Abstract
Cytochrome b5 of the body wall of adult Ascaris suum, a porcine parasitic nematode, is a soluble protein that lacks a C-terminal membrane-anchoring domain, but possesses an N-terminal pre-sequence of 30 amino acids. During the maturation of cytochrome b5, the N-terminal pre-sequence is proteolytically cleaved to form the mature protein of 82 amino acid residues. A. suum cytochrome b5 is a basic protein containing more lysine residues and exhibiting a higher midpoint redox potential than its mammalian counterparts. We developed an expression system for the production of the recombinant nematode cytochrome b5, which is chemically and functionally identical with the native protein. Using this recombinant protein, we have determined the X-ray crystal structure of A. suum cytochrome b5 at 1.8 Å (1 Å=0.1 nm) resolution, and we have shown that this protein is involved in the reduction of nematode body-wall metmyoglobin. The crystal structure of A. suum cytochrome b5 consists of six α-helices and five β-strands. It differs from its mammalian counterparts by having a head-to-tail disulphide bridge, as well as a four-residue insertion in the vicinity of the sixth ligating histidine, which forms an additional α-helix, α4A, between helices α4 and α5. A. suum cytochrome b5 exists predominantly as a haem-orientation B isomer. Furthermore, the haem plane is rotated approx. 80° relative to the axis formed by haem-Fe and Nϵ atoms of the two histidine residues that are ligated to haem-Fe. The charge distribution around the haem crevice of A. suum cytochrome b5 is remarkably different from that of mammalian cytochrome b5 in that the nematode protein bears positively charged lysine residues surrounding the haem crevice. Using immunohistochemistry, we found that A. suum cytochrome b5 is present in the nematode hypodermis. Based on this histochemical and structural information, the physiological function of A. suum cytochrome b5 and its interaction with nematode metmyoglobin can be hypothesized.
Keywords: Ascaris suum, b5-type cytochrome, metmyoglobin reduction, parasitic nematode, X-ray crystal structure
Abbreviations: Em, midpoint redox potential; MAD, multiwavelength anomalous dispersion; rmsd, root mean square deviation
INTRODUCTION
Cytochromes b5 are widely distributed haemoproteins that catalyse a variety of biological redox reactions [1,2]. Two forms of cytochrome b5 have been isolated: a membrane-bound form and a soluble form. The former is localized in microsomes and the outer membranes of mitochondria, whereas the latter is present in erythrocytes. Membrane-bound cytochrome b5 is composed of two distinct domains: a hydrophilic N-terminal domain of approx. 100 amino acids and a hydrophobic C-terminal domain of approx. 30 amino acids. The N-terminal domain contains a protohaem that catalyses electron transfer, whereas the C-terminal domain consists of a hydrophobic membrane-anchoring portion (approx. 20 residues) and a terminal hydrophilic portion (approx. ten residues), with the latter containing sufficient information to target the cytochrome to the endoplasmic reticulum and/or mitochondrial outer membranes [3,4]. Erythrocyte cytochrome b5 is composed of a single domain, which is almost identical with the hydrophilic N-terminal domain of the microsomal cytochrome b5. In contrast, the erythrocyte protein lacks the C-terminal membrane-anchoring domain, thus allowing the protein to be soluble in erythrocytes. Functionally, microsomal cytochrome b5 serves as an electron carrier between NAD(P)H-cytochrome b5 reductase and various enzymes, including stearyl-CoA desaturase [5] and cytochromes P450 [2]; mitochondrial cytochrome b5 is involved in the reduction of monodehydroascorbate to ascorbate [6]; and erythrocyte cytochrome b5 participates in the NADH-dependent reduction of methaemoglobin to haemoglobin [7].
We previously isolated a novel type of cytochrome b5 from the body wall of adult Ascaris suum, a porcine parasitic nematode [8]. This cytochrome b5, which was co-extracted with mitochondrial cytochrome c, was shown to be a soluble protein of 82 amino acids, but it had no equivalent to the C-terminal hydrophobic domain of microsomal cytochrome b5. Thus nematode cytochrome b5 appears to be a counterpart of erythrocyte cytochrome b5, although it was extracted from the nematode's body wall, which includes muscle tissues. A. suum cytochrome b5, however, can be distinguished from other forms of this protein by several properties. First, nematode cytochrome b5 has a much higher Em (midpoint redox potential) (+78 mV) than do mammalian proteins (−102 mV for cytochrome b5 from mitochondrial outer membrane and from −12 to +20 mV for microsomal cytochrome) [1,9,10]. Secondly, in contrast with its mammalian counterpart, which is an acidic protein (pI 4.3) [9] with a number of aspartate and glutamate residues surrounding the exposed edge of the haem group, A. suum cytochrome b5 abounds in lysine residues (14 of the 82 amino acids), making it extremely basic (calculated pI 9.55). Finally, A. suum cytochrome b5 possesses an N-terminal extension (pre-sequence) of 30 amino acids, which is not present in the protein purified from the nematode body wall. To date, the only cytochrome b5 reported to have a pre-sequence is that from the nematode. This cytochrome b5 is probably synthesized as a precursor protein of 112 amino acids, and the pre-sequence is later cleaved, forming mature 82-amino-acid cytochrome b5.
During the course of studying the precursor protein, we established an expression system producing the mature cytochrome b5, which was chemically and functionally identical with the native protein [11]. Using this synthesized protein, we have performed an X-ray crystallographic study to determine the structure of A. suum cytochrome b5, as well as further studies of its physiological function.
EXPERIMENTAL
Crystallization and data collection
Recombinant A. suum cytochrome b5 was prepared from cells transformed with pb5wt as described in [11]. Compared with native protein prepared from nematode body walls, the recombinant cytochrome b5 was indistinguishable in molecular mass, spectral properties and reducibility by microsomal cytochrome b5 reductase [11]. To determine the functional integrity of recombinant cytochrome b5, we measured its Em; the value obtained, +75 mV, was in good agreement with the +78 mV reported for native cytochrome b5 [8].
Crystallization of A. suum cytochrome b5 was carried out at 278 K by a hanging-drop vapour-diffusion method using 10 mg/ml protein solution and a reservoir solution containing 3.2 M ammonium sulphate and 100 mM sodium phosphate buffer (pH 6.8). Plate-shaped red crystals grew to approx. 0.3 mm×0.2 mm×0.05 mm within a couple of weeks. These crystals were transferred to the reservoir solution supplemented with 15% (v/v) glycerol immediately before flash cooling with a nitrogen gas stream at 100 K.
X-ray diffraction data were collected to 1.8 Å (1 Å=0.1 nm) resolution at 100 K using an R-AXIS IIc imaging-plate detector and graphite-monochromatic CuKα radiation from a RIGAKU RU-300 rotating-anode generator operated at 50 kV and 100 mA. The data for MAD (multiwavelength anomalous dispersion) phasing were collected at three different wavelengths (0.17395, 0.17390 and 0.17000 nm) from a single crystal, using synchrotron radiation at the BL18B station in Photon Factory (Tsukuba, Japan) with ADSC Quantum 4R CCD (charge-coupled device) detector systems at 100 K. All data were processed and scaled using MOSFLM and SCALA of the CCP4 program suite [12].
The crystals of A. suum cytochrome b5 were found to belong to a monoclinic space group C2 with cell parameters of a=103.2 Å, b=41.8 Å, c=49.8 Å and β=111.1°. Assuming two molecules in an asymmetric unit, the Matthews coefficient VM was calculated to be 2.71 Å3/Da, which indicated solvent content of approx. 54.6%, a value typical for protein crystals [13].
Structure determination and refinement
The crystallographic analysis of A. suum cytochrome b5 was carried out with MAD methods based on the Fe absorption edge. The datasets collected at 0.17390 and 0.17000 nm were scaled to that at 0.17395 nm using program SCALEIT from the CCP4. Refinement of the heavy-atom (haem-Fe) parameters and calculations of the initial phases were performed with program SOLVE [14]. Solvent flattering with program RESOLVE [14] significantly improved electron density. The mean figure-of-merit reached 0.697 at 20–2.4 Å resolution. The map was of good quality, and the initial atomic model of A. suum cytochrome b5 was gradually built into the 2.4 Å resolution maps using program O [15].
The structure of A. suum cytochrome b5 was subsequently refined by a round of simulated annealing and iterative energy minimization with program CNX [16], using intensity data collected with R-AXIS IIc from 20 to 2.0 Å resolution. This yielded an Rfactor of 26.7% and an Rfree of 32.6%. The haem in cytochrome b5 has two possible conformations because of rotation about the porphyrin α,γ-meso axis [17]. One isomer was adopted, which was considered predominant from an Fo-Fc difference omit map. The resolution limit was finally extended to 1.8 Å, and water molecules were picked up from the difference maps on the basis of peak heights and distance criteria. Water molecules with a temperature factor above 50 Å2 after refinement were excluded from the atomic model. A reciprocal-space or Babinet bulk solvent correction [18] and anisotropic overall B-factor refinement were applied throughout. Further model building and refinement cycles resulted in an Rfactor of 19.2% and an Rfree of 23.6%, using 17891 reflections between 20 and 1.8 Å resolution. Data collection and refinement statistics are summarized in Table 1.
Table 1. Data collection and refinement statistic.
Values in parentheses refer to the last resolution shell Rmerge=ΣhklΣi|Ihkl−〈Ihkl〉|/ΣhklΣi·Ihkl, where Ihkl is observed intensity and 〈Ihkl〉 is average intensity for multiple measurements.
| (a) Data collection | ||||
|---|---|---|---|---|
| R-axis IIc | Peak | Edge | Remote | |
| Space group | C2 | C2 | C2 | C2 |
| Lattice parameter | ||||
| a (Å) | 103.2 | 103.0 | 103.0 | 103.0 |
| b (Å) | 41.77 | 42.30 | 42.30 | 42.30 |
| c (Å) | 49.76 | 49.67 | 49.67 | 49.67 |
| β (°) | 111.1 | 111.2 | 111.2 | 111.2 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Wavelength (nm) | CuKα | 0.17395 | 0.17390 | 0.17000 |
| Resolution range (Å) | 50–1.80 | 30–2.20 | 30–2.20 | 30–2.20 |
| (1.86–1.80) | (2.32–2.20) | (2.23–2.20) | (2.23–2.20) | |
| Number of reflections | ||||
| Total | 127761 | 52427 | 54801 | 52935 |
| Unique | 17915 | 10286 | 10296 | 10291 |
| (1691) | (1490) | (1489) | (1484) | |
| Completeness (%) | 96.7 | 99.3 | 99.9 | 99.9 |
| (93.5) | (99.8) | (99.9) | (99.9) | |
| Bijvoet pairs | 9466 | 9479 | 9495 | |
| Completeness (%) | 99.3 | 99.3 | 99.5 | |
| Rmerge | 5.1 | 7.5 | 7.5 | 6.9 |
| (17.9) | (13.9) | (13.9) | (13.1) | |
| Mean I/σ(I) | 42.1 | 6.8 | 6.8 | 7.5 |
| (9.5) | (4.2) | (4.2) | (4.5) | |
| (b) Refinement | ||||
| Parameter | Value | |||
| Resolution limit (Å) | 20–1.8 | |||
| Rfactor (%) | 19.2 | |||
| Rfree (%) | 23.6 | |||
| Deviations | ||||
| Bond lengths (Å) | 0.005 | |||
| Bond angles (°) | 1.26 | |||
| B-factor | ||||
| Average main chains (Å2) | 16.2 | |||
| Average side chains (Å2) | 19.0 | |||
| Average haem groups (Å2) | 18.9 | |||
| Average waters (Å2) | 30.2 |
The current model consists of two protein molecules, each of 82 amino acids, two haem groups, one sulphate anion and 227 water molecules. The average B-factors of the main-chain atoms in molecules I and II are 15.1 and 17.2 Å2 respectively, and the maximum B-factors in molecules I and II are 35.6 and 34.0 Å2 respectively, corresponding to a carbonyl oxygen as the C-terminal residue in both molecules. An analysis of stereochemistry with PROCHECK from the CCP4 showed that all the non-glycine residues fell into the allowed region of the Ramachandran plot, with 128 residues (88.9%) in the most favoured region and 16 residues (11.1%) in an additionally allowed region.
Preparation of myoglobin and metmyoglobin from A. suum body wall
Myoglobin of A. suum body wall was prepared essentially as described in [19,20]. The ratio of the absorbance of the purified protein at 412 nm to that at 280 nm was 2.18, essentially the same value as described previously [20]. The concentration of purified oxymyoglobin was spectrophotometrically determined at 543 nm using an absorption coefficient of 12.5 mM−1·cm−1 [19]. To prepare A. suum metmyoglobin, a 10-fold molar excess of K3Fe(CN)6 was added to the purified myoglobin (226 nmol) in 1 ml of phosphate buffer. The mixture was dialysed for 5 h against 1 litre of 20 mM potassium phosphate buffer, pH 6.0, and overnight with one change of the buffer. The dialysed sample was loaded on to a CM Cellulofine column (1.5 cm×16 cm), equilibrated previously with 20 mM potassium phosphate buffer, pH 6.0, and the metmyoglobin was eluted as a dark-brown band with 30 mM potassium phosphate buffer, pH 7.0. The absorption spectrum of the purified metmyoglobin showed maxima at 405, 499 and 635 nm, characteristic of acid body-wall metmyoglobin, as described previously [19].
Immunohistochemistry
A female A. suum (approx. 15 cm length), freshly brought to the laboratory, was cut transversely into 5-mm-thick sections, which were fixed for 48 h in 0.38% (w/v) formaldehyde and 2% (v/v) ethanoic (acetic) acid, mounted on to slides, and immobilized with paraffin. Following removal of the paraffin with xylene, the specimens were washed three times with absolute ethanol, immersed in a 1:100 solution of 30% H2O2/methanol for 30 min, and washed with water and PBS. Following incubation with 5% normal goat serum (Wako) for 20 min to block non-specific binding, the slides were incubated with affinity-purified anti-(A. suum cytochrome b5) antibody for 30 min and washed with PBS. The slides were subsequently incubated with 25 μg/ml anti-(rabbit IgG) biotin conjugate (Sigma) for 30 min, washed with PBS, and incubated with peroxidase-conjugated avidin–biotin complex (VECTASTAIN® Elite ABC kit) for 30 min. The slides were subsequently developed with 50 mM Tris/HCl, pH 7.6, containing 0.006% H2O2 and 25 mg/ml 3,3′-diaminobenzidine tetrahydrochloride, as described in the manufacture's protocol. Colour development was monitored under the microscope (Zeiss Axioplan 2), and development was halted by gentle washing with flowing water, after which photographs were taken. As a control, the series of treatments was performed using non-immune rabbit serum instead of anti-(A. suum cytochrome b5) antibody.
Enzyme assays
NADH-metmyoglobin reductase activity was determined spectrophotometrically at 25 °C by monitoring the increase in myoglobin. The reaction mixture contained 60.2 μg of NADH-cytochrome b5 reductase (the microsomal fraction of A. suum muscle) [11], 0.2 mM NADH, 50 mM potassium phosphate buffer, pH 7.0, 6.55 μM A. suum metmyoglobin, and various concentrations of nematode cytochrome b5 (0, 0.384, 0.769 and 1.54 μM) in a total volume of 1 ml. The reaction was started by the addition of NADH, and the increase in absorbance at 578 nm was recorded in dual-wavelength mode using 600 nm as the reference wavelength. To confirm that myoglobin was produced, difference spectra were recorded between the reaction mixture with and without NADH in dual-wavelength mode using 600 nm as the reference wavelength by repeated scanning between 460 nm and 700 nm at a scanning rate of 120 nm·min−1. A molar absorption coefficient (ϵ578) of 11.1 mM−1·cm−1 was used for the calculation [19].
Others
All absorption spectra were recorded on a dual-wavelength spectrophotometer (Hitachi Model 557). The Em of recombinant A. suum cytochrome b5 was determined at pH 7.0 by redox titration techniques [21]. Protein concentration was determined using a modification of the Lowry method [22], using BSA as the standard.
RESULTS AND DISCUSSION
Overall structure
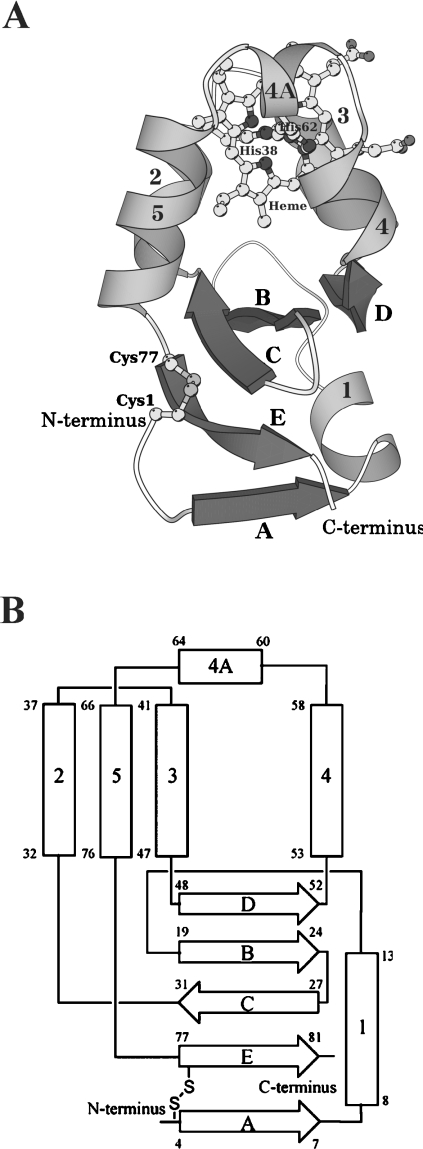
A schematic drawing and a topology diagram of A. suum cytochrome b5 are shown in Figure 1, with the secondary-structure assignment made using program DSSP [23]. Structure diagrams were drawn using MOLSCRIPT [24], POVScript+ [25] and RASTER3D [26] programs. Two molecules in the asymmetric unit, labelled I and II, can be superimposed with an rmsd (root mean square deviation) of 0.45 Å, thus indicating that the structures of the two independent molecules are essentially identical. Small differences, however, were observed in the N-terminal and haem-surrounding regions. In the N-terminal region, the differences were found at Gly2 and Asp3, with the Cα distances between the superimposed molecules being 2.9 and 2.3 Å respectively. These differences probably result from flipping the main chain carbonyl group of Cys1 to avoid collisions. In the haem-surrounding area, a major difference was observed at the Cα position of Gly40, giving a distance of 1.8 Å, which is probably caused by a change in backbone conformation at Pro39 between the two molecules. The haem-binding site in molecule I, including Pro39, was found to be located close to molecule II in a neighbouring asymmetric unit. Therefore the intermolecular contacts, caused by packing forces in the crystal lattice, apparently resulted in minor differences between molecules I and II. In the following sections, we primarily discuss the structure of molecule I, because it has a lower average B-factor than molecule II.
Figure 1. Overall structure (A) and topology diagram (B) of A. suum cytochrome b5.
The protein is folded in six helices and five β strands, comprising a β-pleated sheet. (A) Ribbon and ball-and-stick drawing of the overall structure. The six helices are numbered 1, 2, 3, 4, 4A and 5 in grey, while the five β strands are denoted A–E in dark grey. The haem and His38, His62, Cys1 and Cys77 are drawn as ball-and-stick models. (B) α-Helices and β-strands are represented as rectangles and broad arrows respectively, with residue numbers showing their starting and ending positions. Two histidine residues (His38 and His62) ligating the haem iron and the disulphide bond between Cys1 and Cys77 are also shown. The nomenclature of α-helices and β-strands is the same as in (A).
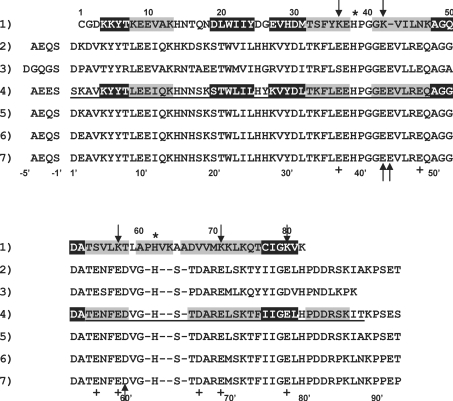
We found that the polypeptide chain of A. suum cytochrome b5 is folded into six α-helices and five β-strands (Figure 1), as is mammalian cytochrome b5. However, the order of formation of α-helices and β-strands differs between the nematode and mammalian cytochromes b5 in their C-terminal regions, with the former being βAα1βBβCα2α3βDα4α4Aα5βE and the latter being β1α1β2β3α2α3β4α4α5β5α6 from the N-terminus to the C-terminus [27]. It is noteworthy that helices 2 and 3 are categorized as a 310 helix rather than an ordinary α-helix. The haem-binding site of mammalian cytochrome b5 is composed of four α-helices (helices 2, 3, 4 and 5). While a region between helices 4 and 5 in A. suum cytochrome b5 (Leu59–Asp67) is four residues longer than the same region in bovine cytochrome b5 (Val61′–Thr65′) (Figure 2), resulting in the formation of a new short α-helix 4A (Figure 1). As a consequence, a five-helix bundle is formed, consisting of α-helices 2, 3, 4, 4A and 5.
Figure 2. Structure-based alignment of the amino acid sequence of A. suum cytochrome b5 with the catalytic domains of several mammalian cytochromes b5.
Residues composing the α-helices and β-strands are represented as letters in grey and black boxes respectively. Sequence 1, A. suum cytochrome b5 (mature, 82 residues) [8]. Sequence 2, rat microsomal cytochrome b5 (97 residues) [48]. Sequence 3, rat mitochondrial outer membrane cytochrome b5 (92 residues) [49]. Sequence 4, bovine microsomal cytochrome b5 (97 residues) [27]; the underlined residues, 1′–93′, are those identified by X-ray crystallographic analyses of the lipase-treated protein. Sequence 5, porcine microsomal cytochrome b5 (97 residues) [48]. Sequence 6, human cytochrome b5 (97 residues) [48]. Sequence 7, human erythrocyte cytochrome b5 (97 residues) [50]. Except for sequence 5, the first residues of alanine are acetylated. For comparison, the residue numbers for mammalian cytochromes b5 are shown beneath as numbers with a prime, based on the numbering system for bovine cytochrome b5. The two histidine residues that are ligated to the haem iron are denoted by an asterisk. Up arrows indicate the acidic residues of mammalian cytochrome b5, which electrostatically interact with the lysine residues of their reaction partner proteins. The acidic residues of mammalian cytochrome b5 that are replaced by other residues in A. suum cytochrome b5 are marked by +, and those replaced by lysine residues are indicated by down arrows. Hyphens indicate the positions of deletions or insertions.
We also found that the haem-Fe atom in A. suum cytochrome b5 is electrostatically bound to each Nϵ atoms of His38 and His62, which correspond to residues His39′ and His63′ respectively in bovine cytochrome b5, as determined from amino acid sequence alignment [8]. The histidine residues that interact with haem-Fe atoms are located in a hydrophobic pocket created by an α-helix bundle, and β-strands B and C and are strictly conserved by all cytochromes b5 [1]. His62 is located on helix 4A in A. suum, while the equivalent residue in the bovine cytochrome, His63′, is in the loop between α-helices 4 and 5.
Another unique feature of A. suum cytochromes b5 is that Cys1 forms a disulphide bond with Cys77 in the β-strand E. This disulphide bond was confirmed from a 2Fo-Fc omit map calculated without these cysteine residues and the surrounding residues. Cys1 is on the surface of the protein, covering Cys77, and contacts Glu27 (β-strand C), Lys80 (β-strand E) and Asp3* (N-terminal region of another molecule) respectively. The formation of a head-to-tail disulphide bond is unique to A. suum cytochrome b5, as this feature has not been found in any other eukaryotic cytochromes b5 so far investigated. The nature and mechanism of formation of this unique disulphide bond in A. suum cytochrome b5 are not clear. Because the recombinant protein used in the present study was purified from the periplasm of Escherichia coli [11], it is reasonable to assume that the disulphide bond was formed in vivo by the thiol–sulphide oxidoreductase system, which catalyses thiol–disulphide exchange reactions [28]. In eukaryotes, protein disulphide bonds are formed in the endoplasmic reticulum by enzymatic reactions similar to those that occur in prokaryotes. Since the disulphide bridge between Cys1 and Cys77 is formed far from the redox centre, haem, and is located on the periphery of the molecule (Figure 1A), it is not likely that forming or breaking this bridge would have a major effect on the redox properties of A. suum cytochrome b5 or on its interaction with its redox partners, metmyoglobin and NADH-cytochrome b5 reductase. Rather, it is more likely that this disulphide bridge would increase the stability of the molecule. The nematode hypodermis, in which A. suum cytochrome b5 is localized (see below), is located immediately below the outermost cuticle and is therefore easily affected by toxic ions, gasses and probably proteases, which are derived from the host's small intestine. The head-to-tail disulphide bond may function to counteract the effects of these molecules that cause fluctuations in protein structure.
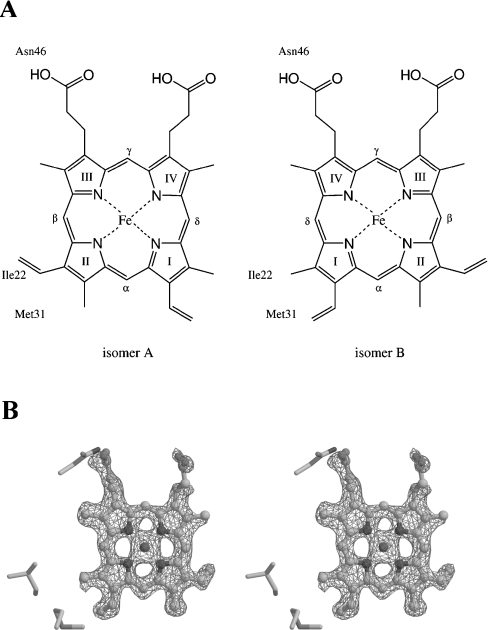
Haem orientation
Cytochrome b5 is generally known to be a mixture of two isomers, in which the haem group is bound in two orientations, called isomers A and B (Figure 3A), which are related to each other by a 180° rotation about the porphyrin α,γ-meso axis [17,29]. The two isomers have been found to be slowly interconverted, and the ratio of the two isomers at equilibrium was observed to differ among cytochromes b5 from various sources. Isomer A is the major component in bovine, chicken and rat microsomal cytochromes b5, with A to B ratios of 8.9:1, 20:1 and 1.6:1 respectively [30]. In contrast, isomer B has been reported to be dominant in wild-type and in the A18S/L47R/I32L triple mutant of rat mitochondrial outer membrane cytochrome b5 after incubation at 65 °C for several hours, with ratios of 1:1.2 and 1:3 respectively [31,32].
Figure 3. Haem conformation in A. suum cytochrome b5.
(A) Two haem conformations about the α,γ-meso axis are denoted as isomers A and B. The haem-Fe atom is bound to His38 and His62 from the front and back sides respectively. Three residues, Ile22, Met31 and Asn46, which are in close contact with the haem group, are shown in the respective Figure. (B) Stereo view of an Fo-Fc omit map for haem at 1.8 Å resolution. The haem group and residues within 10 Å of the haem are excluded from structure factor calculations and difference electron density is contoured at the 3σ level. Isomer B, the predominant haem form of A. suum cytochrome b5, is drawn as a ball-and-stick model.
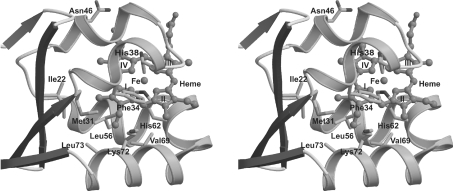
A difference Fourier map showed that A. suum cytochrome b5 is predominantly isomer B (Figure 3B). It is quite likely, however, that there was a small fraction of isomer A. The ratio of isomers A and B was estimated to be 1:2.3 after structural refinement that considered the occupancy of isomers A and B. In isomer B, the vinyl group of pyrrole I is accommodated in a hydrophobic pocket composed of the side chains of Met31, Phe34, Leu56, Val69, Lys72 and Leu73 (Figure 4). The methyl group of pyrrole I faces Ile22 and Met31, with the distance between the methyl group and these two residues being 4.13 Å and 4.24 Å respectively. In isomer A, however, the methyl group of pyrrole II is directed toward the hydrophobic pocket, and the vinyl group interacts with Ile22 and Met31. The void space surrounding Ile22 and Met31 is not large enough to accommodate the vinyl group, and the bond angle C=C-C in the group has to be distorted to 159° from its ideal value, 120°. Therefore the unfavourable angle distortion in isomer A probably explains why isomer B is the predominant form of A. suum cytochrome b5.
Figure 4. Stereo diagram of the haem-binding site.
The haem moiety in isomer B is drawn as a ball-and-stick model. Met31, Phe34, Leu56, Val69, Lys72 and Leu73 form a hydrophobic pocket, which accommodates the vinyl group of haem pyrrole I.
In mammalian cytochrome b5, Em was found to be +0.8 and −26.2 mV for the pure A and B haem orientations respectively [29]: net effects on the redox potential of isomerization different by only 27 mV. Therefore the predominance of B-isomer cannot explain the high redox potential of the nematode protein.
A. suum cytochrome b5 also differs from its mammalian counterparts in haem orientation. In the nematode protein, the haem plane is rotated by approx. 80° against its normal axis, which is formed by haem-Fe and Nϵ atoms of His38 and His62. As a consequence, the two propionate groups in the haem are oriented toward helices 3 and 4. Interestingly, one of the carboxy groups of the propionates is hydrogen-bonded to the Nδ atom of Asn46, with a distance of 2.8 Å. Since the redox potential of cytochrome b5 is affected by the carboxy group of the haem propionate [33], the highest Em value of A. suum cytochrome b5 could be explained by the rotation of the haem, together with the hydrophobic interaction between the haem moiety and three of the residues, Ala60, Pro61 and Val63, located on the newly introduced helix 4A (see below).
Structural comparison of A. summ cytochrome b5 with other related proteins
Three-dimensional structures of the cytochrome b5 catalytic domain from various species have been solved by X-ray crystallography or NMR. Structural studies of wild-type (PDB codes 1B5M and 1EUE) and mutant (V45L/V61L, PDB code 1AWP, and A18S/I32L/L47R, PDB code 1ICC) forms of rat outer mitochondrial membrane cytochrome b5, and wild-type (PDB code 1CYO) and V61H mutant (PDB code 1EHB) forms of bovine microsomal cytochrome b5 [27,32–36] have shown them to be similar in overall structure and haem-binding motifs. As rmsd values of Cα atoms are 0.17–0.38 Å when these proteins, except for the N- and C- terminal residues, are superimposed by the least-squares method, wild-type bovine cytochrome b5 (PDB code 1CYO) has been chosen as the reference for structural comparison with the A. suum protein.
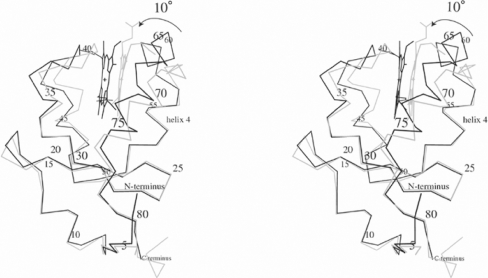
A comparison of the primary and secondary structure of A. suum cytochrome b5 with the bovine protein showed that the former has a one-residue deletion in helix 3 and a four-residue insertion in helix 4A (Figure 2). When the overall structures of the A. suum and bovine cytochromes b5 were superimposed using the corresponding Cα atoms of 76 residues, except for two residues at the N-terminus, the rmsd was 1.46 Å, and the maximum displacement was 6.50 Å. Significant differences were observed at Phe34–Asn46 and Leu59–Gln75; the former comprises helix 2, helix 3 and the loop connecting the two, and the latter comprises helix 4A, helix 5 and the interconnecting loop. The structure of mammalian cytochrome b5 is composed of two cores: the haem-binding site consisting of helices 2, 3, 4 and 5, and a β-sheet with helix 1. The Cα positions of the β-sheet core between A. suum and bovine proteins were well superimposed, with an rmsd of 0.28 Å and a maximum displacement of 1.42 Å. In contrast, the helix bundle core showed an rmsd of Cα atoms of 1.2 Å and a maximum displacement of 5.2 Å, caused by a 10° twist with helix 4 as the hinge (Figure 5).
Figure 5. Stereo diagram showing an overlap of the α-carbon backbone and the haem group of A. suum (heavy line) and bovine (pale line) cytochrome b5 structures.
The α-carbon atom of every fifth residue of the A. suum protein is labelled with a sequence number. Both structures are well superimposed with their β-sheet cores, while the A. suum helical bundle is twisted by 10° from its position in the bovine protein.
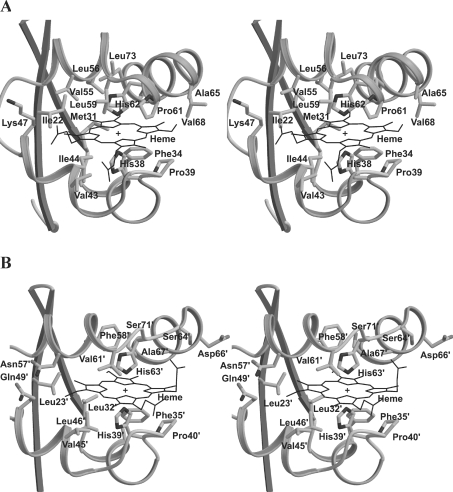
When the haem-binding sites of the A. suum and bovine proteins were superimposed by the least-squares method for haem and haem-interacted residues (Figure 6), it turned out that the conformations of the haem and the periphery of His62 (helix 4A) clearly differed between two proteins, although they were similar in the haem-binding mode. At a glance, Phe34, Pro39 and Gly40, and Phe35′, Pro40′ and Gly41′, all of which are conserved in the primary structure, are well superimposed. The distance between the Cα atoms of the two ligating histidines (His38 and His62 for A. suum, and His39′ and His63′ for bovine) differed in that the Cα distance for the A. suum protein was 10.95 Å, or 0.71 Å shorter than that for the bovine protein, 11.66 Å. The two proteins did not differ significantly, however, in the distance between the haem-Fe atom and each Nϵ atom of the two histidine residues. The hydrophobic pocket accommodating the haem moiety comprises Ile22, Met31, Phe34, Pro39, Gly40, Val43, Ile44, Lys47, Leu56, Leu59, Pro61, Ala65, Val68, Val69 and Leu73. As stated above, Phe34, Pro39 and Gly40 fit well the corresponding residues in bovine protein. Although the Cα positions of Ile22, Met31, Val43, Ile44, Lys47, Leu56, Leu59, Val69 and Leu73 differ from those in the bovine protein (Leu23′, Leu32′, Val45′, Leu46′, Gln49′, Phe58′, Val61′, Ala67′ and Ser71′), their side chains occupy close positions in the superimposed proteins. Pro61, Ala65 and Val68 of A. suum cytochrome b5 form novel hydrophobic interactions with the haem moiety, which arise from the formation of the unique helix 4A. Since three residues are found at the entrance of the haem-binding crevice, these residues, especially Pro61, narrow the width of the crevice entrance compared with that in the bovine protein. It is therefore possible that the existence of residues narrowing the crevice width is one of the factors defining the unique haem orientation in A. suum cytochrome b5.
Figure 6. Stereo view of ribbon-and-stick model for A. suum (A) and bovine (B) cytochrome b5.
Although the haem-binding motifs in the two proteins are similar, the porphyrin ring in the A. suum protein is rotated by 80° about a bond between the haem-Fe and two histidines, His38 and His62.
Immunohistochemical localization of A. suum cytochrome b5 in the nematode body wall
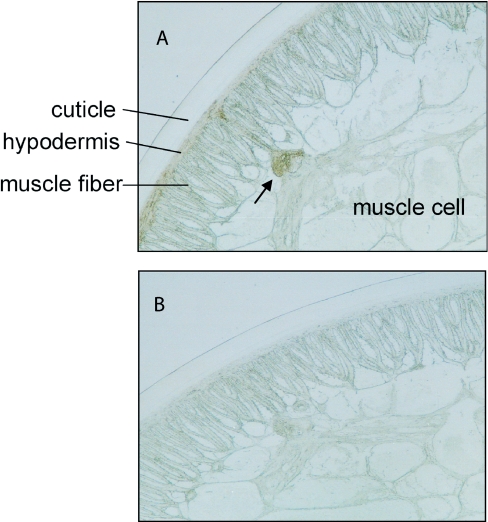
What is the physiological oxidant of A. suum cytochrome b5 in the body wall? In bovine heart, the presence of NADH-metmyoglobin reductase and the in vitro requirement of the trypsin fragment of microsomal cytochrome b5 for the reductase activity have been reported, although no cytochrome b5 can be detected in heart tissues [37,38]. Since nematode cytochrome b5 was extracted from its body wall, it is conceivable that this protein may be involved in the reduction of A. suum body-wall metmyoglobin. Body-wall myoglobin, which is at its highest concentration in the hypodermis immediately below the cuticle, in the ventral, dorsal and lateral lines and in the nerve ring, is also present, but at lower concentrations, in the muscle [19]. When we assayed the distribution of cytochrome b5 in A. suum body wall by immunohisto-chemistry, using anti-(A. suum cytochrome b5) antibody, we found that nematode hypodermis, muscle fibre and ventral nerve were consistently stained in sample sections (Figure 7A), but not in control sections (Figure 7B), indicating that A. suum cytochrome b5 co-localizes with body-wall myoglobin. Since A. suum cytochrome b5 is a soluble protein, lacking a C-terminal domain that would target it to the endoplasmic reticulum or mitochondria, this protein is probably localized to the cytosol. When we performed subcellular fractionation followed by immunoblotting, we found that A. suum cytochrome b5 was present in the cytosolic fraction, but not in either the microsomal or mitochondrial fraction (M. Hashimoto and S. Takamiya, unpublished work). Although the physiological functions of the signal peptide are not known, it may target the protein to the hypodermis after translation at other sites. Further studies are now in progress to determine the role of the signal peptide.
Figure 7. Immunohistochemical localization of A. suum cytochrome b5 in the body wall of adult nematode.
(A) Cross-section of adult A. suum reacted with anti-(A. suum cytochrome b5) antibody showing the staining in the hypodermis and muscle fibre. Note that the ventral nerve (shown by an arrow), which projects into the body cavity from the hypodermis, was also heavily stained. (B) Control section reacted with normal rabbit serum showing no significant staining.
A. suum cytochrome b5 facilitates reduction of metmyoglobin of the nematode body wall
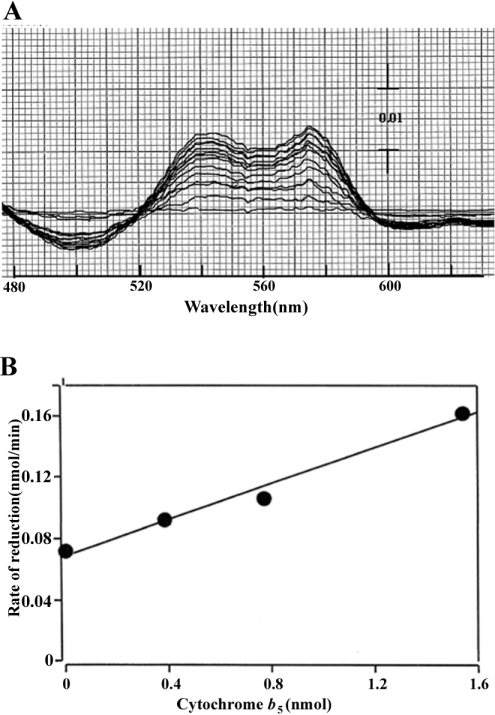
We then tested the effects of A. suum cytochrome b5 on metmyoglobin reductase activity. This reaction required NADH-cytochrome b5 reductase to supply electrons to cytochrome b5. Since A. suum cytochrome b5 is a cytosolic protein, A. suum NADH-cytochrome b5 reductase is probably also cytosolic, enabling it to interact with cytochrome b5. However, we used the microsomal, rather than the cytosolic, fraction to supply reductase activity, primarily because endogenous compounds in the cytosolic fraction severely disturbed the metmyoglobin reductase assay. We found that nematode cytochrome b5 stimulated NADH-dependent metmyoglobin reductase, with an increase in absorption at both 543 and 578 nm, peaks characteristic of nematode oxymyoglobin (Figure 8A), when the reaction was initiated by the addition of NADH. In addition, the rate of oxymyoglobin produced increased linearly with the amount of cytochrome b5 added (Figure 8B). Although non-physiological reduction was observed without added cytochrome b5, the cytochrome b5-dependent increment of the metmyoglobin reduction rate showed that electrons from reduced cytochrome b5 were transferred to metmyoglobin. These results, together with information on localization, show that A. suum cytochrome b5 probably functions as a bona fide electron carrier, shuttling between NADH-cytochrome b5 reductase and nematode body-wall myoglobin. To understand the detailed mechanism of nematode metmyoglobin reduction, further studies are required using NADH-cytochrome b5 reductase purified from the cytosolic fraction. To our knowledge, the present paper is the first report showing the co-existence of myoglobin and cytochrome b5 in animal muscle tissues.
Figure 8. Stimulation by A. suum cytochrome b5 of nematode cytochrome b5 reductase-catalysed reduction of nematode body-wall metmyoglobin.
(A) Difference spectra obtained by repeated scanning show the time course of metmyoglobin reduction in the presence of 0.384 μM cytochrome b5. Time-dependent increase in absorptions at 543 and 578 nm was observed during 13 cycle scanning. (B) Dose-dependent relationship between reduction rate and concentration of added cytochrome b5. Experimental details are described in the Experimental section.
Charge distribution surrounding the haem crevice
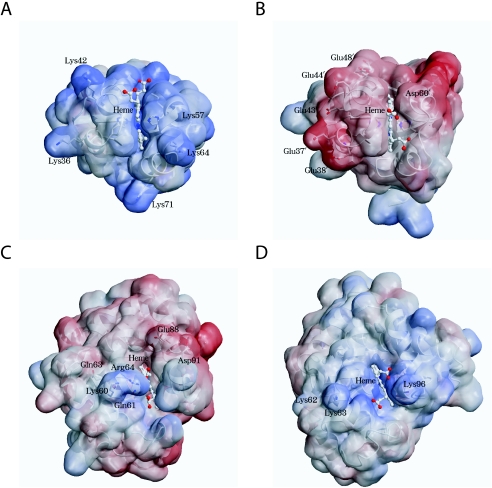
The interaction of mammalian cytochromes b5 with reaction partners is known to take place through hydrogen bonding of negatively charged residues on cytochrome b5 and positively charged residues on the partner protein [39]. For example, the cytochrome b5 residues Glu44′, Glu43′ and Asp60′ (Figure 2, upward arrows) are thought to interact with the methaemoglobin (α-chain) residues Lys56, Lys60 and Lys90 respectively, forming a 1:1 complex. These charged residues are located on the edges of the haem crevices of the respective proteins and are conserved among a variety of species. This does not occur, however, in A. suum cytochrome b5. Structural alignment (Figure 2) shows that the A. suum residues corresponding to mammalian Glu43′ and Asp60′ are Lys42 and Thr58, which are positively charged and hydrophilic residues respectively, whereas the residue corresponding to mammalian Glu44′ is deleted in nematode cytochrome b5. Furthermore, several other acidic residues of the mammalian protein, i.e. Glu37′, Glu48′, Glu56′, Glu59′, Asp66′, Glu69′ and Glu78′ (Figure 2, indicated by +) are replaced by lysine (Figure 2, indicated by downward arrows) or hydrophobic (Val68)/hydrophilic (Ser54 and Asn46) residues in the nematode protein. Except for Glu78′, all of these mammalian and nematode protein residues are located on the haem crevices of their respective molecules. Thus the charge distribution on haem crevices is remarkably different between A. suum and mammalian cytochromes b5, presenting a striking contrast in the electrostatic charge on the molecular surfaces (Figures 9A and 9B).
Figure 9. Comparison of the electrostatic molecular surfaces around the haem crevices of A. suum (A) and bovine (B) cytochrome b5, and A. suum model (C) and horse metmyoglobins (D) (PDB code 1WLA).
The electrostatic potential mapped on the protein surface was calculated using APBS [51]. A positive potential is indicated in blue, and a negative one is indicated in red.
Interaction between A. suum cytochrome b5 and metmyoglobin
Since A. suum cytochrome b5 reacts with nematode body-wall metmyoglobin, it is interesting to survey the residues of the metmyoglobin that interact with cytochrome b5. Although there are no crystallographic data on A. suum myoglobin, we built a tentative docking model based on the three-dimensional structure of A. suum perienteric haemoglobin domain 1 (PDB code 1ASH), which has a 41% sequence identity with nematode myoglobin [40]. Computer modelling suggests that A. suum cytochrome b5 residues Lys42 and Lys64 interact with nematode metmyoglobin Gln63, Glu88 and Asp91 (Figure 9C), forming hydrogen bonds (results not shown). Thus, in A. suum, the positively charged residues on cytochrome b5 interact with the negatively charged residues on myoglobin, which is the opposite to what occurs in mammalian myoglobin–cytochrome b5 couples (Figures 9B and 9D). There is also contact between the carboxy group of haem propionate on cytochrome b5 and two metmyoglobin residues, Lys60 and Arg64, in that the rotated haem configuration of cytochrome b5 makes it possible to form hydrogen bonds. It should be pointed out that cytochrome b5 residue Lys64 is located on helix 4A, which is unique to the A. suum protein, and that the metmyoglobin residues that interact with nematode cytochrome b5 (Lys60, Gln63 and Arg64) are members of tripeptide motif Gln-Gly-Gln [40], which is highly conserved at E6–E8 in nematode globin, or the residues just next to the motif, which form the sequence Lys60-Gln61-Gly62-Gln63-Arg64. Thus the additional helix 4A and rotated haem configuration, both of which are unique to A. suum cytochrome b5, appear to be essential for interactions with their reaction partner, nematode metmyoglobin, and the residues of the latter that interact with A. suum cytochrome b5 are also nematode-specific.
Physiological function
A. suum is a parasitic nematode species, the adult stage of which lives in the micro-aerobic environment of the pig intestine, whose oxygen tension is very low. To adapt to this hypoxic environment, adult nematodes exploit a unique anaerobic energy metabolism, especially in their body-wall mitochondria, where, instead of oxygen, fumarate functions as the terminal electron acceptor during ATP production (see [41] for a review). Nevertheless, the adult nematode possesses at least two types of globin associated with haem molecules, haemoglobin and myoglobin, which are localized in the pseudocoelomic (perienteric) fluid and body wall tissues of the nematode respectively. Since A. suum adult nematodes do not use molecular oxygen for ATP production, the physiological roles of the nematode globins have been the subject of many debates [42,43]. In fact, the pseudocoelomic and body wall globins in A. suum both exhibit unusual physicochemical features. For example, the oxygen affinities of nematode haemoglobin and myoglobin are respectively approx. 13000- and 20-fold higher than that of their mammalian counterparts, as measured by their p50O2 (partial pressure of O2 producing 50% saturation) values [20]. Previously, A. suum pseudocoelomic haemoglobin was reported to be a nitric oxide-dependent deoxygenase that uses endogenously produced NO to detoxify oxygen [44]. There is also evidence suggesting that body-wall myoglobin functions as an oxygen carrier in the biosynthesis of molecules that directly incorporate oxygen. One of these is protocollagen proline hydroxylase, which catalyses the hydroxylation of proline residues in A. suum protocollagen and is an oxygenase that uses molecular oxygen as substrate [45]. Furthermore, in vivo deoxygenation of body-wall myoglobin has been observed to render the worms inactive, suggesting that a supply of oxygen is needed for muscle movement and that globin functions as an oxygen carrier [46]. In any case, oxidation of myoglobin to metmyoglobin is likely to be harmful to A. suum nematodes. Thus our results show that A. suum cytochrome b5 is involved in the reduction of body-wall metmyoglobin, which is produced during parasitic life in the small intestine, where the nematodes are exposed to exogenous oxidants, including various nitrogen oxides [47].
Acknowledgments
The MAD experiment was performed at BL18B station at Photon Factory (Tsukuba, Japan) with the assistance of Foundation for Advancement of International Science (FAIS). This work was supported in part by the ‘National Project on Protein Structural and Functional Analyses’ run by the Japanese Ministry of Education, Culture, Sports, Science and Technology, a Grant-in-Aid for Scientific Research (C) (Nos. 12670241 and 14570220) from the Ministry of Education, Science, Sports and Culture of Japan, and a research grant from the Institute for Environmental and Gender-specific Medicine, Juntendo University School of Medicine. We thank Emiko Sato for her excellent technical assistance.
References
- 1.Lederer F. The cytochrome b5-fold: an adaptable module. Biochimie. 1994;76:674–692. doi: 10.1016/0300-9084(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 2.Vergeres G., Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 1995;77:604–620. doi: 10.1016/0300-9084(96)88176-4. [DOI] [PubMed] [Google Scholar]
- 3.Mitoma J., Ito A. The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J. 1992;11:4197–4203. doi: 10.1002/j.1460-2075.1992.tb05513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuroda R., Ikenoue T., Honsho M., Tsujimoto S., Mitoma J., Ito A. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 1998;273:31097–31102. doi: 10.1074/jbc.273.47.31097. [DOI] [PubMed] [Google Scholar]
- 5.Oshino N., Omura T. Immunochemical evidence for the participation of cytochrome b5 in microsomal stearyl-CoA desaturation reaction. Arch. Biochem. Biophys. 1973;157:395–404. doi: 10.1016/0003-9861(73)90655-3. [DOI] [PubMed] [Google Scholar]
- 6.Ito A., Hayashi S., Yoshida T. Participation of a cytochrome b5-like hemoprotein of outer mitochondrial membrane (OM cytochrome b) in NADH-semidehydroascorbic acid reductase activity of rat liver. Biochem. Biophys. Res. Commun. 1981;101:591–598. doi: 10.1016/0006-291x(81)91300-0. [DOI] [PubMed] [Google Scholar]
- 7.Hultquist D. E., Passon P. G. Catalysis of methaemoglobin reduction by erythrocyte cytochrome b5 and cytochrome b5 reductase. Nat. New Biol. 1971;229:252–254. doi: 10.1038/newbio229252a0. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y., Yamasaki H., Kita K., Takamiya S. Purification and molecular characterization of a novel b5-type cytochrome of the parasitic nematode, Ascaris suum. Arch. Biochem. Biophys. 1996;328:165–172. doi: 10.1006/abbi.1996.0157. [DOI] [PubMed] [Google Scholar]
- 9.Abe K., Sugita Y. Properties of cytochrome b5 and methemoglobin reduction in human erythrocytes. Eur. J. Biochem. 1979;101:423–428. doi: 10.1111/j.1432-1033.1979.tb19735.x. [DOI] [PubMed] [Google Scholar]
- 10.Lloyd E., Ferrer J. C., Funk W. D., Mauk M. R., Mauk A. G. Recombinant human erythrocyte cytochrome b5. Biochemistry. 1994;33:11432–11437. doi: 10.1021/bi00204a005. [DOI] [PubMed] [Google Scholar]
- 11.Takamiya S., Yamasaki H., Hashimoto M., Taka H., Murayama K., Tagaya M., Aoki T. Heterologous expression of Ascaris suum cytochrome b5 precursor protein: a histidine-tagged full-length presequence is correctly processed to transport the mature protein to the periplasm of Escherichia coli. Arch. Biochem. Biophys. 2003;413:253–261. doi: 10.1016/s0003-9861(03)00124-3. [DOI] [PubMed] [Google Scholar]
- 12.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 13.Matthews B. W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 14.Terwilliger T. C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones T. A., Zou J.-Y., Cowan S. W., Kjeldgaard M. Improved methods for building of protein models in electron density maps and location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 16.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewski J., Nilges N., Pannu N. S., et al. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 17.La Mar G. N., Burns P. D., Jackson J. T., Smith K. M., Langry K. C., Strittmatter P. Proton magnetic resonance determination of the relative haem orientations in disordered native and reconstituted ferricytochrome b5: assignment of haem resonances by deuterium labeling. J. Biol. Chem. 1981;256:6075–6079. [PubMed] [Google Scholar]
- 18.Tronrud D. E. TNT refinement package. Methods Enzymol. 1997;277:306–319. doi: 10.1016/s0076-6879(97)77017-4. [DOI] [PubMed] [Google Scholar]
- 19.Okazaki T., Wittenberg B. A., Briehl R. W., Wittenberg J. B. The hemoglobin of Ascaris body walls. Biochim. Biophys. Acta. 1967;140:258–265. doi: 10.1016/0005-2795(67)90466-7. [DOI] [PubMed] [Google Scholar]
- 20.Wittenberg J. B. Facilitated oxygen diffusion: the role of leghemoglobin in nitrogen fixation by bacteroids isolated from soybean root nodules. J. Biol. Chem. 1974;249:4057–4066. [PubMed] [Google Scholar]
- 21.Takamiya S., Kita K., Matsuura K., Furushima R., Oya H. Oxidation–reduction potentials of cytochromes in Ascaris muscle mitochondria: high-redox-potential cytochrome b558 in complex II (succinate–ubiquinone reductase) Biochem. Int. 1990;21:1073–1080. [PubMed] [Google Scholar]
- 22.Markwell M. A. K., Haas S. M., Bieber L. L., Tolbert N. E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 25.Fenn T. D., Ringe D., Petsko G. A. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J. Appl. Crystallogr. 2003;36:944–947. [Google Scholar]
- 26.Merritt E. A., Murphy M. E. P. Raster3D Version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 27.Durley R. C. E., Mathews F. S. Refinement and structural analysis of bovine cytochrome b5 at 1.5 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996;52:65–76. doi: 10.1107/S0907444995007827. [DOI] [PubMed] [Google Scholar]
- 28.Sevier C. S., Kaiser C. A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 29.Walker F. A., Emrick D., Rivera J. E., Hanquet B. J., Buttlaire D. H. Effect of heme orientation on the reduction potential of cytochrome b5. J. Am. Chem. Soc. 1988;110:6234–6240. doi: 10.1021/ja00226a045. [DOI] [PubMed] [Google Scholar]
- 30.Lee K.-B., La Mar G. N., Kehres L. A., Fujinari E. M., Smith K. M., Pochapsky T. C., Sligar S. G. 1H NMR study of the influence of hydrophobic contacts on protein-prosthetic group recognition in bovine and rat ferricytochrome b5. Biochemistry. 1990;29:9623–9631. doi: 10.1021/bi00493a017. [DOI] [PubMed] [Google Scholar]
- 31.Silchenko S., Sippel M. L., Kuchment O., Benson D. R., Mauk A. G., Altuve A., Rivera M. Hemin is kinetically trapped in cytochrome b5 from rat outer mitochondrial membrane. Biochem. Biophys. Res. Commun. 2000;273:467–472. doi: 10.1006/bbrc.2000.2968. [DOI] [PubMed] [Google Scholar]
- 32.Altuve A., Silchenko S., Lee K.-H., Kuczera K., Terzyan S., Zhang X., Benson D. R., Rivera M. Probing the differences between rat liver outer mitochondrial membrane cytochome b5 and microsomal cytochrome b5. Biochemistry. 2001;40:9469–9483. doi: 10.1021/bi010636i. [DOI] [PubMed] [Google Scholar]
- 33.Rivera M., Seetharaman R., Girdhar D., Wirtz M., Zhang X., Wang X., White S. The reduction potential of cytochrome b5 is modulated by its exposed heme edge. Biochemistry. 1998;37:1485–1494. doi: 10.1021/bi972390g. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Maranon M. J., Qiu F., Stark R. E., White S. P., Zhang X., Foundling S. I., Rodriguez V., Schilling C. L., III, Bunce R. A., Rivera M. 13C NMR spectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c. Biochemistry. 1996;35:16378–16390. doi: 10.1021/bi961895o. [DOI] [PubMed] [Google Scholar]
- 35.Wirtz M., Oganesyan V., Zhang X., Studer J., Rivera M. Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5. Faraday Discuss. 2000;116:221–234. doi: 10.1039/b001520m. [DOI] [PubMed] [Google Scholar]
- 36.Wu J., Gan J.-H., Xia Z.-X., Wang Y.-H., Wang W.-H., Xue L.-L., Xie Y., Huang Z.-X. Crystal structure of recombinant trypsin-solubilized fragment of cytochrome b5 and the structural comparison with Val61His mutant. Proteins. 2000;40:249–257. doi: 10.1002/(sici)1097-0134(20000801)40:2<249::aid-prot70>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 37.Hagler L., Coppes R. I., Jr, Herman R. H. Metmyoglobin reductase: identification and purification of a reduced nicotinamide adenine dinucleotide-dependent enzyme from bovine heart which reduces metmyoglobin. J. Biol. Chem. 1979;254:6505–6514. [PubMed] [Google Scholar]
- 38.Livingston D. J., McLachlan S. J., La Mar G. N., Brown W. D. Myoglobin:cytochrome b5 interactions and the kinetic mechanism of metmyoglobin reductase. J. Biol. Chem. 1985;260:15699–15707. [PubMed] [Google Scholar]
- 39.Poulos T. L., Mauk A. G. Models for the complexes formed between cytochrome b5 and the subunits of methemoglobin. J. Biol. Chem. 1983;258:7369–7373. [PubMed] [Google Scholar]
- 40.Blaxter M. L., Vanfleteren J. R., Xia J., Moens L. Structural characterization of an Ascaris myoglobin. J. Biol. Chem. 1994;269:30181–30186. [PubMed] [Google Scholar]
- 41.Kita K., Takamiya S. Electron-transfer complexes in Ascaris mitochondria. Adv. Parasitol. 2002;51:95–131. doi: 10.1016/s0065-308x(02)51004-6. [DOI] [PubMed] [Google Scholar]
- 42.Sherman D. R., Guinn B., Perdok M. M., Goldberg D. E. Components of sterol biosynthesis assembled on the oxygen-avid hemoglobin of Ascaris. Science. 1992;258:1930–1932. doi: 10.1126/science.1470914. [DOI] [PubMed] [Google Scholar]
- 43.Blaxter M. L. Nemoglobins: divergent nematode globins. Parasitol. Today. 1993;9:353–360. doi: 10.1016/0169-4758(93)90082-q. [DOI] [PubMed] [Google Scholar]
- 44.Minning D. M., Gow A. J., Bonaventura J., Braun R., Dewhirst M., Goldberg D. E., Stamler J. S. Ascaris haemoglobin is a nitric oxide-activated ‘deoxygenase’. Nature (London) 1999;401:497–502. doi: 10.1038/46822. [DOI] [PubMed] [Google Scholar]
- 45.Fujimoto D., Prockop D. J. Protocollagen proline hydroxylase from Ascaris lumbricoides. J. Biol. Chem. 1969;244:205–210. [PubMed] [Google Scholar]
- 46.Davenport H. E. The haemoglobins of Ascaris lumbricoides. Proc. R. Soc. London Ser. B. 1949;136:255–270. doi: 10.1098/rspb.1949.0024. [DOI] [PubMed] [Google Scholar]
- 47.Green L. C., Wagner D. A., Glogowski J., Skipper P. L., Wishnok J. S., Tannenbaum S. R. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal. Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 48.Ozols J. Structure of cytochrome b5 and its topology in the microsomal membrane. Biochim. Biophys. Acta. 1989;997:121–130. doi: 10.1016/0167-4838(89)90143-x. [DOI] [PubMed] [Google Scholar]
- 49.Lederer F., Ghrir R., Guiard B., Cortial S., Ito A. Two homologous cytochrome b5 in a single cell. Eur. J. Biochem. 1983;132:95–102. doi: 10.1111/j.1432-1033.1983.tb07330.x. [DOI] [PubMed] [Google Scholar]
- 50.Abe K., Kimura S., Kizawa R., Anan F. K., Sugita Y. Amino acid sequences of cytochrome b5 from human, porcine, and bovine erythrocytes and comparison with liver microsomal cytochrome b5. J. Biochem. 1985;97:1659–1668. doi: 10.1093/oxfordjournals.jbchem.a135224. [DOI] [PubMed] [Google Scholar]
- 51.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. Electrostatics of nanosystems: application to microtubules and ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]