is underlined in white. An additional putative leucine-rich NES site (L568EELELRM575) predicted using NetNES 1.1 (http://www.cbs.dtu.dk/services/NetNES/) is indicated by downward-pointing triangles downstream of the X domain. The dileucine (L1060L1061) and a mammalian sorting YXXΦ motif (Y1063SRI1066) is indicated with closed circles and diamonds respectively under the sequence at the C-terminal tail. (C) The numbers show the starting and finishing amino acid for each domain. The bold line indicates the N-terminal extension and two triangles represent the inserts in the X-Y domain linker. The peptide used to generate polyclonal antibody against TgPI-PLC (Antigen) is indicated above the Y domain. The protein size is not to scale. The consensus NES of the EF hand domain is compared with those of other δ-type PLCs. The important hydrophobic residues are in black boxes. Ψ represents a hydrophobic residue (isoleucine, valine or methionine); X represents any amino acid.
The polyclonal antibody against TgPI-PLC was generated in guinea-pigs by Cocalico Biologicals according to standard protocols. Briefly, guinea-pigs were subcutaneously injected with 100 μg of antigen emulsified in Freund's complete adjuvant after pre-serum was collected. At 2, 3, 7 and 11 weeks after the initial injection, guinea-pigs were boosted with 100 μg of antigen. The final bleed serum was divided into aliquots and was stored at −80 °C. The affinity-purification of anti-TgPI-PLC serum was performed using CNBr-activated matrices (Sigma). Briefly, 5 mg of purified TgPI-PLC antigen was coupled with CNBr-activated resin in coupling buffer (0.5 M NaCl and 0.1 M NaHCO3, pH 8.3). After blocking with 0.2 M glycine (pH 8.0) for 3 h, the resin was extensively washed with four cycles of coupling buffer and acetate buffer (0.1 M sodium acetate and 0.5 M NaCl, pH 4.0). The anti-TgPI-PLC serum was diluted in PBS and was incubated directly with the coupled resin overnight at 4 °C, and the bound antibody was eluted with 0.1 M glycine (pH 2.5–3.0) and 150 mM NaCl, and immediately neutralized with 1 M Tris/HCl (pH 8.0). The fractions with higher antibody activity were pooled, dialysed against PBS overnight at 4 °C using a Slide-A-Lyzer dialysis cassette (Pierce) and concentrated with a Microcon centrifugal filter device.
Immunofluorescence microscopy
To localize TgPI-PLC within intracellular parasites, indirect immunofluorescence assays were performed as described previously [21]. HFF monolayers grown on coverslips in six-well plates were infected with tachyzoites (RH strain; 1×105 cells per well) for 20 h at 37 °C. The coverslips were washed extensively with cold PBS to remove any unattached parasites, and the remaining serum was immediately fixed with 2.5% (w/v) formaldehyde and 0.02% (w/v) glutaraldehyde, freshly prepared in ice-cold PBS for 1 h. After blocking with PBS containing 1% fish gelatin, 5% goat serum, 3% BSA and 50 mM NH4Cl, pH 7.2, for 1 h, the parasites were permeabilized with PBS containing 0.05% saponin (Sigma) and 10% (v/v) foetal bovine serum for 30 min. After washing with PBS, the parasites were stained with affinity-purified guinea-pig anti-TgPI-PLC antibody (at a dilution of 1:200 in PBS containing 1% BSA), and incubated with mouse monoclonal antibody against T. gondii SAG1 (surface antigen 1) [22] (kindly provided by Dr David Alexander from Dr John Boothroyd's laboratory, Stanford University, Stanford, CA, U.S.A.) (at a dilution of 1:3000) for 1 h. After five washings with PBS, coverslips were incubated with FITC-conjugated AffiniPure goat anti-guinea-pig IgG (H+L) at a dilution of 1:200 and Alexa Fluor® 546-conjugated goat anti-mouse IgG (Molecular Probes) at a dilution of 1:1000. To view the parasite nucleus, coverslips were incubated with 0.1 μg/ml DAPI (4,6-diamidino-2-phenylindole) in PBS for 10 min. Slides were then mounted with ProLong gold anti-fade reagent (Molecular Probes). Confocal images were captured with a Leica laser-scanning confocal microscope (TCS SP2) using a 63× Plan-Apo objective with NA (numerical aperture) 1.32 (Beckman Institute, University of Illinois at Urbana-Champaign).
Cell-surface biotinylation and immunoprecipitation
Freshly lysed tachyzoites were harvested and washed three times with ice-cold PBS (pH 8.0) at 2000 g for 10 min. The cell pellets were immediately resuspended in 1 mM sulpho-NHS-biotin (sulpho-N-hydroxysuccinimidobiotin) (Pierce Biotechnology) freshly prepared in 1 ml of PBS (pH 8.0). After incubating the cells at room temperature for 30 min, they were washed five times with ice-cold PBS supplemented with 100 mM glycine (pH 8.0).
For immunoprecipitation, all procedures were carried out at 4 °C. The labelled cells were lysed in RIPA (radioimmunoprecipitation analysis) buffer (150 mM NaCl, 1% Nonidet P40, 0.5% deoxycholic acid, 50 mM Tris base, pH 7.5, 0.02% sodium azide and protease inhibitor mixture Set III from Sigma) on a rocking platform with gentle agitation for 1 h. The cell lysates were clarified by centrifugation at 20000 g for 20 min to remove debris, and the supernatants were pre-absorbed with Protein A–agarose (Roche Applied Science) overnight with gentle agitation. After centrifugation at 6000 g for 30 s, the supernatants were divided into three 0.4 ml aliquots in fresh tubes and immunoprecipitated for 4 h with guinea-pig polyclonal antibody against TgPI-PLC, mouse monoclonal antibody against T. gondii SAG1, and mouse monoclonal antibody against E. coli β-galactosidase respectively. Immunocomplexes were allowed to bind to Protein A–agarose overnight with gentle agitation. The agarose beads were washed sequentially, twice with RIPA buffer, twice with high-salt buffer (500 mM NaCl, l50 mM Tris base, pH 7.5, 0.1% Nonidet P40 and 0.05% deoxycholic acid), and once with low-salt buffer (50 mM Tris base, pH 7.5, 0.1% Nonidet P40 and 0.05% deoxycholic acid). The bound proteins were eluted with boiling 2×Laemmli sample (SDS) buffer, applied to an SDS/10% polyacrylamide gel and transferred on to a nitrocellulose membrane (Osmonics). The blot was probed with the horseradish-peroxidase-conjugated ImmunoPure streptavidin (Pierce Biotechnology), and the protein bands were visualized on blue-sensitive X-ray films (Midwest Scientific) using the ECL® detection kit described above. A similar procedure was followed to immunoprecipitate TgPI-PLC from T. gondii lysates and bacterial fractions obtained as described above to demonstrate that the antibody could precipitate the protein, which was then probed with the same antibody (at a dilution of 1:2000) following a similar protocol to the one described above.
RESULTS
Isolation of the TgPI-PLC gene and sequence analysis
The complete coding region of TgPI-PLC was established as described under the Materials and methods section, and the translation of the ORF of 3294 bp yielded a polypeptide of 1097 amino acids with a predicted molecular mass of 121 kDa. Comparison of the TgPI-PLC cDNA sequence with its genomic sequence revealed that the TgPI-PLC gene comprised 22 exons (the size varying from 58 to 665 bp) interrupted by 21 introns (the size varying from 135 to 830 bp) (Figure 1A). The deduced amino acid sequence was analysed to identify similarities with other genes. This analysis revealed several domains characteristic of PI-PLCs (Figure 1B). Besides the catalytic X and Y domains, TgPI-PLC also contains an apparent N-terminal PH (pleckstrin homology) domain, an EF hand motif and a C-terminal C-2 domain (Figures 1B and 1C). TgPI-PLC shares a high level of sequence identity with mammalian and plant δ-type PLCs (31–35% identity). TgPI-PLC has an additional N-terminal extension and two insertions in the central region that are not present in other PI-PLCs and has 200–500 amino acids more than other δ-type PI-PLCs. In addition, there is a NES (nuclear export signal) within the EF hand domain (Figures 1B and 1C) and typical mammalian sorting motifs YXXΦ (Φ is an amino acid with a bulky hydrophobic side chain) (-Y1063SRI1066-) and dileucine (LL) in the C-terminal tail (Figure 1B). Comparisons of whole sequences showed only 21–25% identities with some protozoan PI-PLCs such as those from Plasmodium falciparum (GenBank® accession number AAN35330), Trypanosoma cruzi (GenBank® accession number AAD12583), Trypanosoma brucei (GenBank® accession number AY157307), and Leishmania major (genome database temporary ID number LmjF35.0040). Interestingly, multiple sequence alignments revealed that the X and Y domains of TgPI-PLC share higher identities with these domains of other parasite PI-PLCs (41–49% and 43–61% identities respectively). Of four parasite PI-PLCs, the X and Y domains of P. falciparum PI-PLC share the highest identity with those of TgPI-PLC (49% and 61% respectively). Southern blot analysis of parasite genomic DNA suggested that the TgPI-PLC gene is present as a single-copy gene in the haploid Toxoplasma genome (results not shown). Searches of the T. gondii genome database (http://ToxoDB.org/) in addition to the genome sequencing project and the OrthoMCL-DB (http://orthomcl.cbil.upenn.edu) [23] predict the presence of only one PI-PLC in T. gondii. This is not uncommon in unicellular organisms as, for example, D. discoideum [9] and Trypanosoma cruzi [11].
Expression, purification and catalytic activity of recombinant TgPI-PLC
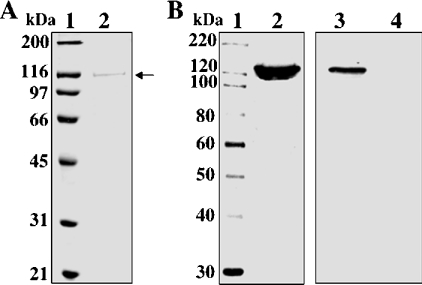
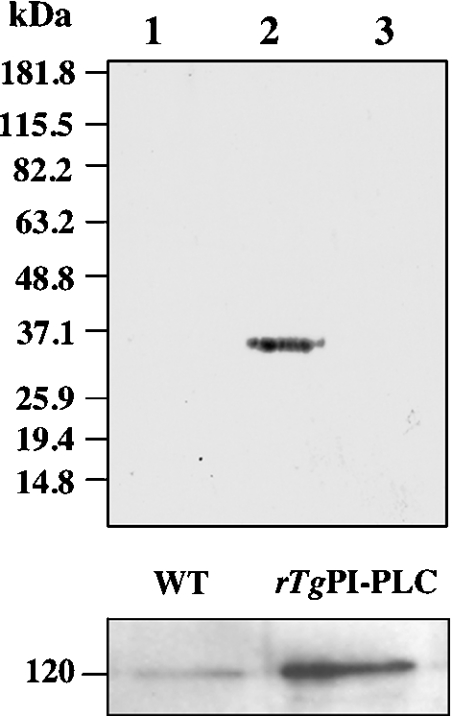
TgPI-PLC was expressed in E. coli BL21-CodonPlus(DE3)-RIPL as a fusion protein with an N-terminal polyhistidine tag. Affinity chromatography on a nickel-chelated agarose column permitted simple one-step protein purification. Enzyme purity was judged by using SDS/10% PAGE with Coomassie Blue staining (Figure 2A). The recombinant TgPI-PLC was recognized by the anti-TgPI-PLC antibody (Figure 2B, lane 2). The expressed TgPI-PLC appeared as a protein with an approximate size of 120 kDa, which is very close to the size predicted by its amino acid sequence (121 kDa). Figure 2(B) shows Western blot analyses of crude lysates obtained from E. coli transformed with the TgPI-PLC/pET28a recombinant plasmid (lane 3) and from the non-transformed bacteria (lane 4). The purified recombinant TgPI-PLC was subsequently used for further characterization of the enzyme.
Figure 2. Expression and affinity purification of recombinant TgPI-PLC from E. coli.
(A) Protein samples were separated by SDS/10% PAGE and visualized by Coomassie Brilliant Blue staining. Lane 1, standard protein size markers (MagicMark™ XP standards) with sizes given in kDa. Lane 2, nickel column-purified fraction (2.5 μg of protein/lane). The position of TgPI-PLC (120 kDa) is shown by the arrow. (B) Detection of TgPI-PLC by immunoblot using an affinity-purified polyclonal antibody against the peptide shown in Figure 1(C). Proteins were separated by SDS/10% PAGE, transferred on to nitrocellulose membranes and probed with affinity-purified anti-TgPI-PLC antibody (1:2000). Lane 1, protein standards (sizes are given in kDa); lane 2, recombinant TgPI-PLC (0.5 μg/lane) is shown by the arrow; lane 3, crude total cell lysate of E. coli BL21 CodonPlus(DE3)-RIPL transformed with TgPI-PLC/pET28a recombinant plasmid; lane 4, crude total cell lysate of non-transformed E. coli BL21 CodonPlus(DE3)-RIPL.
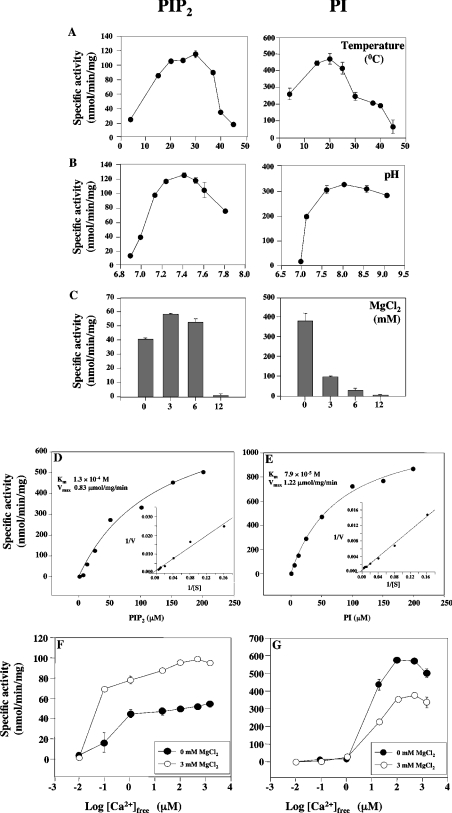
Optimal levels of PIP2 hydrolysis were obtained at 30 °C (Figure 3A), and between pH 7.2 and 7.5 (Figure 3B), while optimal levels of PI hydrolysis were obtained at 20 °C (Figure 3A), and between pH 7.5 and 8.5 (Figure 3B). MgCl2 slightly stimulated PIP2 hydrolysis, but inhibited PI hydrolysis (Figure 3C). Standard procedures were used to determine kinetic parameters. The enzyme reaction was carried out at a free Ca2+ concentration of 250 μM and pH 7.5. As shown in Figures 3(D) and 3(E), maximum activity was achieved at approx. 200 μM PIP2 or PI respectively. Km and Vmax values were obtained using a non-linear regression fit of the data to the Michaelis–Menten equation (SigmaPlot for Windows, version 3.06). When the rate of PIP2 and PI hydrolysis by the recombinant enzyme were measured, Km values of 130±33.9 and 79±9.3 μM and Vmax values of 0.83±0.11 and 1.22±0.06 μmol/mg per min respectively were calculated (Figures 3D and 3E).
Figure 3. PI-PLC activity of recombinant TgPI-PLC as a function of temperature (A), medium pH (B), Mg2+ concentration (C), substrate concentration (D, E) and free Ca2+ concentration (F, G).
Experimental conditions were as described under the Materials and methods section with 250 μM free Ca2+ and 40 μM PIP2 or 40 μM PI, adjusted to different temperatures (A), pH values (B), MgCl2 concentration (C), different concentrations of PIP2 (D) or PI (E) and 250 μM Ca2+, or in the same buffer with different free Ca2+ concentrations and 40 μM PIP2 (F) or 40 μM PI (G), with (open symbols) or without (closed symbols) 3 mM MgCl2. Insets in (D) and (E) represent the linear transformation, by double-reciprocal plot, of the curve. The results are representative of three experiments, each one carried out in duplicate.
The effect of free Ca2+ concentration was examined using both PIP2 and PI as substrates. The enzyme activity was stimulated by free Ca2+ in both cases. When PI was used as a substrate, the enzyme activity increased with increasing free Ca2+ concentrations above 1 μM, and the activity decreased when free Ca2+ concentration exceeded 1 mM (Figure 3F). When PIP2 was used as substrate, the enzyme was activated at lower Ca2+ concentrations and its activity increased with increasing free Ca2+ concentration (Figure 3F). Addition of 3 mM MgCl2 stimulated PIP2 hydrolysis (Figure 3F) and inhibited PI hydrolysis (Figure 3G). These results showed that TgPI-PLC preferred PI to PIP2 as a substrate in the presence of high Ca2+ concentrations.
Effect of PI-PLC inhibitors
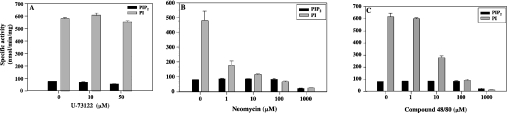
The effect of known PI-PLC inhibitors was also tested. Both neomycin and compound 48/80 inhibited PI and PIP2 hydrolysis in a concentration-dependent manner, these effects being more pronounced when PI was used as substrate (Figures 4B and 4C). However, U-73122 did not significantly affect hydrolysis of either PI or PIP2 at concentrations between 0 and 50 μM (Figure 4A).
Figure 4. Effect of PI-PLC inhibitors on the activity of recombinant TgPI-PLC.
TgPI-PLC activity was measured as described in the Materials and methods section in the presence of 40 μM substrate (PI, grey bars; PIP2, black bars) and 250 μM Ca2+, pH 7.5, and different concentrations of U-73122 (A), neomycin (B), and compound 48/80 (C).
PI-PLC activity in T. gondii membranes
Since a higher activity with PI rather than with PIP2 as substrate was unusual for a PI-PLC, we investigated whether this behaviour was also observed with the native enzyme obtained from a membrane fraction of T. gondii tachyzoites. A crude membrane fraction was prepared as described in the Materials and methods section, and the activity was measured following a similar protocol to that used for the recombinant enzyme. At 600 μM Ca2+, PI-PLC activity was higher when PI was used as substrate (160.5±2.1 with PI and 30.5±7.7 pmol/min per mg of protein with PIP2 as substrate respectively) and the inhibitor U-73122 partially inhibited PIP2 hydrolysis (42±5%), but not PI hydrolysis (7±3%). Figure 5 shows that, as occurs with the recombinant enzyme (Figures 3F and 3G), although Ca2+ stimulated PI-PLC activity with both PI (Figure 5A) and PIP2 (Figure 5B) as substrate, the activity was much higher when PI was the substrate.
Figure 5. PI-PLC activity of a T. gondii membrane fraction as a function of free Ca2+ concentration.
Freshly isolated parasites were ruptured by freezing and thawing at low osmolarity, and the homogenate was centrifuged at 15000 g. The pellet fraction was used to measure PI-PLC activity using 40 μM PI (A) or 40 μM PIP2 (B) as substrate, with different free Ca2+ concentrations. Other experimental details were as described in the Materials and methods section. The results are representative of three experiments, each one performed in duplicate.
Localization of TgPI-PLC
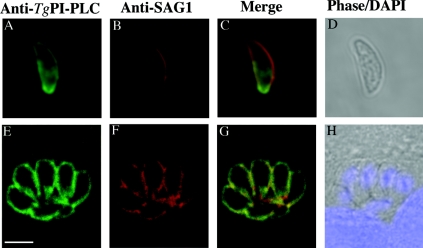
To investigate the subcellular localization of TgPI-PLC, polyclonal antibodies were raised in a guinea-pig against an antigenic polypeptide (amino acids 613–907, shown in Figure 1C) and were purified by affinity chromatography. These antibodies gave a strong reaction with the recombinant protein as demonstrated by Western blot analysis (Figure 2B), while no detectable band was observed using pre-immune serum (results not shown). The localization of TgPI-PLC was determined by indirect immunofluorescence microscopy using the affinity-purified anti-TgPI-PLC antibodies. HFFs infected with tachyzoites were fixed and permeabilized with saponin before antibody binding. TgPI-PLC was detected in the plasma membrane of tachyzoites, but not in the host cells (Figure 6E) and co-localized with SAG1 (Figure 6F), a known plasma membrane marker [22]. No detectable signal was observed when pre-immune serum was used (results not shown). Similar results were obtained when extracellular tachyzoites were treated in a similar way (Figure 6A). TgPI-PLC was also shown to co-localize with SAG1 (Figures 6B and 6C).
Figure 6. Distribution of TgPI-PLC in intracellular and extracellular tachyzoites.
Cells were permeabilized with 0.05% saponin for 30 min. The Figure shows the co-localization of TgPI-PLC (A, E) with SAG1 (B, F) in the plasma membrane. (C) and (G) show the overlap of (A) and (B), and (E) and (F) respectively. (D) and (H) are phase images. DAPI (4,6-diamidino-2-phenylindole) staining is shown in (H). Scale bar, 5 μm.
To investigate whether TgPI-PLC is located in the extracellular phase of the plasma membrane, recently released tachyzoites of the 2F1 strain (expressing β-galactosidase) were analysed following biotinylation. The surface proteins of tachyzoites were labelled with sulpho-NHS-biotin, a reagent that couples biotin to lysine residues of exposed proteins. The cells were incubated with the reagent and lysed, and TgPI-PLC was immunoprecipitated with the polyclonal antibody. As controls, the lysates were also immunoprecipitated with antibodies against β-galactosidase, which is cytosolic, or with monoclonal antibodies against SAG1, which is GPI-anchored to the plasma membrane and therefore exposed to the outer surface of the cells [22]. The precipitated proteins were electrophoresed and blotted, and biotinylated proteins were visualized using peroxidase-conjugated streptavidin and ECL®. A 30 kDa polypeptide corresponding to SAG1 was detected when lysates were immunoprecipitated with anti-SAG1 antibodies (Figure 7, upper panel, lane 2), while no bands were detected with the negative control immunoprecipitated with β-galactosidase antibodies (Figure 7, upper panel, lane 3) or with the lysates immunoprecipitated with anti-TgPI-PLC antibodies (Figure 7, upper panel, lane 1). The lower panel of Figure 7 shows the immunoprecipitation by the anti-TgPI-PLC antibody of the native PI-PLC from a T. gondii lysate (left-hand lane) and of the recombinant TgPI-PLC (right-hand lane), as probed with the same antibody. Taken together, these results indicate that TgPI-PLC is not localized on the outer surface of tachyzoites.
Figure 7. Biotinylation of cell-surface proteins in tachyzoites.
Upper panel, tachyzoites of the 2F1 strain were incubated with 2 mM sulpho-NHS-biotin for 30 min. After lysis of the cells with RIPA buffer, the lysates were immunoprecipitated by the affinity-purified anti-TgPI-PLC polyclonal antibody, and the immunoprecipitates were subjected to Western blot analysis. Detection of biotinylation was carried out using streptavidin–peroxidase conjugate and ECL®. No band was detected in the immunoprecipitates with anti-PI-PLC (lane 1). Lane 2 shows a positive control with anti-SAG1 antibody instead of anti-TgPI-PLC antibody for immunoprecipitation. Lane 3 shows a negative control with anti-β-galactosidase antibody for immunoprecipitation. Migration of molecular-mass standards (in kDa) is shown to the left of the gels. Lower panel, the positive control experiment shows that the anti-TgPI-PLC antibody can immunoprecipitate both the native PLC from T. gondii (left-hand lane) and the recombinant TgPI-PLC (right-hand lane), as probed with the guinea-pig anti-TgPI-PLC antibody. The position of a 120 kDa protein is indicated.
DISCUSSION
In the present work, we have demonstrated that a gene, TgPI-PLC, encoding a functional PI-PLC, is present in the T. gondii genome. We have also shown that the T. gondii enzyme has several peculiarities that distinguish it from other known PI-PLCs. Both the recombinant and the native enzyme have PI- and PIP2-hydrolysing activities. It is interesting to note that the PI-hydrolysing activity was almost 10-fold higher than the PIP2-hydrolysing activity in the absence of Mg2+ (Figure 3C). In the presence of 6 mM MgCl2, however, the PIP2-hydrolysing activity was twice as high as the PI-hydrolysing activity (Figure 3C). As shown in Figure 3(F), between 1 and 10 μM Ca2+ and in the presence of millimolar Mg2+ concentrations, the enzyme hydrolyses mainly PIP2. It is possible that TgPI-PLC functions mainly as a PIP2-hydrolysing enzyme under the physiological millimolar concentrations of Mg2+ that are present inside the cell. However, we cannot rule out a preferential hydrolysis of PI if the enzyme reaches the extracellular medium, which is rich in Ca2+ and poor in Mg2+.
The temperature is also another important variable, since it has been shown that Ca2+-stimulated microneme secretion is temperature-sensitive [21]. PIP2-hydrolysing activity was higher at the physiological temperature to which tachyzoites are exposed (37 °C), while the PI-hydrolysing activity was higher at 20 °C. Furthermore, at Ca2+ levels below 1 μM, rTgPI-PLC had high activity with PIP2 and negligible activity with PI as substrate respectively. The PI-hydrolysing activity increased at Ca2+ concentrations above 10 μM and reached its maximum activity at 100–200 μM Ca2+. Taken together, these results suggest that TgPI-PLC prefers PIP2 as substrate under the physiological conditions that are present in the parasite. However, under certain conditions, such as the high Ca2+ concentration induced by treatment of T. gondii-infected cells with Ca2+ ionophore A23187, a process known to stimulate parasite egress [16], the PI-hydrolysing activity could be more relevant. In this regard, the escape of Listeria monocytogenes [24] and Trypanosoma cruzi [25] from their parasitophorous vacuoles are known to be mediated by PLCs (phospholipase C), and such a role has also been postulated in the case of T. gondii on the basis of inhibitor studies [16].
Immunofluorescence results suggest that TgPI-PLC is predominantly localized in the internal leaflet of the plasma membrane (Figures 6A and 6E). This is supported by surface biotinylation studies (Figure 7).
Sequence comparisons of TgPI-PLC with other PI-PLCs suggest that it is most closely related to the mammalian δ isoform and to PI-PLCs found in yeasts, D. discoideum and plants in terms of sequence identity and arrangement of conserved domains. In addition, among other lower eukaryotic and mammalian PI-PLCs, TgPI-PLC is one of the largest, with 1097 amino acids. When compared with other δ-type PI-PLCs, TgPI-PLC has an extended N-terminal region and two insertions between the X and Y domains. However, TgPI-PLC conserves most of the amino acid residues that have been found, in mammalian PI-PLC-δ1 [26], to be in contact with IP3 and Ca2+ in the catalytic domains (His420, Asn421, Glu450, Asp452, His465, Glu502, Lys547, Lys549, Ser814, Arg841 and Tyr843) (Figure 1B). This conservation of amino acid residues that are important for substrate and Ca2+ binding explains the similar characteristics and Ca2+ requirement of TgPI-PLC as compared with mammalian δ-type PI-PLCs (Figure 3). In addition, similar to the PI-PLC-δ isoenzymes from other organisms, TgPI-PLC contains an EF hand motif upstream of the X domain as well as an apparent PH domain at the N-terminal region. However, in contrast with all δ-type PI-PLCs reported to date [26–28], it has a short C-terminal extension following the C2 domain. The presence of unusual clusters of negatively charged and mixed-charged amino acids between the X and Y catalytic domains is not common in PI-PLCs of lower eukaryotes or in mammalian δ- or γ-type PI-PLCs. However, mammalian β-type PI-PLCs also contain highly charged clusters of amino acids between the X and Y domain. This is essential for activation by βγ subunits of G-proteins. D. discoideum δ-type PI-PLC is also postulated to be activated in this manner [29].
The presence of NESs approx. 14-amino-acids long and rich in leucine, suggests the possibility that the enzyme is imported to the nucleus and that this sequence is probably responsible for its export from this organelle. This has been demonstrated to occur with the mammalian PLC-δ1 [30]. Blocking its NES-dependent nuclear export results in its nuclear accumulation, although it is not known by which mechanism the enzyme is transported into the nucleus [30].
In conclusion, TgPI-PLC has distinct peculiarities when compared with mammalian PI-PLCs, but its sequence and the organization of its different domains are similar to those of δ-type isoenzymes. The meaning of these differences in the physiological context of host–parasite interaction is being explored at present.
Acknowledgments
The monoclonal antibody against E. coli β-galactosidase was developed by Joshua Sanes and was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD (National Institute of Child Health and Human Development) and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242, U.S.A. We thank David Alexander and John Boothroyd (Stanford University) for the monoclonal antibody against SAG1, L. David Sibley (Washington University School of Medicine) for T. gondii 2F1 strain, Maria Laura Salto for help in measuring PLC activity, Peter Rohloff for help with immunofluorescence imaging, and Linda Brown for her assistance with the growth of T. gondii. This work was supported in part by U.S. National Institutes of Health Grant AI-43614 to S. N. J. M. This investigation was conducted in part in a facility constructed with support from Research Facility Improvement Grant Number C06 RR16515-01 from the National Center for Research Resources, U.S. National Institutes of Health.
References
- 1.Berridge M. J. Inositol trisphosphate and diacylglycerol: two interacting second messengers. Annu. Rev. Biochem. 1987;56:159–193. doi: 10.1146/annurev.bi.56.070187.001111. [DOI] [PubMed] [Google Scholar]
- 2.Nishizuka Y. The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature (London) 1988;334:661–665. doi: 10.1038/334661a0. [DOI] [PubMed] [Google Scholar]
- 3.Nakahara M., Shimozawa M., Nakamura Y., Irino Y., Morita M., Kudo Y., Fukami K. A novel phospholipase C, PLCη2, is a neuron-specific isozyme. J. Biol. Chem. 2005;280:29128–29134. doi: 10.1074/jbc.M503817200. [DOI] [PubMed] [Google Scholar]
- 4.Irvine R. Phospholipid signalling: taking stock of PI-PLC. Nature (London) 1996;380:581–583. doi: 10.1038/380581a0. [DOI] [PubMed] [Google Scholar]
- 5.Hodson E. A., Ashley C. C., Hughes A. D., Lymn J. S. Regulation of phospholipase C-δ by GTP-binding proteins-rhoA as an inhibitory modulator. Biochim. Biophys. Acta. 1998;1403:97–101. doi: 10.1016/s0167-4889(98)00028-7. [DOI] [PubMed] [Google Scholar]
- 6.Flick J. S., Thorner J. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:5861–5876. doi: 10.1128/mcb.13.9.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne W. E., Fitzgerald-Hayes M. A mutation in PLC1, a candidate phosphoinositide-specific phospholipase C gene from Saccharomyces cerevisiae, causes aberrant mitotic chromosome segregation. Mol. Cell. Biol. 1993;13:4351–4364. doi: 10.1128/mcb.13.7.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoko-o T., Matsui Y., Yagisawa H., Nojima H., Uno I., Toh-e A. The putative phosphoinositide-specific phospholipase C gene, PLC1, of the yeast Saccharomyces cerevisiae is important for cell growth. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1804–1808. doi: 10.1073/pnas.90.5.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drayer A. L., van Haastert P. J. Molecular cloning and expression of a phosphoinositide-specific phospholipase C of Dictyostelium discoideum. J. Biol. Chem. 1992;267:18387–18392. [PubMed] [Google Scholar]
- 10.Bominaar A. A., Van Haastert P. J. Phospholipase C in Dictyostelium discoideum: identification of stimulatory and inhibitory surface receptors and G-proteins. Biochem. J. 1994;297:189–193. doi: 10.1042/bj2970189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuya T., Kashuba C., Docampo R., Moreno S. N. A novel phosphatidylinositol-phospholipase C of Trypanosoma cruzi that is lipid modified and activated during trypomastigote to amastigote differentiation. J. Biol. Chem. 2000;275:6428–6438. doi: 10.1074/jbc.275.9.6428. [DOI] [PubMed] [Google Scholar]
- 12.Salto M. L., Furuya T., Moreno S. N., Docampo R., de Lederkremer R. M. The phosphatidylinositol-phospholipase C from Trypanosoma cruzi is active on inositolphosphoceramide. Mol. Biochem. Parasitol. 2002;119:131–133. doi: 10.1016/s0166-6851(01)00392-9. [DOI] [PubMed] [Google Scholar]
- 13.Okura M., Fang J., Salto M. L., Singer R. S., Docampo R., Moreno S. N. A lipid-modified phosphoinositide-specific phospholipase C (TcPI-PLC) is involved in differentiation of trypomastigotes to amastigotes of Trypanosoma cruzi. J. Biol. Chem. 2005;280:16235–16243. doi: 10.1074/jbc.M414535200. [DOI] [PubMed] [Google Scholar]
- 14.Carruthers V. B., Moreno S. N., Sibley L. D. Ethanol and acetaldehyde elevate intracellular [Ca2+] and stimulate microneme discharge in Toxoplasma gondii. Biochem. J. 1999;342:379–386. [PMC free article] [PubMed] [Google Scholar]
- 15.Lovett J. L., Marchesini N., Moreno S. N., Sibley L. D. Toxoplasma gondii microneme secretion involves intracellular Ca2+ release from inositol 1,4,5-triphosphate (IP3)/ryanodine-sensitive stores. J. Biol. Chem. 2002;277:25870–25876. doi: 10.1074/jbc.M202553200. [DOI] [PubMed] [Google Scholar]
- 16.Moudy R., Manning T. J., Beckers C. J. The loss of cytoplasmic potassium upon host cell breakdown triggers egress of Toxoplasma gondii. J. Biol. Chem. 2001;276:41492–41501. doi: 10.1074/jbc.M106154200. [DOI] [PubMed] [Google Scholar]
- 17.Moreno S. N., Zhong L. Acidocalcisomes in Toxoplasma gondii tachyzoites. Biochem. J. 1996;313:655–659. doi: 10.1042/bj3130655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kissinger J. C., Gajria B., Li L., Paulsen I. T., Roos D. S. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 20.Ghosh S., Pawelczyk T., Lowenstein J. M. Phospholipase C isoforms δ1 and δ3 from human fibroblasts: high-yield expression in Escherichia coli, simple purification, and properties. Protein Expression Purif. 1997;9:262–278. doi: 10.1006/prep.1996.0682. [DOI] [PubMed] [Google Scholar]
- 21.Carruthers V. B., Giddings O. K., Sibley L. D. Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell. Microbiol. 1999;1:225–235. doi: 10.1046/j.1462-5822.1999.00023.x. [DOI] [PubMed] [Google Scholar]
- 22.Burg J. L., Perelman D., Kasper L. H., Ware P. L., Boothroyd J. C. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J. Immunol. 1988;141:3584–3591. [PubMed] [Google Scholar]
- 23.Chen F., Mackey A. J., Stoeckert C. J., Roos D. S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marquis H., Hager E. J. pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 2000;35:289–298. doi: 10.1046/j.1365-2958.2000.01708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall B. F., Webster P., Ma A. K., Joiner K. A., Andrews N. W. Desialylation of lysosomal membrane glycoproteins by Trypanosoma cruzi: a role for the surface neuraminidase in facilitating parasite entry into the host cell cytoplasm. J. Exp. Med. 1992;176:313–325. doi: 10.1084/jem.176.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Essen L. O., Perisic O., Cheung R., Katan M., Williams R. L. Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature (London) 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 27.Paterson H. F., Savopoulos J. W., Perisic O., Cheung R., Ellis M. V., Williams R. L., Katan M. Phospholipase Cδ1 requires a pleckstrin homology domain for interaction with the plasma membrane. Biochem. J. 1995;312:661–666. doi: 10.1042/bj3120661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagisawa H., Sakuma K., Paterson H. F., Cheung R., Allen V., Hirata H., Watanabe Y., Hirata M., Williams R. L., Katan M. Replacements of single basic amino acids in the pleckstrin homology domain of phospholipase C-δ1 alter the ligand binding, phospholipase activity, and interaction with the plasma membrane. J. Biol. Chem. 1998;273:417–424. doi: 10.1074/jbc.273.1.417. [DOI] [PubMed] [Google Scholar]
- 29.Brazill D. T., Lindsey D. F., Bishop J. D., Gomer R. H. Cell density sensing mediated by a G protein-coupled receptor activating phospholipase C. J. Biol. Chem. 1998;273:8161–8168. doi: 10.1074/jbc.273.14.8161. [DOI] [PubMed] [Google Scholar]
- 30.Ochocka A. M., Pawelczyk T. Isozymes δ of phosphoinositide-specific phospholipase C and their role in signal transduction in the cell. Acta Biochim. Pol. 2003;50:1097–1110. [PubMed] [Google Scholar]