Abstract
Claudin-1 is an integral membrane protein component of tight junctions. The Snail family of transcription factors are repressors that play a central role in the epithelial–mesenchymal transition, a process that occurs during cancer progression. Snail and Slug members are direct repressors of E-cadherin and act by binding to the specific E-boxes of its proximal promoter. In the present study, we demonstrate that overexpression of Slug or Snail causes a decrease in transepithelial electrical resistance. Overexpression of Slug and Snail in MDCK (Madin–Darby canine kidney) cells down-regulated Claudin-1 at protein and mRNA levels. In addition, Snail and Slug are able to effectively repress human Claudin-1-driven reporter gene constructs containing the wild-type promoter sequence, but not those with mutations in two proximal E-box elements. We also demonstrate by band-shift assay that Snail and Slug bind to the E-box motifs present in the human Claudin-1 promoter. Moreover, an inverse correlation in the levels of Claudin-1 and Slug transcripts were observed in breast cancer cell lines. E-box elements in the Claudin-1 promoter were found to play a critical negative regulatory role in breast cancer cell lines that expressed low levels of Claudin-1 transcript. Significantly, in invasive human breast tumours, high levels of Snail and Slug correlated with low levels of Claudin-1 expression. Taken together, these results support the hypothesis that Claudin-1 is a direct downstream target gene of Snail family factors in epithelial cells.
Keywords: Claudin-1, E-cadherin, Slug, Snail, tight junction, tumour
Abbreviations: c, canine; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; EMT, epithelial–mesenchymal transition; ERK, extracellular-signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; GST, glutathione S-transferase; HMEC, human mammary epithelial cell; m, mouse; MDCK, Madin–Darby canine kidney; pAb, polyclonal antibody; TEER, transepithelial electrical resistance; TJ, tight junction; ZO-1, zonula occludens-1
INTRODUCTION
TJs (tight junctions) constitute continuous circumferential seals around cells and serve as the primary barrier, preventing solutes and water from passing freely through the paracellular pathway [1]. One of the most important functions of TJs is to maintain the differentiated state of epithelial cells. An absence of TJs, or defective TJ formation, has been associated with the development of neoplastic phenotypes in normal epithelial cells [2–4]. Among the proteins present in TJs, Claudin-1 is a major constituent [5,6]. Several lines of evidence suggest that Claudin-1 is directly involved in the barrier and fence functions of TJs [1,7].
Human Claudin-1 was identified by differential display, comparing the mRNA of proliferating, early passage, and normal HMECs (human mammary epithelial cells) with the mRNA of normal senescent HMECs [8]. cDNA levels proved low in proliferating HMECs compared with the high expression detected in senescent HMECs; interestingly, the absence or significantly reduced expression of Claudin-1 has been observed in several established breast cancer cell lines [8]. Comparison of the expression profile of Claudin-1 in non-malignant cells with that in tumour-derived cells reveals this gene to be a key player in tumorigenesis, primarily by acting as a suppressor of mammary epithelial proliferation [8]. Analysis of the coding region of Claudin-1 in sporadic tumour cells and hereditary breast cancer patients did not reveal a clear relationship between alterations in Claudin-1 and its expression pattern. Furthermore, mutational analysis of the Claudin-1 gene and its putative promoter in breast cancer cell lines did not indicate any apparent modification [9].
Snail family members encode zinc-finger transcription factors that are essential for mesoderm formation in several organisms, from flies to mammals [10]. More recently, this role in promoting cell movement has been elucidated further to include more generalized phenomena such as EMT (epithelial–mesenchymal transition), a process that occurs at defined stages of embryonic development and during cancer progression [11–13]. EMT involves the conversion of an epithelial cell into a mesenchymal cell, one characterized by a more motile, invasive and aggressive phenotype. These changes allow some tumour cells to migrate through the extracellular matrix and colonize lymph/blood vessels in the first steps of the metastatic process. In the last few years, great advances have been made in understanding the EMT process and several critical molecules have been identified. Snail and Slug have now been firmly established as repressors of E-cadherin, one of the key molecules in the EMT process, both in early development and in cancer progression [11,12]. However, additional target genes are most likely required to explain the role of Snail in cell migration and cancer development, such as the recently identified mucin-1, collagen IIa1 or MMP-2 (matrix metalloproteinase-2) genes [14–16].
In the present study, we show that overexpression of Slug or Snail in MDCK (Madin–Darby canine kidney) cells led to a dramatic down-regulation of Claudin-1 protein levels and a significant reduction of Claudin-1 mRNA. The E-boxes in human Claudin-1 promoter are responsible for the Slug- and Snail-induced repression of its promoter activity, exerting both a critical and negative regulatory role in breast cancer cell lines that express low levels of the Claudin-1 transcript. Significantly, in invasive human breast tumours, high levels of Snail and Slug have been correlated with low levels of Claudin-1 expression. These observations suggest that the Snail family of transcription factors are strong candidates for mediating the repression of Claudin-1 expression in epithelial cells.
EXPERIMENTAL
Antibodies, recombinant proteins and cells
Reagents were purchased from Sigma, unless stated otherwise. Rabbit pAbs (polyclonal antibodies) were used to detect Claudin-1 and ZO-1 (zonula occludens-1) (Zymed). E-cadherin monoclonal antibody was purchased from Transduction Laboratories. Polyclonal antibodies against Snail (E-18) and Slug (H-140) were purchased from Santa Cruz Laboratories and polyclonal antibodies against GFP (green fluorescent protein) were from Clontech. MDCK cells stably transfected with Snail, Slug or pcDNA3 (control) have been described previously [11,17]. Primary fibroblast and the human breast cancer cell lines MDA-MB231, MDA-MB435, MDA-MB468, MCF-7 and T47D were obtained from the Cell Line Collection of Barcelona University (EucellBank). Cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) foetal calf serum. All experiments were performed with at least two clones. Transwell polycarbonate membrane inserts were purchased from Costar. Radioactive products were obtained from Amersham Biosciences. GST (glutathione S-transferase)–Snail and GST–Slug were produced as described previously [12,17].
Measurement of TEER (transepithelial electrical resistance)
For TEER measurements, 1×105 transfected cells were plated on Transwell polycarbonate membrane inserts with a pore size of 0.4 μm and an area of 1.1 cm2. A Millicell-ERS volt–ohmmeter (Millipore) was used to determine the TEER value. The Millicell-ERS system was used in accordance with the manufacturer's instruction. Calculations for ohm×cm2 were made by subtracting values of blank inserts from all samples and multiplying by the area of the monolayer. TEER values were determined after 48 h in culture in DMEM supplemented with 10% (v/v) foetal calf serum. TEER experiments were performed in triplicate for each transfected cell.
Immunofluorescence and Western blot analysis
Cells grown on coverslips were rinsed with PBS, fixed with 3% (w/v) paraformaldehyde for 20 min at room temperature (22 °C), and were permeabilized using 0.5% (v/v) Triton X-100. Monolayers were processed for indirect immunofluorescence with pAbs against Claudin-1 (1:100), ZO-1 (1:100) or E-cadherin (1:100), incubated with FITC-conjugated anti-rabbit or anti-mouse IgGs (Dako) and were analysed using confocal microscopy [18]. For Western blotting, 20 μg of proteins (whole extracts) were separated by SDS/7.5% PAGE and transferred on to nitrocellulose membranes (Schleicher & Schuell), and protein expression was analysed as described previously [18].
RT (reverse transcription)–PCR analysis
Total RNA was isolated from the different transfected cell lines using an RNA purification kit (Invitrogen). Mouse and canine PCR products were obtained after 30–35 cycles of amplification with an annealing temperature of 60–65 °C using a SuperScript One-Step RT–PCR kit (Invitrogen). Primer sequences were as follows. For canine Claudin-1 (cClaudin-1): forward, 5′-CGGTTCTGCGTCTCAGTTC-3′, and reverse, 5′-GTTGCCCATGACTCGCTC-3′; the primer pair amplified a fragment of approx. 250 bp. Database searches with the putative cClaudin-1 sequence revealed 96% identity with human Claudin-1. For canine GAPDH (glyceraldehyde-3-phosphate dehydrogenase): forward, 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′, and reverse, 5′-CATGTAGGCCATGAGGTCCACCAC-3′; the primer pair amplified a fragment of approx. 900 bp. For canine Snail (cSnail): forward, 5′-CCCAAGCCCAGCCGATGAG-3′, and reverse, 5′-CTTGGCCACGGAGAGCCC-3′ (amplified a fragment of approx. 200 bp). For mouse Snail (mSnail): forward, 5′-GGCGGATCCACCATGCCGCGCTCCTTCCTGGTC-3′, and reverse, 5′-CCGGATATCCGCGAGGGCCTCCGGAGCA-3′ (amplified a fragment of approx. 800 bp). For canine Slug (cSlug): forward, 5′-AGTGATTATTTCCCCATATCTCTATGA-3′, and reverse, 5′-GTAGTCTTTCCTCTTCATCACTAATGG-3′ (amplified a fragment of approx. 260 bp). For mouse Slug (mSlug): forward, 5′-CGCGAATTCCCGCCCGCAGCCACC-3′, and reverse, 5′-ACTCTCGAGCTAGTGTCAATGGGCGAC-3′ (amplified a fragment of approx. 850 bp).
Quantitative real-time PCR
Total RNA (1 μg) from each breast cancer cell line was reverse-transcribed in a final volume of 50 μl using Taqman reverse transcription reagents (Applied Biosystems). Real-time PCR on 20 ng of cDNA was performed for each of the following genes using assays-on-demand from Applied Biosystems: human Claudin-1 (hs00221623), Snail (hs00195591), Slug (hs00161904) and GAPDH (hs99999905). All PCRs were performed using an ABI Prism 7700 sequence detection system. For any sample, the expression levels of Claudin-1, Snail and Slug were normalized to the housekeeping gene GAPDH. Relative mRNA expression levels were determined using the comparative threshold cycle method [19]. The graphics represent [2−CT (target)/2−CT (housekeeping)] cell line/[2−CT (target)/2−CT (housekeeping)] fibroblast cells.
Isolation of promoter fragments, mutagenesis and reporter assays
A human Claudin-1 (−748 to +252) promoter fragment was amplified by PCR from genomic DNA of Caco2 cells and cloned into the pGL3 vector (Promega) upstream of firefly luciferase. PCR products were obtained after 40 cycles of amplification with an annealing temperature of 65 °C. Primer sequences were as follows: forward, 5′-GGAAACTACAGTCCCAGCGA-3′, and reverse, 5′-GATGTTGTCGCCGGCATA-3′ (amplified a fragment of 1384 bp). The fragment was digested using NheI/PvuII and cloned into NheI and SmaI sites of pGL3 vector (large promoter). The short construct (−82 to +236) was made by PCR-based site-directed mutagenesis. The PCR products were gel-purified and cloned using standard procedures. These reporter constructs (300 ng) were transfected into cells cultured in 24-well plates, in the absence or presence of the indicated amounts of pcDNA3 (control), pcDNA3-Snail or pcDNA3-Slug vectors; the total amount of transfected DNA was normalized with empty pcDNA3 plasmid. At 24 h after transfection, firefly luciferase (Luc) activity was measured using the Luciferase Reporter Assay system (Promega) according to the manufacturer's instructions. In all experiments, luciferase activity was normalized by taking into account the co-transfection efficiency of a GFP vector. The percentages of GFP-positive cells were analysed in a FACScalibur microflow cytometer (Becton Dickinson).
To mutate each E-box sequence in the Claudin-1 promoter, a QuikChange Site-Directed Mutagenesis kit (Stratagene) was used. The core sequence, 5′-CA(G/C)(G/C)TG-3′, was mutated to 5′-TG(G/C)(G/C)TA-3′. The insert vector, deletions and each combination of mutations were confirmed by sequencing.
EMSA (electrophoretic mobility-shift assay)
The double-stranded oligonucleotides used as probes for gel retardation corresponded to the following sequence in the Claudin-1 human promoter (+165 to +201): 5′-GTTGCCCACCTGCAAACTCTCCGCCTTCTGCACCTGCACCCC-3′ (E-boxes are indicated in bold). The probe was end-labelled with [α-32P]CTP using Klenow enzyme. Sequences of oligonucleotides used as competitors were as follows: E1 box (+153 to +192), 5′-TCGGGAGTCCGGGTTGCCCACCTGCAAACTCTCCGCCTTC-3′; E2 box (+176 to +215), 5′-GCAAACTCTCCGCCTTCTGCACCTGCCACCCCTGAGCCAG-3′. In mutated probes, the core sequence 5′-CACCTG-3′ of E1 and E2 boxes was mutated to 5′-TGCCTG-3′.
Gel-retardation assays were performed as described previously [12]. Briefly, 10 ng of recombinant proteins were incubated for 30 min with 2.5 ng of radiolabelled oligonucleotides. Binding buffer contained 20 mM Hepes, pH 7.6, 150 mM KCl, 3 mM MgCl2, 4% Ficoll, 0.1% Nonidet P40, 1 mM dithiothreitol, 1.5 μM ZnCl2 and 0.5 mg/ml BSA. The reactions were supplemented as indicated in Figure 5. For the competition experiments, unlabelled oligonucleotides were added 10 min before the labelled ones. To detect band supershifts, antibodies (anti-Snail, anti-Slug or non-immune serum) were added after this step for 15 min at room temperature. Complexes were resolved on 4% acrylamide gels (19:1 acrylamide/bisacrylamide) prepared in 22 mM Tris/borate.
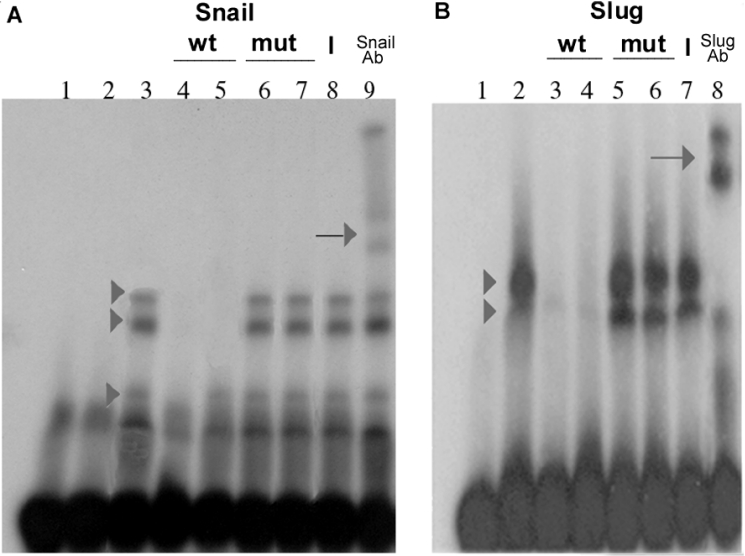
Figure 5. Direct binding of Snail and Slug to the E-boxes of the Claudin-1 promoter.
EMSA for the interaction of Snail and Slug with the E-box sequence. (A) Snail binding. Affinity-purified GST (lane 2) or GST–Snail (lanes 3–9) (10 ng) was incubated with double-stranded 32P-labelled oligonucleotides (2.5 ng) (+165 to +201) corresponding to the sequence containing two E-boxes of the Claudin-1 promoter; in lane 1, no protein was added. GST–Snail (arrowheads in lane 3), but not GST (lane 2), formed DNA complexes. Competitions were performed with a 200-fold excess of E1 (+153 to +192) and E2 (+176 to +215) wild-type probes (unlabelled oligonucleotides) (lanes 4 and 5), or mutant E1 and E2 boxes (lanes 6 and 7). Complex formation was affected by unlabelled wild-type E1 and E2 boxes, but not by unlabelled mutated boxes. Supershift experiments were performed by adding 200 ng of anti-Snail (lane 9) or non-specific (lane 8) antibody. Arrows in lane 9 indicate the supershifted retarded complexes. (B) Slug binding. Affinity-purified GST–Slug (lanes 2–8) (10 ng) was incubated with double-stranded 32P-labelled oligonucleotides (2.5 ng), containing E1 and E2 boxes (+165 to +201); in lane 1, no protein was added. GST–Slug formed two DNA complexes (arrowheads in lane 2). Competitions were performed with a 200-fold excess of wild-type E1 and E2 (lanes 3 and 4), or mutant E1 and E2 boxes (lanes 5 and 6). Results were very similar to those shown in (A). Supershift experiments were analysed by adding 200 ng of anti-Slug (lane 8) or non-specific (lane 7) antibody. Representative results from six independent experiments are shown. wt, wild-type; mut, mutant; Ab, antibody.
Microarray analysis
Using microarray analysis and samples from the fresh-frozen tissue bank of the Netherlands Cancer Institute, primary invasive breast carcinomas from a series of 295 consecutive women with breast cancer were studied by van de Vijver et al. [20]. Briefly, total RNA from each tumour was isolated and used to generate cRNA, which was then labelled and hybridized into microarrays containing approx. 25000 human genes. Fluorescence intensities of scanned images were quantified and normalized. The ratio was calculated with respect to the intensity of a reference pool made up of equal amounts of cRNA from all tumours [20]. From this dataset, we selected the entries containing the expression vectors for our genes of interest: Snail, Slug and Claudin-1. In order to identify trends in the expression levels of these genes, complete linkage hierarchical clustering was conducted on the normalized median-centred expression values using Euclidian distance [21].
RESULTS
Overexpression of Slug or Snail induces a disruption of TJs
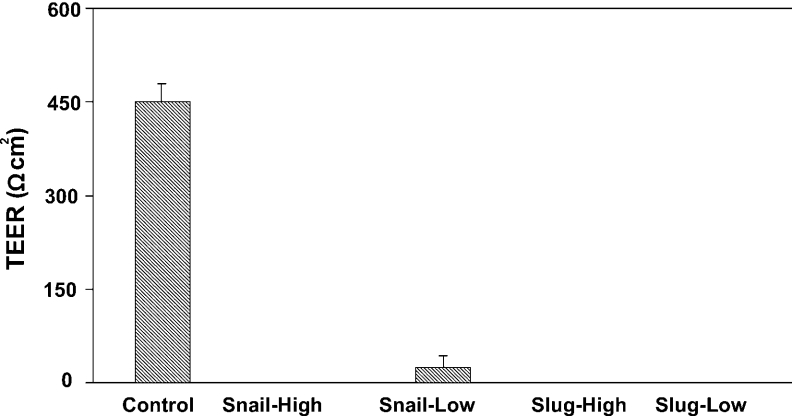
To determine whether Slug and/or Snail may contribute to the increased permeability of TJs during EMT, we measured the TEER of Slug/Snail-transfected MDCK cells (Figure 1). Slug- and Snail-expressing clones exhibited a complete abolition of TEER values, independently of the number of days in culture (results not shown). The reduction in TEER values proved to be unrelated to the levels of Snail and Slug expression.
Figure 1. Snail and Slug induce a disruption of TJs in epithelial cells.
Snail-, Slug- and pcDNA3- (control) stably transfected MDCK cells (1×105) were plated on Transwell polycarbonate membrane inserts with an area of 1.1 cm2. TEER values were determined after 48 h in culture in DMEM supplemented with 10% (v/v) foetal calf serum. Snail- and Slug-MDCK overexpressing clones (Snail-High, Snail-Low, Slug-High and Slug-Low) exhibited a complete abolition of TEER values when compared with control cells, independent of transcription factor expression levels. Results are means±S.D. for three independent experiments, each performed in triplicate.
Overexpression of Slug or Snail modifies Claudin-1 expression
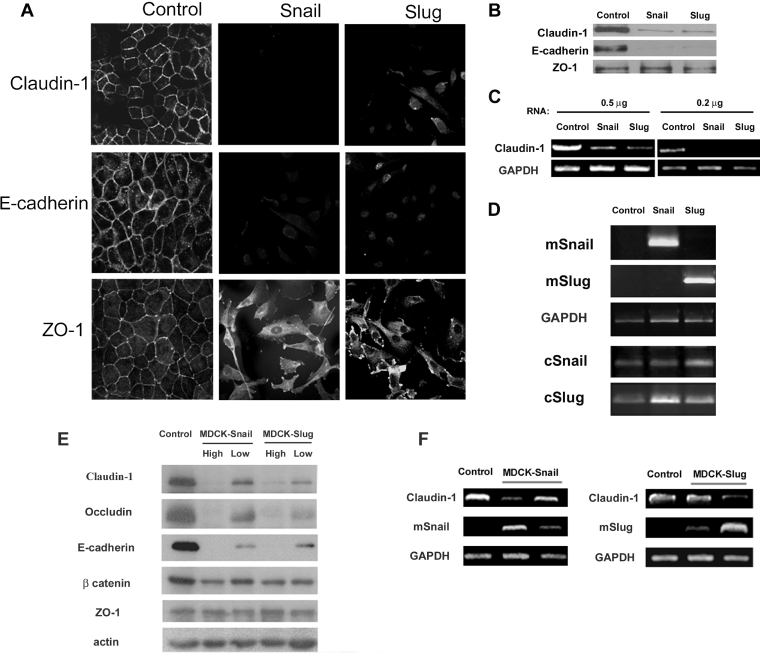
Using immunofluorescence microscopy, we examined Claudin-1 expression in MDCK (control) and MDCK clones derived after stable transfection with murine Slug or Snail [11,17]. As shown in Figure 2(A), control cells exhibited a regular linear labelling of Claudin-1, E-cadherin and ZO-1 at sites of cellular contact. However, both Slug- and Snail-transfected cells presented almost undetectable immunoreactivity for Claudin-1 and E-cadherin. The reduction in the labelling resulted from a dramatic decrease in protein levels of Claudin-1 and E-cadherin (Figure 2B). Consistent with previous observations [22], ZO-1 staining was redistributed from TJs in control MDCK cells to the cytoplasm in Snail- and Slug-transfectants, while protein levels remained unchanged in both cell types (Figures 2A and 2B). To clarify the mechanism underlying Claudin-1 down-regulation, we performed semi-quantitative RT–PCR to detect the expression of cClaudin-1 mRNA. In control cells, Claudin-1 transcripts were abundantly expressed, while, in Slug- and Snail-transfected cells, they were down-regulated (Figure 2C). To investigate further the potential influence of Snail and Slug levels in the regulation of TJs, protein Western blot and immunofluorescence analyses of Claudin-1, Occludin and ZO-1 were conducted in different MDCK-Snail and -Slug clones. As controls, we also included E-cadherin and β-catenin, members of adherens junctions (Figure 2E). Interestingly, while we observed a dose-dependent inhibition of Claudin-1, Occludin and E-cadherin expression, ZO-1 and β-catenin levels remained almost unchanged. The subcellular distribution of TJ proteins changed from cellular contact in control MDCK cells to the cytoplasm in Snail- and Slug-transfectants cells (Supplementary Figure 1S at http://www.BiochemJ.org/bj/394/bj3940449add.htm). In addition, the reduction of Claudin-1 transcription levels also occurred in a dose-dependent manner (Figure 2F).
Figure 2. Overexpression of Snail and Slug are associated with a full repression of Claudin-1 expression.
(A) Control (pcDNA3), Snail- and Slug-transfected MDCK clones were analysed by immunofluorescence and confocal microscopy for the expression of Claudin-1, E-cadherin and ZO-1. In Snail- and Slug-transfected cells, Claudin-1 and E-cadherin became almost undetectable. (B) Western blot analysis of whole-cell extracts for the indicated protein was conducted. Snail and Slug transfection caused a reduction in the protein levels of Claudin-1. (C) The reduction in the protein levels of Claudin-1 was caused by a reduction in the transcript. The presence of cClaudin-1 transcripts in control, Slug- and Snail-transfected clones was analysed by RT–PCR using different amounts of RNA. The expression of GAPDH was analysed in the same sample as a control. (D) RT–PCR for mSnail, mSlug cSnail, cSlug and GAPDH expression in control, Snail and Slug clones. (E) Expression patterns of TJ and adherens junction proteins in control and transfected cells with different levels of Snail and Slug expression. (F) RT–PCR for cClaudin-1, mSnail, mSlug and GAPDH expression in control and different Snail and Slug clones. Representative data from three independent experiments are shown.
Slug and Snail function as direct repressors of the Claudin-1 gene
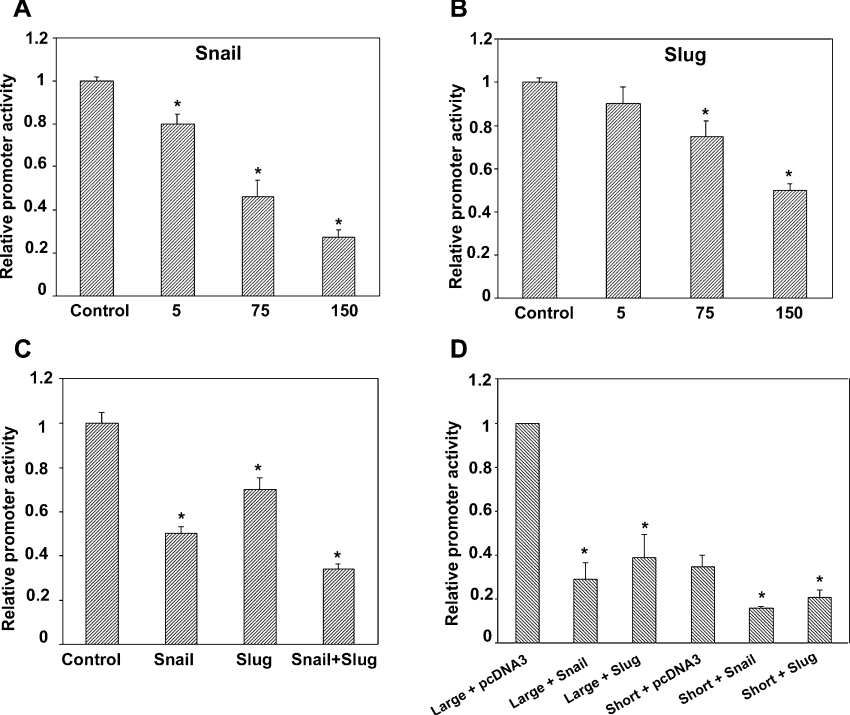
We subsequently examined whether or not the Claudin-1 gene is directly regulated by the Snail family members Slug and Snail (Figure 3). To this end, we isolated the promoter region of the human Claudin-1 gene (−748 to +252) [9] and fused it to the luciferase reporter cDNA. Transient transfection assays in the prototypic epithelial cell line MDCK in the presence of pcDNA3-Slug or pcDNA3-Snail demonstrated that both Slug and Snail are able to repress Claudin-1 promoter activity in a dose-dependent manner, although with apparent distinct efficiencies, inducing a 50% and 70% repression respectively, at 150 ng (Figures 3A and 3B). An additive effect was observed in the presence of 75 ng of each transcription factor (Figure 3C). These findings indicated that the transcription promoter activity of human Claudin-1 was regulated directly by both Slug and Snail factors. To account for the apparent discrepancies between the study published by Ohkubo and Ozawa [23] and our own findings, we obtained a short promoter of human Claudin-1 gene (−82 to +236) [23]. Both promoters were tested in MDCK cells. Consistent with our RT–PCR analysis, the shorter promoter of Claudin-1 was repressed by Snail and Slug in a manner similar to that of the large promoter (Figure 3D).
Figure 3. Snail- and Slug-induced repression of Claudin-1 promoter activity.
Luciferase reporter constructs carrying the human Claudin-1 promoter (−748 to +252) were transfected in MDCK cells (300 ng) with empty vector (pcDNA3) (control), or together with Snail or Slug, or a combination of both factor expression constructs. (A, B, C) When Snail or Slug was co-expressed in MDCK cells, the activity of the Claudin-1 reporter construct was repressed in a dose-dependent manner, an additive effect that was observed when both factors were co-transfected (C). (D) Luciferase reporter constructs carrying a large or short fragment of the human Claudin-1 promoter (300 ng) were transfected in MDCK cells, together with (150 ng) Snail or Slug expression constructs, or with an empty vector (pcDNA3). A shorter fragment was also sensitive to Snail and Slug repression. Results are means±S.D. for three independent experiments, each performed in quadruplicate. *, P<0.05.
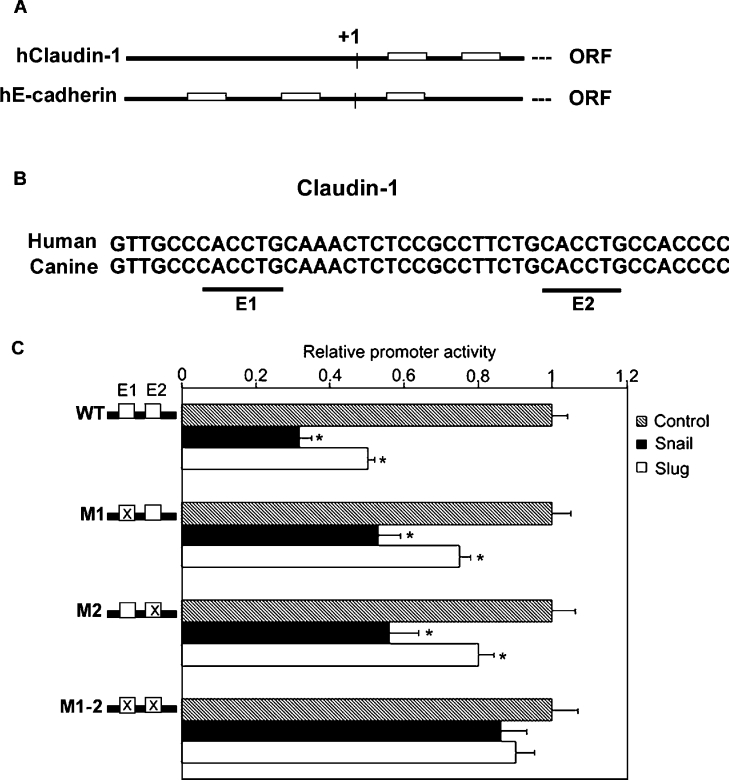
To clarify further the molecular mechanism underlying the Slug- and Snail-induced repression of Claudin-1 transcription, we examined the human Claudin-1 promoter in detail and found two E-box motifs (Figures 4A and 4B) conforming to the Snail-binding E-boxes (CACCTG) at +171 and +189. Interestingly, the two E-boxes and adjacent sequences were fully conserved between human and canine Claudin-1 genes (Figure 4B). We generated a series of reporter constructs that carried individual or double mutations in the E-boxes of the human Claudin-1 promoter. Tellingly, constructs with a single mutated E-box (M1 or M2) became less sensitive to Slug and Snail repression. In addition, an additive effect was observed when both E-boxes were mutated, the double-mutated construct (M1-2) becoming insensitive to Slug and Snail repression, thereby suggesting that the two proximal E-boxes in the Claudin-1 promoter are responsible for Slug- and Snail-induced repression (Figure 4C).
Figure 4. Impairment of Snail- and Slug-induced repression of Claudin-1 promoter activity by mutations in the E-boxes.
(A) Schematic representation of the E-boxes in the promoter region of human Claudin-1 and E-cadherin: open box, E box; +1, putative transcription star point; ORF, open reading frame. (B) Comparison between the 5′-flanking region of human and canine Claudin-1. Alignment of the sequences revealed the presence of conserved E-boxes (underlined). (C) Mutational analyses. The core 5′-CA(G/C)(G/C)TG-3′ sequence of the E-boxes (E1, E2 and E1-2) was mutated to 5′-TG(G/C)(G/C)TG-3′, in various combinations (shadowed boxes). Luciferase reporter constructs carrying wild-type or mutated boxes were transfected in MDCK cells (300 ng), together with (150 ng) Snail or Slug expression constructs or the empty vector (pcDNA3). Luciferase activity in cells co-transfected with wild-type reporter constructs and the pcDNA3 empty vector was defined as 1. Results are means±S.D. for three independent experiments, each performed in quadruplicate. *, P<0.05.
Snail and Slug bind directly to E-boxes in the Claudin-1 promoter
We confirmed that Slug and Snail proteins interact directly with their putative binding sites in the Claudin-1 promoter by using an EMSA (Figure 5). Recombinant mSlug and mSnail fused to GST, efficiently binding the proposed target sequences (boxes E1 and E2). GST–Snail formed three retarded complexes, while GST–Slug formed two complexes. Competitions were performed with a 200-fold excess of wild-type or mutant E1 and E2 cold probes. The specificity of each retarded complex was demonstrated by the fact that their formation was affected by unlabelled wild-type oligonucleotides, but not by unlabelled mutated oligonucleotides. Indeed, mutation of either of the two E-boxes was sufficient to block competition of the complexes generated by GST–Snail or GST–Slug with the labelled oligonucleotide, suggesting that both E-boxes are required for effective binding of the recombinant factors. Supershift experiments were performed by adding 200 ng of anti-Snail, anti-Slug or non-specific antibody. The presence of supershifted bands that appeared when incubating the complexes with an antibody directed against Snail or Slug, but not in the negative-control antibody, indicates that Snail and Slug are present in the complex.
Expression of Snail, Slug and Claudin-1 in human breast cancer cell lines and in human breast tumour samples
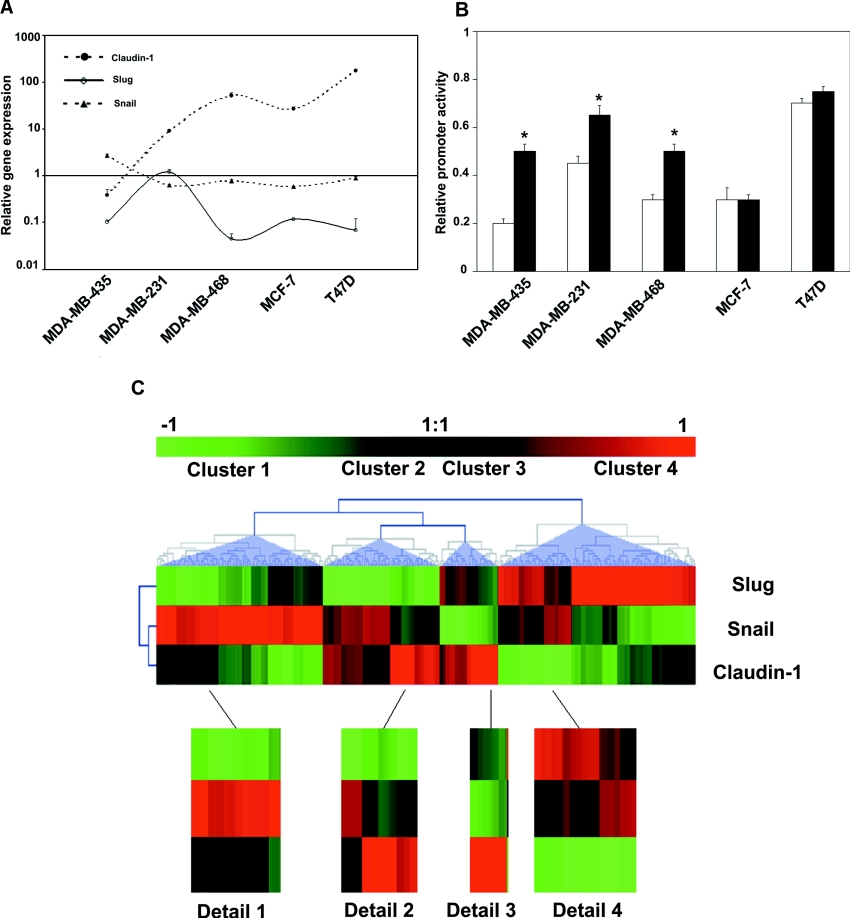
To define the putative repressor role of Snail and Slug in Claudin-1 expression in tumours of epithelial origin, real-time PCRs of these genes were conducted using a panel of epithelial breast cancer cell lines (Figure 6A). The low levels of Claudin-1 transcripts in these cell lines inversely correlated with Slug expression (Spearman coefficient, r=−0.64; P=0.042), while for Claudin-1 and Snail the Spearman coefficient indicated an inverse correlation, though it did not reach statistical significance (r=−0.15; P=0.6). The lowest levels of Claudin-1 expression corresponded to the invasive metastatic cell lines, MDA-MB435 and MDA-MB-231, which also expressed the highest levels of the Slug transcript. However, the non-metastatic cells lines MCF-7 and T47D expressed the highest levels of Claudin-1 and lower levels of Slug. To assess the putative role of Snail and Slug in the repression of Claudin-1 in breast cancer cell lines, luciferase experiments were conducted with the wild-type or E-box-mutated constructs. A strong reduction in the wild-type promoter was observed in almost all of the cell lines tested. Inhibition was dependent on the E-box elements of the Claudin-1 promoter in three of five breast cancer cell lines tested (Figure 6B). We then determined whether or not expression of Snail and Slug correlated with Claudin-1 in clinical breast tumour samples. To this end, we analysed a microarray gene expression data set from 295 invasive human breast tumours produced by van de Vijver et al. [20]. Figures of Merit analysis applied to the sample category suggested the presence of four main clusters. Figure 6(C) represents a hierarchical clustering of the samples, where each row corresponds to a gene of interest and each column represents the relative level of gene expression in a given tumour sample. Red indicates a high level of mRNA expression in the tumour, compared with the reference mRNA level, and green indicates a low level of expression. Blue triangles were used to delimit the behavioural clusters.
Figure 6. Expression of Snail, Slug and Claudin-1 in breast cancer cell lines and in human breast tumour samples.
(A) Total RNA from a panel of breast cancer cell lines was reverse-transcribed into cDNA. Results show the cell line/fibroblast ratio of normalized mRNA levels of Claudin-1, Snail and Slug using the comparative threshold cycle method. Claudin-1 compared with Slug: Spearman r=−0.64; P=0.042. Claudin-1 compared with Snail: Spearman r=−0.15; P=0.6. (B) Luciferase reporter constructs carrying wild-type or mutated boxes (300 ng) were transfected in a panel of breast cancer cell lines. Luciferase activity in MDCK co-transfected with wild-type reporter constructs was defined as 1. Results are means±S.D. for three independent experiments. *, P<0.05. (C) Gene cluster assay of 295 human primary invasive breast carcinoma samples using three genes: Snail, Slug and Claudin-1. For each gene, the ratio was calculated with respect to the intensity of a reference pool made up of equal amounts of cRNA from all tumours [20].
The first group of tumours was characterized by a high expression of Snail when compared with the rest of the samples. A clear negative correlation between the levels of Slug and Snail expression can be observed. Despite the small variation observed in the expression of Snail, maximal Claudin-1 expression was attained in those samples with the lowest expression of both transcription factors (Figure 6C, Detail 1). Low levels of Slug and intermediate levels of Snail characterized the second group identified. Here again, Claudin-1 expression inversely correlated with the expression of Snail and Slug, proving maximal when both factors were at lower levels (Figure 6C, Detail 2). The third cluster behaved like the previous two. This cluster comprised the samples with lower Snail and Slug expression within the whole dataset. Interestingly, Claudin-1 levels were at their highest. The fourth group contained samples in which Slug expression was at higher levels. Claudin-1 expression was again inversely proportional to the expression of the transcription factors. Interestingly, this cluster contained the samples where a concomitant high expression of the transcription factors was found, correlating with the lowest levels of Claudin-1 of the whole data set.
Taken as a whole, hierarchical clustering highlights the elevated heterogeneity within the studied tumours in the expression of the transcription factors Snail and Slug. Both factors seem to inversely correlate with the expression of Claudin-1, as their relative contribution, while constant, proved highly dependent on the tumour ‘type’. Extreme effects on Claudin-1 expression were observed when both factors were at their maximal or minimal levels, with any other combination generating only intermediate effects.
DISCUSSION
Several lines of evidence support an association between abnormalities in TJs and neoplasia [3,9,24]. To begin with, alterations in the number, appearance and permeability of TJs have been demonstrated in various cancer types [3,9,24,25]. It has been hypothesized that abnormalities in TJ permeability disrupt the concentration of luminal growth factors, allowing them to cross the epithelium and bind to receptors on the basolateral surface or on other cell types, triggering cell proliferation [4,7]. The loss of expression of Claudin-1 has been demonstrated in several mammary carcinoma cell lines [8]. Furthermore, analysis of primary breast tumours has revealed a similar loss of Claudin-1 expression in contrast with the broad expression spectrum found in other epithelial tissues [9]. However, mutation analysis of the Claudin-1 gene and its putative promoter has provided no further insight into the loss of Claudin-1 expression in breast cancer cell lines [9]. In addition, analysis of Claudin-1 expression not only revealed that the Ras/MEK [mitogen-activated protein kinase/ERK (extracellular-signal-regulated kinase) kinase]/ERK pathway is not involved in the dysregulated TJ formation observed in breast tumour cells, but also indicated that the elevated activity of Ras might not be of great importance for the disruption of such structures [26].
The Snail superfamily of zinc-finger transcription factors has emerged in recent years as an important regulator of EMT [11]. Snail and Slug have now been firmly established as repressors of E-cadherin in early development and in different murine and human carcinoma and melanoma cell lines and tumours [11,12,17,27]. Recently, it has also been shown that Snail is able to repress the expression of the TJ proteins Claudin-3, -4 and -7 and Occludin [22].
In the present study, we have extended the analyses on the regulation of TJ components to Slug, another member of the Snail family, providing evidence for the direct repressor effect on human Claudin-1 gene expression. Our results suggest a new mechanism for Snail/Slug-induced TJ down-regulation in epithelial cells, and indicate that elevated levels of Slug and Snail may be important for the disruption of TJ structures in epithelial cells. We examined the behaviour of Claudin-1 in Slug- and Snail-transfected MDCK cells and found that, similar to the Snail- and Slug-mediated down-regulation of E-cadherin transcription, Slug and Snail not only repress endogenous Claudin-1 expression, but also induce a dramatic loss of TEER. The Slug- and Snail-induced repression of Claudin-1 in MDCK cells is exerted at the transcriptional level and is dependent on the direct binding of Snail/Slug to proximal E-boxes as confirmed in in vitro and in vivo binding assays. In fact, the integrity of the proximal E-boxes of the human Claudin-1 promoter is a requirement for the binding activity and repression effects of Snail and Slug, as confirmed by promoter activity and band-shift assays. In the proximal E-box cluster, Slug- and Snail-mediated repression of Claudin-1 appears to occur primarily through the co-ordinated action of E1 and E2 boxes, although the requirement of additional regulatory elements to achieve this repression cannot be discarded at present.
The present results differ from a recent report [23] showing post-transcriptional down-regulation of Claudin-1 by Snail and the apparent absence of transcriptional regulation in MDCK cells. Such absences may be explained by differences in the levels of Snail expression in the transfected cell line used in both studies, as we demonstrated that there is an inverse dose-dependent correlation between Snail and Slug levels and the repression of the Claudin-1 gene. In addition, we demonstrated that a shorter promoter sequence is sensitive to the repression effects of Snail and Slug factors in MDCK cells. In the A431 cell line, the model used by Ohkubo and Ozawa [23] in the luciferase reporter assay, the basal activities of both constructs were dramatically reduced. In addition, although Snail and Slug were able to repress the large promoter, this repression proved smaller than in MDCK, as the small promoter was insensitive to repression (Supplementary Figure 2S at http://www.BiochemJ.org/bj/394/bj3940449add.htm). Real-time PCR analysis of the Snail and Slug genes in the A431 cell line demonstrated high levels of Slug expression and low levels of Snail expression (Supplementary Figure 3S at http://www.BiochemJ.org/bj/394/bj3940449add.htm). We speculated that in A431, a carcinoma cell line, the activities of both promoters were probably already repressed by Slug.
The data reported in the present paper support the idea that, when overexpressed, Snail and Slug can behave as potent repressors of Claudin-1 in epithelial cells. While the regulatory mechanism underlying the formation and destruction of TJs have been examined extensively, promoter analyses of Claudins and Occludin are only just beginning. Recently, a repressor effect of Snail on Claudin-3, -4 and -7 and Occludin was reported [22]. The findings of the present study are the first to indicate that Slug can also control the expression of TJ proteins, in particular Claudin-1. As assessed by real-time PCR, an inverse correlation between the expression levels of Claudin-1 and Slug factor was found in breast cancer cell lines. We observed that Claudin-1 expression levels were lowest in the invasive and metastatic cell lines MDA-MB 231 and MDA-MB-435. Moreover, in these cell lines, the expression levels of Slug were higher than in non-metastatic tumour cell lines, such as MCF7 and T47D. Slug expression is increasingly being recognized as an alteration in mesenchymal tumours, suggesting that Slug, like Snail, may be a critical invasion factor [28]. The microarray gene expression data set from the 295 invasive human breast tumours reviewed in the present study demonstrated an inverse correlation between Snail and Slug expression and Claudin-1. We identified four groups of tumours, based on the expression levels of each protein analysed. Tellingly, when one of the transcription factors is expressed at very low levels, such as in groups 1 and 3, small changes in the expression levels of the other are able to induce a dramatic decrease in the Claudin-1 levels. The highest levels of Claudin-1 expression were observed in the second group, which corresponded to tumours with low levels of Snail and Slug expression. Interestingly, group 4 contained tumours with the lowest levels of Claudin-1 and the highest levels of Snail and Slug expression.
Our data indicate that both Slug and Snail may serve as potential repressors of Claudin-1 in epithelial tumour cells. This could explain the loss of Claudin-1 expression in breast epithelial tumour cell lines and primary tumours. The existence of both Slug and Snail in non-tumorigenic epithelial cells suggests the presence of additional factors that contribute to their inhibitory effect in tumour epithelial cells. The specific role of each, or their potential co-operation, in specific cellular contexts and in different types of tumour cells remains to be fully elucidated.
Online data
Acknowledgments
We thank Antonio García De Herrero for providing reagents, Merce Martín for helpful advice with the band-shift experiments, and Robin Rycroft for the revision of the typescript. This work was supported by the Spanish Ministry of Science and Technology (SAF2001-3602).
References
- 1.Tsukita S., Furuse M., Itoth M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 2.Mullin J. M., Marano C. W., Laughlin K. V., Nuciglio M., Stevenson B. R., Soler P. Different size limitations for increased transepithelial paracellular solute flux across phorbol ester and tumor necrosis factor-treated epithelial cell sheets. J. Cell. Physiol. 1997;171:226–233. doi: 10.1002/(SICI)1097-4652(199705)171:2<226::AID-JCP14>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Soler A. P., Miller R. D., Laughlin K. V., Carp N. Z., Klurfeld D. M., Mullin J. M. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20:1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 4.Mullin J. M., Laughlin K. V., Ginanni N., Marano C. W., Clarke H. M., Peralta S. A. Increased tight junction permeability can result from protein kinase C activation/translocation and act as a tumor promotional event in epithelial cancers. Ann. N.Y. Acad. Sci. 2000;915:231–236. doi: 10.1111/j.1749-6632.2000.tb05246.x. [DOI] [PubMed] [Google Scholar]
- 5.Tsukita S., Furuse M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann. N.Y. Acad. Sci. 2000;915:129–135. doi: 10.1111/j.1749-6632.2000.tb05235.x. [DOI] [PubMed] [Google Scholar]
- 6.Morita K., Furuse M., Fujimoto K., Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. U.S.A. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuse M., Sasaki H., Fujimoto K., Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell. Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swisshelm K., Machl A., Planitzer S., Robertson R., Kubbies M., Hosier S. SEMP1, a senescence-associated cDNA isolated from human mammary epithelial cells, is a member of an epithelial membrane protein superfamily. Gene. 1999;226:285–295. doi: 10.1016/s0378-1119(98)00553-8. [DOI] [PubMed] [Google Scholar]
- 9.Kramer F., White K., Kubbies M., Swisshelm K., Weber B. H. Genomic organization of claudin-1 and its assessment in hereditary and sporadic breast cancer. Hum. Genet. 2000;107:249–256. doi: 10.1007/s004390000375. [DOI] [PubMed] [Google Scholar]
- 10.Nieto M. A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell. Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 11.Cano A., Perez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 12.Batlle E., Sancho E., Franci C., Dominguez D., Monfar M., Baulida J., Garcia D. H. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 13.Thiery J. P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 14.Guaita S., Puig I., Franci C., Garrido M., Dominguez D., Batlle E., Sancho E., Dedhar S., De Herreros A. G., Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J. Biol. Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 15.Seki K., Fujimori T., Savagner P., Hata A., Aikawa T., Ogata N., Nabeshima Y., Kaechoong L. Mouse Snail family transcription repressors regulate chondrocyte, extracellular matrix, type II collagen, and aggrecan. J. Biol. Chem. 2003;278:41862–41870. doi: 10.1074/jbc.M308336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama K., Kamata N., Fujimoto R., Tsutsumi S., Tomonari M., Taki M., Hosokawa H., Nagayama M. Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int. J. Oncol. 2003;22:891–898. [PubMed] [Google Scholar]
- 17.Bolos V., Peinado H., Perez-Moreno M. A., Fraga M. F., Esteller M., Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Estrada O. M., Villa A., Breviario F., Orsenigo F., Dejana E., Bazzoni G. Association of junctional adhesion molecule with calcium/calmodulin-dependent serine protein kinase (CASK/LIN-2) in human epithelial caco-2 cells. J. Biol. Chem. 2001;276:9291–9296. doi: 10.1074/jbc.M006991200. [DOI] [PubMed] [Google Scholar]
- 19.Lu C., Schwartzbauer G., Sperling M. A., Devaskar S. U., Thamotharan S., Robbins P. D., McTiernan C. F., Liu J. L., Jiang J., Frank S. J., Menon R. K. Demonstration of direct effects of growth hormone on neonatal cardiomyocyte. J. Biol. Chem. 2001;276:22892–22900. doi: 10.1074/jbc.M011647200. [DOI] [PubMed] [Google Scholar]
- 20.van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 21.Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 22.Ikenouchi J., Matsuda M., Furuse M., Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J. Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 23.Ohkubo T., Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J. Cell Sci. 2004;117:1675–1685. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- 24.Mullin J. M., O'Brien T. G. Effects of tumor promoters on LLC-PK1 renal epithelial tight junctions and transepithelial fluxes. Am. J. Physiol. 1986;251:C597–C602. doi: 10.1152/ajpcell.1986.251.4.C597. [DOI] [PubMed] [Google Scholar]
- 25.Liebner S., Fischmann A., Rascher G., Duffner F., Grote E. H., Kalbacher H., Wolburg H. Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol. 2000;100:323–331. doi: 10.1007/s004010000180. [DOI] [PubMed] [Google Scholar]
- 26.Macek R., Swisshelm K., Kubbies M. Expression and function of tight junction associated molecules in human breast tumor cells is not affected by the Ras-MEK1 pathway. Cell Mol. Biol. 2003;49:1–11. [PubMed] [Google Scholar]
- 27.Hajra K. M., Chen D. Y., Fearon E. R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 28.Perez-Mancera P. A., Gonzalez-Herrero I., Perez-Caro M., Gutierrez-Cianca N., Flores T., Gutierrez-Adan A., Pintado B., Sanchez-Martin M., Sanchez-Garcia I. Slug in cancer development. Oncogene. 2005;24:3073–3082. doi: 10.1038/sj.onc.1208505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.