Abstract
The African swine fever (ASF) virus polyprotein pp220 is processed at Gly-Gly-X sites by a virally encoded SUMO-like protease to produce matrix proteins p150, p37, p34, and p14. Four Gly-Gly-X sites are used to produce the matrix proteins, but the polyprotein contains an additional 15 sites potentially recognized by the protease. This study shows that cleavage occurs at many, if not all, Gly-Gly-X sites, and at steady state, p150 and p34 are minor products of processing. Significantly, only the final structural proteins, p150 and p34, were found in mature virions, suggesting that there is a mechanism for excluding incorrectly processed forms. ASF virus is assembled on the cytoplasmic face of the endoplasmic reticulum, and the distribution of pp220 products between membranes and cytosol was studied. Incorrectly processed forms of p34 were recovered from both the cytosol and membrane fractions. Interestingly, p34 was only detected in the membrane fraction, and of the many processed forms bound to membranes, only p34 was protected from trypsin, suggesting envelopment. The majority of the incorrectly processed forms of p150 were recovered from the cytosol. Again, the correct product of processing, p150, was selectively recruited to membranes. Sucrose density centrifugation showed that membrane-associated forms of p34 and p150 assembled into large structures suggestive of a viral matrix, while cytosolic and/or incorrectly processed forms of pp220 did not. Taken together, these results suggest that association with cellular membranes is important for regulating the correct processing of pp220 and the packaging of matrix proteins into virions.
African swine fever (ASF) virus and the closely related Iridoviridae are among the largest icosahedral viruses assembled in animal cells (17, 26). It has been proposed recently that they have a common evolutionary origin with other large nucleocytoplasmic DNA viruses such as the Poxviridae and Phycodnaviridae (23). The ancestral virus may have had a large icosahedral capsid, as seen in ASF virus (35) and the characteristic inner membrane envelopes (31) shared by all members of the group.
We have been studying the assembly of ASF virus as a means of understanding how these highly complex, and possibly very ancient, structures are assembled in cells. In common with poxviruses and iridoviruses, ASF virus is assembled in perinuclear sites called viral factories (7, 27, 28). ASF virus factories lie close to the microtubule-organizing center, and their integrity is dependent on microtubules (22). Interestingly, they are surrounded by intermediate filament (vimentin) cages and closely resemble cellular aggresomes (16). Aggresomes use microtubules to sequester protein aggregates (22, 24), and recent work suggests that ASF virus, the poxviruses, and members of the Reoviridae use elements of the aggresome pathway to concentrate viral proteins at virus assembly sites (22, 29, 30).
ASF virus particles are 200 nm in diameter and contain double-stranded DNA genomes ranging in size from 200 to 350 kbp and at least 50 virally encoded proteins (14, 36). When viewed by electron microscopy, virions appear as hexagons in cross-section and have internal concentric layers of different electron densities (8, 31). These layers represent a central electron-dense nucleoid surrounded by a matrix, internal lipid envelopes and capsid layer (8, 31). The major capsid protein of ASF virus, p73, provides 35% of the protein present in the virion (6). Newly synthesized p73 binds a virally encoded chaperone in the cytosol that prevents capsid aggregation (13). Capsid assembly is initiated by dissociation of the chaperone and the rapid recruitment of p73 onto the endoplasmic reticulum membrane (11, 15). There then follows a lag period of approximately 60 min, after which the capsid protein oligomerizes into a large capsid-like complex of 50,000 kDa, containing between 600 and 700 copies of the capsid protein (12).
Current models of ASF virus assembly propose that protein-protein interactions between the capsid protein, and possibly other viral proteins assembled onto the endoplasmic reticulum, lead to constriction of endoplasmic reticulum cisternae and the bending of the membranes through one- to six-sided angular intermediates, eventually producing icosahedral structures (12, 15, 31). This process is energy dependent and incorporates two envelopes into the virion in a single step (10, 31).
The viral matrix, or inner core shell, of ASF virus lies between the inner membrane and the nucleoprotein core (4, 31). The major proteins of the matrix, p150, p37, p34, and p14, are produced through proteolytic processing of the pp220 virus polyprotein (33) (Fig. 1A) and together provide 25% of the protein mass of the virus and are present in equimolar amounts (6). While the pathway for the delivery of p73 into ASF virus has been studied in some detail, very little is known about the method of recruitment of the pp220 polyprotein or its products into virions. The polyprotein is cleaved at Gly-Gly-X sites by a virally encoded SUMO-1-specific protease encoded by the S273R gene (3, 25). Four sites have been identified for the production of p150, p37, p34, and p14, but the sequence of the pp220 polyprotein reveals a total of 19 Gly-Gly-X sites that could be recognized by the protease (36) (see Fig. 1B). Processing by the viral protease can therefore produce many more proteins than the p150, p37, p34, and p14 proteins packaged into virions.
FIG. 1.
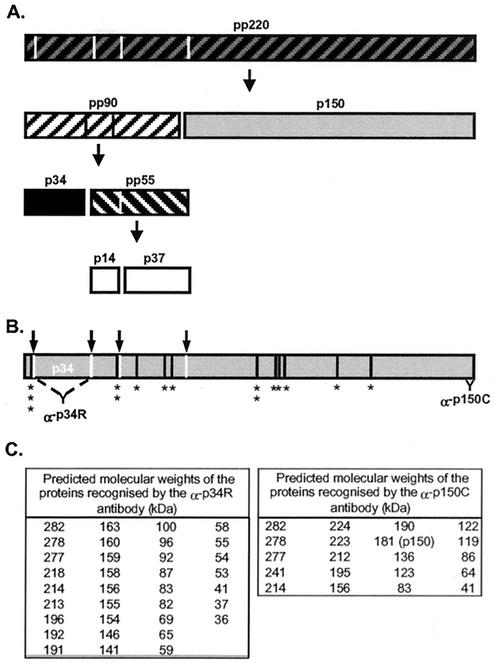
Processing of pp220 polyprotein and location of Gly-Gly-X protease cleavage sites. (A) The ordered processing of pp220 polyprotein as described by Simón-Mateo et al. (33). The hatched blocks show the polyproteins, and the solid boxes depict the final structural proteins. (B) The locations of the 19 Gly-Gly-X sites (*) and the correct cleavage sites (arrows) are shown, as are the binding sites for the antibodies raised against the recombinant p34 protein (α-p34R) and the C-terminal peptide (α-p150C). (C) Tables displaying the predicted molecular masses of proteins recognized by the antibodies if cleavage occurred at all 19 Gly-Gly-X sites.
Here we provide evidence that many of these processed forms are made during infection, and at steady state, p150 and p34 are minor products of processing. But, as seen for the major capsid protein, p73, recruitment onto cellular membranes precedes delivery of the correctly processed forms of pp220 into virions.
MATERIALS AND METHODS
Cells, virus, and antibodies.
Vero (ECACC84113001) cells were grown and infected with the attenuated BA71v strain of ASF virus as previously described (11). The rabbit polyclonal antibody raised against p34 has been described previously (22). The rabbit antibody specific for p150 was raised against a synthetic peptide (TEYGFSITGPSETFSDKQYDSDIR) representing the C terminus of p150, generated by Severn Biotech Ltd.
Cell lysis, fractionation, and virus preparation.
Total cell lysates and postnuclear membrane and cytosolic fractions were prepared as previously described (11). Crude virus preparations were prepared by allowing infected cells to reach an extensive cytopathic effect, and the cellular debris was removed from the medium by centrifugation for 15 min at 500 × g. The virus was collected by pelleting at 30,000 × g for 1 h at 4°C in a Beckman 50Ti rotor. The metabolic labeling of infected cells with [35S]methionine-cysteine was carried out as previously described (11). The proteins were separated by running lysates, under reducing conditions, on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12.5% or 7.5% polyacrylamide) and were visualized either by Western blotting and subsequent detection with specific antibodies or by autoradiography.
Trypsin protection assay.
Postnuclear membrane fractions were incubated with trypsin as previously described (11). Following the assay, the membranes were pelleted by centrifugation at 16,000 × g for 15 min at 4°C, and the proteins were resolved by immunoprecipitation and Western blotting as described above.
Sucrose density sedimentation analysis.
Postnuclear membrane fractions were prepared and floated on a 10-ml gradient of 10 to 40% sucrose layered over a 1-ml 70% sucrose cushion, which were prepared and characterized as previously described (12). The gradients were centrifuged at 200,000 × g for 20 h at 4°C and then collected into 10 fractions. Continuous 10 to 70% gradients were prepared and calibrated as previously described (12) and, following addition of the sample, were centrifuged at 79,000 × g for 2.5 h at 4°C. The gradients were collected into 1.2-ml fractions.
RESULTS
The proteolytic processing of pp220 was first described by Simón-Mateo et al. (33), where ordered processing produced the structural proteins p150, p37, p34, and p14 (Fig. 1A). The molecular sizes designated for these proteins were based on their migration in SDS-polyacrylamide gels. However, translation of the CP2475L reading frame encoding the polyprotein predicts that some proteins are larger than originally estimated (6, 36). The pp220 polyprotein is predicted to be 282 kDa, while the structural protein p150 is 180 kDa. Minor increases in size are also predicted for p37, p34, and p14.
In the following experiments, recombinant proteins and synthetic peptides were used to raise antibodies recognizing p150 and p34. When analyzed with these antibodies, the polyprotein consistently migrated slower than the 250-kDa size marker, suggesting a mass of between 250 and 300 kDa, and p150 migrated between 170 and 180 kDa. We have maintained the pp220/p150/p34 nomenclature described previously for these ASF virus structural proteins (33).
The reading frame of the polyprotein contains 19 Gly-Gly-X sites (36) (Fig. 1B), only 4 of which are utilized to produce the structural proteins identified in virions (33). This raised the possibility that the polyprotein may be processed at several Gly-Gly-X sites in cells infected with ASF virus and that these products may also be packaged into virions. Alternatively, if incorrectly processed forms are not packaged, then either the protease must recognize the four correct sites specifically, or the virus has developed a different mechanism to ensure that only p150, p37, p34, and p14 are incorporated into the matrix of mature virions. Evidence for processing of pp220 at multiple sites was sought by Western blotting of lysates of infected cells and virions with antibodies raised against recombinant p34 protein or antibodies specific for a peptide representing the C terminus of p150.
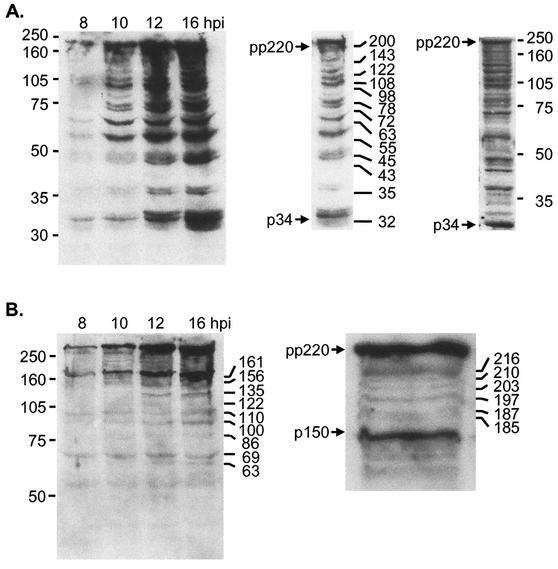
Figure 2A shows that proteins reactive with the antibody raised against the p34 protein were detected in cells 8 h postinfection, and the levels increased throughout the time course. At 10 h, a clear ladder of antibody-reactive proteins was observed, and the intensity of the ladder increased between 12 and 16 h postinfection. The estimated sizes of 13 proteins most easily detected by the antibody are indicated (left inset). The smallest protein migrated between 32 and 35 kDa, and the largest migrated above 200 kDa, at a position consistent with the size of the ASF virus polyprotein.
FIG. 2.
pp220 polyprotein is cleaved at several sites in cells infected with ASF virus. At the indicated times after infection with the BA71v strain of ASF virus, Vero cells were lysed, and the proteins were resolved by SDS-PAGE on a 12.5% (A) or 7.5% (B) polyacrylamide gel, Western blotted, and probed with antibodies specific for p34 (A) or p150 (B). Enlarged views of the 16-h postinfection (hpi) lane and the upper section of the 1-h postinfection lane are also shown in panels A and B, respectively. The positions of the molecular size markers are shown on the left, and the interpolated molecular sizes are shown on the right (in kilodaltons). pp220 and p34 or p150 are indicated. The right insert in panel A shows a Western blot of cells lysed directly in 7.5% SDS at 16 h postinfection.
The Western blots were prepared from cells lysed in mild detergent containing protease inhibitors. Even so, it was possible that the ladder of proteins may have resulted from proteolysis of pp220 after lysis of cells. To test this, infected cells were lysed on the culture dish by direct addition of SDS. The right insert of Fig. 2A shows that the ladder of proteins detected by Western blotting was still present in the sample and argued strongly that they were produced following proteolysis of pp220 inside cells rather than during sample preparation. Figure 2B shows a similar experiment in which cell lysates were probed with an antibody recognizing the C terminus of p150. A major reactive protein migrated above the 250-kDa marker, suggesting recognition of the 280-kDa pp220 polyprotein; a second protein was detected at 170 kDa, indicating the structural protein p150. In addition to these two major proteins, a ladder of six proteins with approximate molecular masses of 185, 187, 197, 203, 210, and 216 kDa appeared at 10 h (inset), while at 12 h postinfection, the antibody also detected several smaller proteins at approximately 63, 86, 122, and 135 kDa.
The detection of multiple proteins by these antibodies suggested proteolysis at many of the Gly-Gly-X sites within the polyprotein. Proteolysis at all 19 Gly-Gly-X sites would produce over 30 proteins reactive with antibodies specific for p34 (Fig. 1C). Significantly, there are no internal Gly-Gly-X sites in p34. If the proteins seen on Western blots were produced by the Gly-Gly-X-specific viral protease rather than by random proteolysis by cellular proteases, then p34 should be the smallest protein recognized by antibodies raised against p34. Inspection of the blot in Fig. 2A shows that this is the case and argues that the reactive proteins are produced by the viral protease.
The antibody raised against p150 binds the C terminus of the polyprotein (Fig. 1B). Figure 1C predicts that 16 proteins would be recognized by the antibody if the polyprotein were cleaved at all Gly-Gly-X sites. Ten proteins would migrate between the polyprotein and p150, and additional smaller proteins would be detected at 120, 86, and 64 kDa. The blot shown in Fig. 2B shows that the sizes of the proteins detected by the antipeptide antibody were again consistent with processing at several Gly-Gly-X sites by the viral protease.
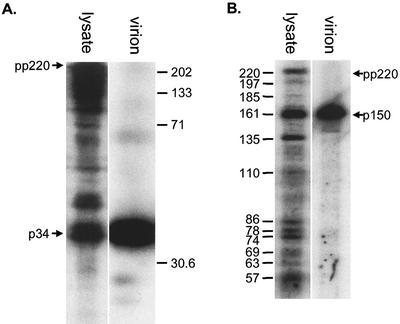
The observation that pp220 could be processed at sites other than those producing the structural proteins p150, p37, p34, and p14 raised the possibility that the incorrectly processed forms could be incorporated into virions. To test this, a crude virus preparation was separated from cellular material by centrifugation, and the distribution of proteins reactive with the antibodies was compared by Western blotting. Figure 3A shows that the cellular material contained several proteins, ranging from 34 kDa to above 220 kDa, that reacted with the antibody raised against p34. The virions contained a single reactive band at 32 to 35 kDa, even when the blot was overexposed. Figure 3B shows the experiment repeated with the antibody specific for p150. Again, the cell lysates contained several proteins reactive with the antibody, but ASF virions contained a single protein, corresponding to the structural protein p150. These data suggested that while the polyprotein was processed at several Gly-Gly-X sites in cells, there was selective packaging of the final p34 and p150 proteins into mature virions.
FIG. 3.
Incorrectly processed forms of pp220 are excluded from virions. Vero cells were infected with BA71v for 16 h or cultured until an extensive cytopathic effect was observed. Following the appearance of cytopathic effect, a crude virus preparation was generated by centrifugation at 500 × g for 15 min to remove cell debris, followed by pelleting at 30,000 × g. Virus pellets (virion) and infected cells (lysate) were lysed in SDS-PAGE sample preparation buffer and resolved by SDS-12.5% PAGE (A) or SDS-7.5% PAGE (B). Gels were Western blotted and probed with antibodies specific for p34 (A) or p150 (B). The molecular masses are shown (in kilodaltons), and pp220 and p34 or p150 are indicated by arrows.
The next experiments were designed to determine the site of segregation of structural proteins from inappropriately processed forms of pp220. ASF virus is assembled on the cytoplasmic face of the endoplasmic reticulum (5, 11), suggesting that membrane association may be important for the segregation of the correct structural proteins. Interestingly, the 220-kDa polyprotein is myristoylated at its N terminus (2, 33), evoking the idea that the myristate may target the polypeptide chain to cellular membranes during assembly (4, 19, 32, 34).
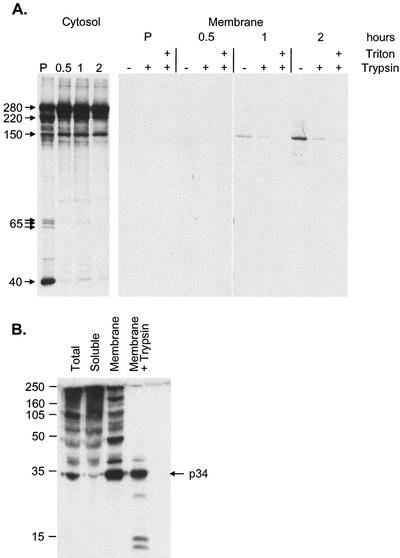
The next experiments used crude membrane fractionation and a protease protection assay to follow the translocation of pp220 onto membranes and subsequent envelopment of processed products into virions. The assay is based on the assumption that enveloped matrix proteins associated with cellular membranes would be protected from proteolysis by trypsin. To do this, Vero cells infected with ASF virus were pulse labeled with [35S]methionine-cysteine and chased for increasing times. Crude cytosol and postnuclear membrane fractions were prepared at each time point, and membrane fractions were incubated with trypsin in the presence or absence of mild detergent. All samples were then solubilized and immunoprecipitated with antibodies specific for p150.
A ladder of proteins between 280 and 150 kDa and a triplet at 65 kDa were immunoprecipitated from the cytosol (Fig. 4A). This pattern was similar to the bands reactive on Western blots (Fig. 2B) suggesting that they were the same proteins. The intensity of the slower-migrating bands (280 and 220) increased between the pulse and the 0.5-h time point. This was routinely seen in these kinetic experiments and may reflect conformational maturation of pp220. A similar maturation has been observed for p73, the major capsid protein of ASF virus (13). A protein of 40 kDa was observed to associate with pp220 in the pulse but was lost during the chase. A prominent band of approximately 220 kDa was also seen between the 280-kDa polyprotein and p150. The nature of this protein is unknown, but it may be a processing intermediate formed during the production of p150.
FIG. 4.
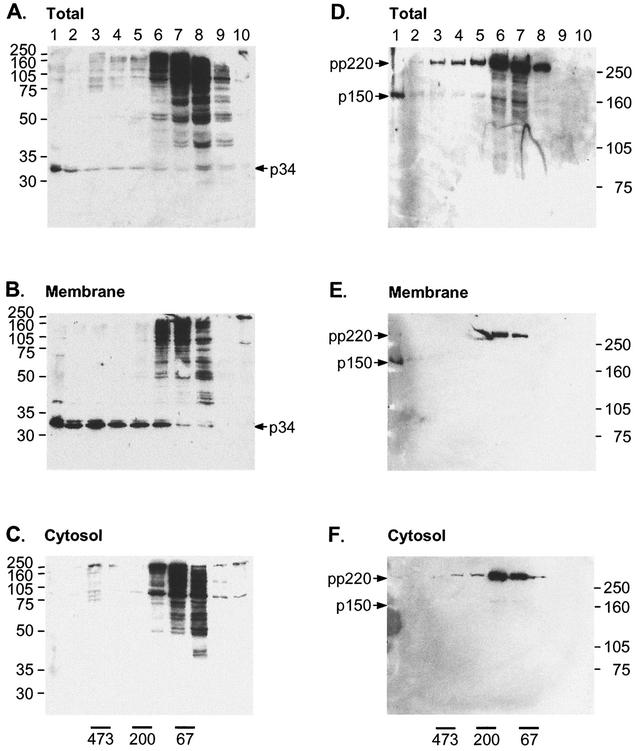
Subcellular distribution of correctly and incorrectly processed forms of pp220. (A) Vero cells infected for 16 h with BA71v were pulse labeled for 10 min (P) and chased for the indicated times (0.5, 1, and 2 h), and then postnuclear cytosolic and membrane fractions were prepared as described previously (11). The membrane fractions were incubated in the absence (−) or presence (+) of 0.4 mg of trypsin per ml and/or 1% Triton X-100 for 30 min at 37°C, as indicated. Proteolysis was then blocked by the addition of 5 mg of trypsin inhibitor per ml. Membrane and cytosol fractions were lysed in immunoprecipitation buffer and immunoprecipitated with an antibody raised against a peptide representing the C terminus of p150. Immunoprecipitated proteins were visualized by SDS-7.5% PAGE under reducing conditions, followed by autoradiography. (B) Vero cells were infected with the BA71v strain of ASF virus for 16 h. Postnuclear membrane and cytosolic fractions were prepared, and the membrane fraction was treated with trypsin as described above. Representative samples of the total cell lysate (total), cytosolic pool (soluble), and membranes before and after incubation with trypsin were resolved by SDS-12.5% PAGE under reducing conditions, Western blotted, and probed with antibodies specific for p34. The positions of molecular size markers are shown (in kilodaltons).
Proteins were first recovered from the membrane fraction at 1 h. Interestingly, even though several proteins were present in the cytosol, only the p150 protein was recovered from the membrane fraction at this time point. The signal was weak but represented at least one third of the p150 produced from pp220 1 h into the chase. Approximately one half of the membrane-associated p150 was protected from trypsin, suggesting packaging into virions. When the chase time was increased to 2 h, greater levels of p150 were seen in the membrane fraction, and minor proteins of approximately 250, 200, and 150 kDa were visible; of these, only the p150 was protected from trypsin, again suggesting selective packaging of the membrane pool of p150.
It was not possible to immunoprecipitate p34 with the antibodies available, so the cellular fractions were analyzed by Western blotting. Figure 4B shows that both the cytosolic and membrane fractions contained a ladder of proteins reactive with the antibody specific for p34. The cytosolic fraction contained reactive proteins ranging from above 220 kDa down to 70 kDa, with minor bands extending to approximately 35 kDa. The membrane fraction contained a less densely compact ladder of reactive proteins ranging from 220 kDa to 34 kDa. The major difference between the membrane and cytosolic fractions was that the great majority of the final product of processing, p34, was found in the membrane fraction. Significantly, when the membrane fraction was incubated with trypsin (Fig. 4B, right lane), the 220-kDa polyprotein and intermediates were lost from membranes and a major band of 34 kDa survived, suggesting selective protection of p34 against trypsin. Minor bands of lower molecular mass were also seen. Since these were only observed after addition of trypsin, it is likely that they are proteolytic products of pp220 or processing intermediates bound to the membrane fraction. Taken together, the results from both experiments suggested that several pp220 processing intermediates were able to bind cellular membranes, but only p34 and p150 were enveloped.
It has been suggested that pp220 may act as a scaffolding protein (6, 33) during the assembly of ASF virus. Scaffold proteins are incorporated into large procapsids and displaced during capsid maturation or processed by proteases during procapsid to capsid transition (9). The results above showed that the 220-kDa polyprotein and several processing intermediates were found on membranes but were absent from mature virions. It was possible that pp220 and the processing intermediates detected on membranes may represent a population of scaffold-like intermediates present in procapsids early during virus assembly. Evidence for the incorporation of pp220 and the other precursors into large structures indicative of procapsids was sought by using sucrose gradients. It was argued that large procapsids would sediment towards the bottom of sucrose gradients, while unassembled precursors would migrate according to the molecular size predicted for monomeric proteins.
Vero cells infected with ASF virus were solubilized in 1% Brij 35 and analyzed on 10 to 40% sucrose gradients, and each fraction was analyzed by Western blotting with antibodies raised against p34 or p150 (Fig. 5). In total cell lysates (Fig. 5A and B), the bulk of the pp220 polyprotein and the processing intermediates were found near the top of the gradient in fractions 6 to 8, suggesting sizes between 70 and 200 kDa. The polyprotein and the incorrectly processed forms therefore did not form large oligomers indicative of a procapsid. However, the products of correct processing, p34 and p150, migrated towards the bottom of the gradient, suggesting incorporation into large complexes.
FIG. 5.
Incorrectly processed forms of pp220 are excluded from large structures indicative of virus precursors. Vero cells were infected with BA71v for 16 h. Total cell lysates and cytosol and membrane fractions were prepared as described for Fig. 4, solubilized in 1% Brij 35 in immunoprecipitation buffer, and centrifuged on a 10 to 40% sucrose gradient at 200,000 × g for 20 h at 4°C. Samples taken from each fraction were resolved by SDS-PAGE with 12.5% (A, B, and C) or 7.5% (D, E, and F) polyacrylamide, Western blotted, and probed with antibodies specific for p34 (A, B, and C) or p150 (D, E, and F). The positions of molecular size markers are shown on the right (in kilodaltons), and pp220 and p34 or p150 are indicated with arrows.
The experiment was repeated for separated membrane and cytosol fractions (Fig. 5B and C). In both gradients, a ladder of proteins reactive with the antibody raised against p34 were present in light fractions 6 to 8. p34 was only recovered from the membrane fraction and migrated in heavier fractions extending to the bottom of the gradient. Similar results were obtained following analysis of p150 (Fig. 5E and F). The pp220 polyprotein precursor and p150 was recovered from both fractions. In the cytosol, both pp220 and p150 migrated in fractions 6 to 8, suggesting a low molecular mass. In contrast, p150 present in the membrane fraction migrated to the bottom of the gradient, suggesting packaging into virus and virus precursors.
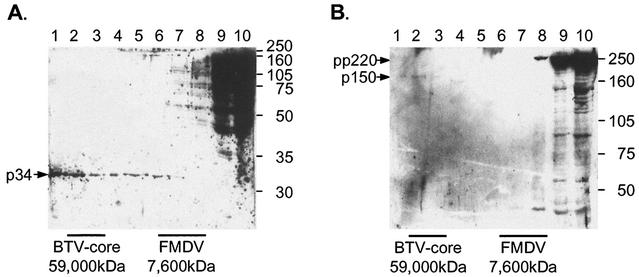
The sizes of complexes containing p34 and p150 were determined in more detail with 10 to 70% sucrose gradients. The gradients were calibrated with foot and mouth disease virus particles and bluetongue disease virus cores with approximate molecular masses of 7,600 and 59,000 kDa, respectively (1, 18, 20). Figures 6A and B compare the migration of these particles with the distribution of proteins detected by the antibodies specific for p34 and p150, respectively. The pp220 polyprotein and processing intermediates remained at the top of the gradients, while p34 and p150 protein were found in fractions 1 to 7. Most of the protein was recovered in fractions 1 and 2, suggesting assembly into a range of oligomers with molecular masses ranging from 7,600 kDa to over 59,000 kDa. This was consistent with the incorporation of p34 and p150 into virion precursors of sizes similar to those described previously for the major capsid protein, p73 (12). The exclusion of pp220 and processing intermediates from large procapsid-like structures suggests that processing occurs before, rather than after, assembly of p34 or p150 into virions.
FIG. 6.
Only correctly processed forms of pp220 assemble into large structures indicative of a viral matrix. Vero cells were infected with BA71v for 16 h. Total cell lysates were solubilized in 1% Brij 35 in immunoprecipitation buffer, centrifuged on a 10 to 70% sucrose gradient, and centrifuged at 79,000 × g for 2.5 h at 4°C. Samples taken from each fraction were resolved by SDS-PAGE with 12.5% (A) or 7.5% (B) polyacrylamide, Western blotted, and probed with antibodies specific for p34 (A) or p150 (B). The positions of molecular size markers are shown on the right (in kilodaltons), and pp220 and p34 or p150 are indicated with arrows. The gradients were calibrated with foot and mouth disease virus (FMDV, 7,600 kDa) and bluetongue disease virus (BTV, 59,000 kDa) cores as described previously (12).
DISCUSSION
ASF virus encodes a protease with specificity for Gly-Gly-X sites (3), which has been shown to cleave the polyprotein pp220 at four Gly-Gly-X sites to produce the four major matrix proteins (3, 33). Interestingly, the pp220 polyprotein contains 19 Gly-Gly-X sites (33, 36), and we have investigated whether the protease cleaves at the other Gly-Gly-X sites and, if so, whether the alternatively processed forms are assembled into virions. Antibodies raised against either p34 or p150 reacted with multiple proteins in cells infected with ASF virus, suggesting proteolysis of the polyprotein at several sites. The many reactive proteins were produced by either random proteolysis of the protein by cellular proteases or specific processing of Gly-Gly-X sites in the polyprotein by the viral protease.
The published sequence of the CP2475L reading frame encoding pp220 was used to predict the sizes of the proteins formed by cleavage at all 19 Gly-Gly-X sites (Fig. 1C) (36), and these were compared with those seen on Western blots. The limitations of the SDS-PAGE system made it difficult to resolve the precise molecular weights of all the proteins, particularly for clusters of proteins of similar size. But if cleavage occurred at all 19 Gly-Gly-X sites, the antibody raised against p34 would be expected to bind to 34 proteins broadly distributed between 280 and 34 kDa. Similarly, the antibody specific for the C terminus of p150 would bind 16 proteins, most of which would be between 280 and 120 kDa. The distributions of proteins recognized by the antibodies on Western blots were in broad agreement with the sizes predicted for processing of Gly-Gly-X sites by the viral protease.
It was also possible to determine the size of the smallest protein that would be reactive with the antibodies, and for both antibodies, the smallest protein seen on Western blots was the size predicted to be produced by proteolysis at the Gly-Gly-X sites nearest the antibody binding site. Taken together, the results suggest that the viral protease cleaves pp220 at multiple Gly-Gly-X sites and that proteins detected on Western blots result from this processing rather than from proteolysis by cellular proteases.
Interestingly, even though infected cells contain many different processed forms of pp220, only the structural proteins p34 and p150 were detected in the mature virions. The structural proteins and their processing intermediates are all derived from the same polyprotein and have the same amino acid sequences. It was interesting, therefore, to ask how the structural proteins are packaged selectively into virions while incorrectly processed forms are excluded. For p34, the problem is slightly less complex than for p150. The p34 protein does not have internal Gly-Gly-X sites, and as long as proteolytic processing was complete, p34 would eventually be generated by the viral protease for packaging into virions. The p150 protein, however, has seven internal Gly-Gly-X sites that need to be protected from the viral protease, while cleavage at the 12th Gly-Gly-X site is needed to release the mature protein.
Subcellular fractionation suggested that membrane association was important for the correct processing and selective recruitment of p34 into virions. Investigation of the steady-state distribution of proteins reactive with the antibody raised against p34 showed that the p34 protein was recovered only from the membrane fraction. In contrast, the processing intermediates reactive with the antibody were found in both the cytosol and membrane fractions. Significantly, of the membrane-bound pool of processed forms, only p34 was protected from trypsin, and only p34 was incorporated into large structures indicative of assembling virions.
The time course of recruitment of p150 onto membranes was studied. The bulk of the intact pp220 polyprotein and the different processed forms remained in the cytosol, but within 1 h of synthesis, approximately one third of the p150 produced was recovered from membranes. At later times, low levels of precursors were also seen in the membrane fraction, but only p150 was protected from trypsin, suggesting incorporation into virions.
The recovery of a large proportion of the cellular pool of the pp220 protein from the cytosol was surprising. The pp220 polyprotein is myristoylated at the N terminus (2, 33), and myristoylation would offer a means of targeting the polyprotein to membranes. This is supported by the observation that pp220 expressed transiently in cells binds membranes (4). Interestingly, proteolysis of the polyprotein at the first Gly-Gly-X site removes myristate and five amino acids from the N terminus (Fig. 1A) of the protein.
One explanation for the presence of a cytosolic pool of pp220 in infected cells is that the newly synthesized pp220 binds to membranes via myristate, but the protein is subsequently released following proteolysis at this first Gly-Gly-X site by the viral protease. This would not happen during transient transfection of cells because the viral protease responsible for processing is absent, and the protein remains attached to membranes. Interestingly, the transiently expressed pp220 aggregates if the myristoylation signal is removed (4). In our experiments, the polyprotein recovered from the cytosolic fraction of infected cells did not aggregate, because it migrated as a soluble protein on sucrose gradients. This suggests that there is a mechanism to prevent the aggregation of non-membrane-bound pp220 in infected cells.
We have shown that the aggregation of p73, the major capsid protein of ASF virus, is prevented by a virally encoded chaperone (13). A similar mechanism may operate for pp220. Interestingly, loss of the first five amino acids after processing by the viral protease means that many of the proteolytic products of pp220 and the final structural proteins p34 and p150 also lack myristate (33, 34). The observation that these and several other processed forms are recovered with the membrane fraction means that a mechanism other than myristate retains them on cellular membranes. This could involve cellular or virally encoded proteins targeted to membranes within virus assembly sites. Another possibility is that the myristate group does not function as a membrane anchor, but plays a different role in virus assembly and/or structure. Several myristoylated viral proteins that have a role in virus assembly/structure but are cytosolic have been described (21, 34).
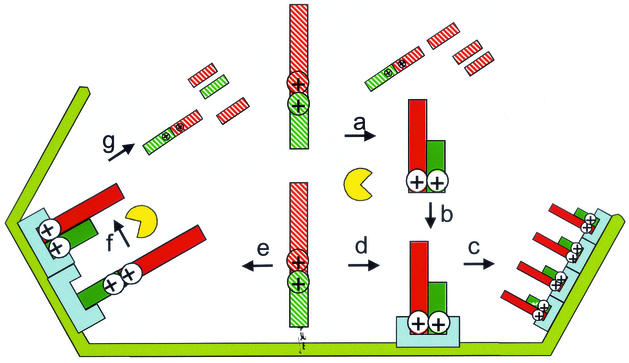
Figure 7 depicts two mechanisms of ensuring the selective packaging of the structural proteins and exclusion of processing intermediates. In each case, the correct processing is predicted to expose structural features that are required for assembly. These structures could allow the proteins to assemble with each other during the formation of the virus matrix layer or facilitate assembly with other structural proteins previously targeted to assembly sites. If these structural features were masked (or missing) in the polyprotein and incorrectly processed forms, these proteins would be excluded from the virion. The polyprotein is translated in the cytoplasm, and processing of the polyprotein could either occur in the cytosol or on the endoplasmic reticulum membrane. Processing would reveal assembly and packaging signals, allowing assembly into virions. In a second possibility, the membrane-bound polyprotein exposes assembly and packaging signals and moves into viral precursors or procapsids, where it acts as a scaffold. Subsequent proteolytic processing in the procapsid allows assembly of the correctly processed forms, while the incorrectly processed forms lack packaging signals and are ejected.
FIG. 7.
Mechanisms for the selective packaging of correctly processed products of pp220. The polyprotein is translated in the cytoplasm; at this point, and assembly sites are masked (hatched ⊕). Proteolytic processing could occur in the cytosol (a and b) or on the membrane (d). This reveals assembly targeting sequences (⊕), and the correctly processed proteins move into the assembling virion (c). In a second model, the polyprotein reveals assembly targeting sequences (⊕) before proteolytic processing and moves into the virion as a scaffold (e) with the viral protease. Correctly processed forms retain assembly targeting sequences and remain in the virion (f), while incorrectly processed forms are ejected (g).
A role for pp220 as a scaffold protein comes from the work of Andrés et al., who found that the products of the polyprotein are found in equimolar concentrations in virions (6), suggesting that the polyprotein is processed after incorporation into the procapsid. Furthermore, the viral protease is packaged into virions (3). Our experiments with sucrose gradient sedimentation were unable to detect a large procapsid complex containing pp220. It is important to point out, however, that if the scaffold-to-capsid transition was very rapid, and a large procapsid structure containing pp220 may have been missed by our assay. The coupling of pp220 processing with scaffold assembly is an attractive possibility because it could allow steric constraints within the procapsid to prevent proteolysis of internal Gly-Gly-X sites within p150.
Acknowledgments
This work was funded by grants from the BBSRC and DEFRA.
REFERENCES
- 1.Acharya, R., E. Fry, D. Stuart, G. Fox, D. Rowlands, and F. Brown. 1989. The 3 dimensional structure of foot and mouth disease virus at 2.9 Å resolution. Nature 337:709-716. [DOI] [PubMed] [Google Scholar]
- 2.Aguado, B., E. Viñuela, and A. Alcamí. 1991. African swine fever virus fatty-acid acylated proteins. Virology 185:942-945. [DOI] [PubMed] [Google Scholar]
- 3.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 4.Andrés, G., R. García-Escudero, M. L. Salas, and J. M. Rodríguez. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J. Virol. 76:2654-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, J. 1960. The initiation of vaccinia infection. Virology 11:603-623. [DOI] [PubMed] [Google Scholar]
- 8.Carrascosa, J. L., J. M. Carazo, A. L. Carrascosa, N. Garcia, A. Santisteban, and E. Viñuela. 1984. General morphology and capsid fine-structure of African swine fever virus particles. Virology 132:160-172. [DOI] [PubMed] [Google Scholar]
- 9.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y.
- 10.Cobbold, C., S. M. Brookes, and T. Wileman. 2000. Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store during envelopment of African swine fever virus. J. Virol. 74:2151-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobbold, C., J. T. Whittle, and T. Wileman. 1996. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J. Virol. 70:8382-8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobbold, C., and T. Wileman. 1998. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J. Virol. 72:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobbold, C., M. Windsor, and T. Wileman. 2001. A virally encoded chaperone specialized for folding of the major capsid protein of African swine fever virus. J. Virol. 75:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteves, A., M. I. Marques, and J. V. Costa. 1986. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected-cells. Virology 152:192-206. [DOI] [PubMed] [Google Scholar]
- 15.García-Escudero, R., G. Andrés, F. Almazán, and E. Viñuela. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Mata, R., Z. Bebök, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goorha, R., and A. Granoff. 1979. Icosahedral cytoplasmic deoxyviruses. Newly characterised vertebrate viruses. Comp. Virol. 14:367-369. [Google Scholar]
- 18.Gouet, P., J. M. Diprose, J. M. Grimes, R. Malby, J. N. Burroughs, S. Zientara, D. I. Stuart, and P. P. C. Mertens. 1999. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97:481-490. [DOI] [PubMed] [Google Scholar]
- 19.Grand, R. J. A. 1989. Acylation of viral and eukaryotic proteins. Biochem. J. 258:625-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 21.Grosenbach, D. W., and D. E. Hruby. 1998. Biology of vaccinia virus acylproteins. Front. Biosci. 3:354-364. [DOI] [PubMed] [Google Scholar]
- 22.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Otín, C., C. Simón-Mateo, L. Martinez, and E. Viñuela. 1989. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 264:9107-9110. [PubMed] [Google Scholar]
- 26.Mathews, R. E. F. 1982. Classification and nomenclature of viruses. Intervirology 17:1-199. [DOI] [PubMed] [Google Scholar]
- 27.Moura-Nunes, J., F. D. Vigario, and A. M. Terrinha. 1975. Ultrastructural study of ASF virus replication in cultures of swine bone marrow cells. Arch. Virol. 49:59-66. [DOI] [PubMed] [Google Scholar]
- 28.Murti, K. G., and R. Goorha. 1983. Interaction of frog virus-3 with the cytoskeleton. 1. Altered organization of microtubules, intermediate filaments, and microfilaments. J. Cell Biol. 96:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Risco, C., J. R. Rodríguez, C. López-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodríguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouiller, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz, A. M., L. E. Henderson, and S. Oroszlan. 1988. Fatty acylation of proteins. Annu. Rev. Cell Biol. 4:611-647. [DOI] [PubMed] [Google Scholar]
- 33.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus—a novel gene-expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towler, D. A., J. I. Gordon, S. P. Adams, and L. Glaser. 1988. The biology and enzymology of eukaryotic protein acylation. Annu. Rev. Biochem. 57:69-99. [DOI] [PubMed] [Google Scholar]
- 35.Viñuela, E. 1985. African swine fever virus. Curr. Top. Microbiol. 116:151-170. [DOI] [PubMed] [Google Scholar]
- 36.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enríquez, J. F. Rodriguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]