Abstract
Human immunodeficiency virus type 1 (HIV-1) entry into target cells requires folding of two heptad-repeat regions (N-HR and C-HR) of gp41 into a trimer of N-HR and C-HR hairpins, which brings viral and target cell membranes together to facilitate membrane fusion. Peptides corresponding to the N-HR and C-HR of gp41 are potent inhibitors of HIV infection. Here we report new findings on the mechanism of inhibition of a N-HR peptide and compare these data with inhibition by a C-HR peptide. Using intact envelope glycoprotein (Env) under fusogenic conditions, we show that the N-HR peptide preferentially binds receptor-activated Env and that CD4 binding is sufficient for triggering conformational changes that allow the peptide to bind Env, results similar to those seen with the C-HR peptide. However, activation by both CD4 and chemokine receptors further enhances Env binding by both peptides. We also show that a nonconservative mutation in the N-HR of gp41 abolishes C-HR peptide but not N-HR peptide binding to gp41. These results indicate that there are two distinct sites in receptor-activated Env that are potential targets for drug or vaccine development.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) mediates virus attachment and fusion to target cells. Binding of the surface subunit (gp120) of Env to the CD4 and chemokine cellular receptors triggers conformational changes in the oligomeric Env complex that activate the membrane fusion activity of the transmembrane subunit (gp41). A detailed understanding of these structural changes in Env would create new opportunities to prevent and treat HIV infection.
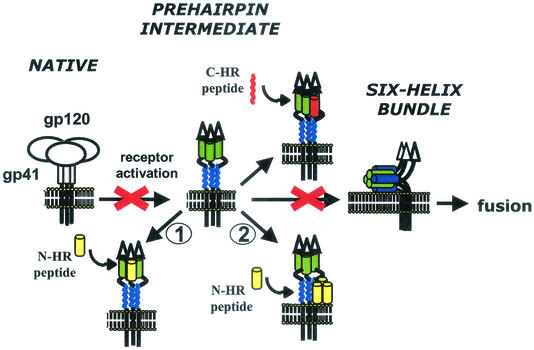
A leading model of HIV entry proposes substantial refolding of Env, in which Env transitions from a metastable, native (prefusion) conformation through a prehairpin fusion intermediate to a thermostable, six-helix bundle structure (Fig. 1) (reviewed in reference 11). The six-helix bundle is created when two heptad repeat motifs (HR) in the ectodomain of gp41 self-assemble into a trimer-of-hairpins (3, 7, 17, 24, 28). The N-terminal HR (N-HR) folds into a parallel, trimeric coiled-coil core, whereas three C-terminal HR (C-HR) form helices that pack in the grooves of the coiled-coil trimer in an antiparallel manner. gp120 binding to cellular receptors loosens its association with gp41, probably resulting in the release of the hydrophobic fusion peptide at the N terminus of gp41 from a sequestered site so that it can insert into the target membrane. With the fusion peptide inserted into the target membrane and the transmembrane embedded in the viral membrane, gp41 likely then folds into the compact six-helix bundle, promoting fusion by bringing the membranes close together and perhaps releasing energy as Env folds to a more stable structure.
FIG. 1.
Model of fusion-inducing conformational changes in Env. Env binding to receptors causes a stepwise transition from native conformation to the prehairpin fusion intermediate to the six-helix bundle. Peptides inhibit entry by preventing formation of the six-helix bundle. N-HR peptides could bind to N-HR of gp41 (pathway 1) and/or to C-HR of gp41 (pathway 2) of the prehairpin fusion intermediate. For simplicity, the N-HR peptide is shown as a monomer, but it is likey to be in a monomer-oligomer equilibrium.
Peptides corresponding to the N-HR and C-HR are potent inhibitors of HIV infection (15, 26, 27; C. Wild, T. Greenwell, and T. Matthews, Letter, AIDS Res. Hum. Retrovir. 9:1051-1053, 1993). It is widely believed that these peptides bind the Env prehairpin fusion intermediate, preventing formation of the six-helix bundle by endogenous HR of gp41 in a dominant-negative manner (reviewed in reference 8). This theory is supported by experimental data showing that a C-HR peptide (DP-178) preferentially binds receptor-activated Env (12) and that viruses resistant to DP-178 have mutations in the N-HR (20). The mechanism of inhibition of N-HR peptides is less clear. Previous biological studies showed that a N-HR peptide (DP-107) does not neutralize native virions (27), suggesting that this peptide binds a receptor-activated form of Env. However, according to the dominant-negative model, the N-HR peptides could bind to either the N-HR or C-HR of gp41 (Fig. 1, inhibitory pathway 1 and 2, respectively). For example, if the N-HR does not exist in an extended coiled coil in the native conformation but forms one after receptor activation in a manner analogous to the spring-loaded mechanism of influenza virus hemagglutinin (HA) (6), then an N-HR peptide might bind the N-HR in gp41 to facilitate a loop-to-helix transition, forming a peptide-gp41 coiled coil. An N-HR peptide could also bind to a preformed N-HR coiled coil during a monomer-to-trimer equilibrium (4), as has been recently proposed for an N-HR peptide that was mutated to preclude binding to the C-HR (2). Alternatively, an N-HR peptide could bind the C-HR of gp41 to mimic interactions in the six-helix bundle. A five-helix gp41 construct with an exposed and stabilized N-HR coiled-coil trimer has been shown to interact with a C-HR peptide in this way (21).
Here we report new data on how N-HR and C-HR peptides bind intact Env under fusogenic conditions. We show that an N-HR peptide (DP-107, T21) preferentially binds receptor-activated Env, similar to a C-HR peptide (DP-178, T20). For both DP-107 and DP-178 peptides, gp120 interactions with CD4 were sufficient and necessary to induce peptide binding to gp41, but peptide binding was clearly enhanced when gp120 was activated by both CD4 and chemokine receptors. Using Envs with point mutations in gp41, we also demonstrate that a nonconservative mutation in the N-HR prevents C-HR peptides from binding gp41 but not an N-HR peptide. These studies indicate that both N- and C-HR regions in the fusion intermediate are accessible to broadly active fusion inhibitors.
MATERIALS AND METHODS
Peptides.
Peptide C34HA (WMEWDREINNYTSLIHSLIEESQNQQEKNEQELLGGGYPYDVPDYAGPG) was synthesized by standard Fmoc (9-fluore- nylmethoxy carbonyl) chemistry and purified by reversed-phase high-performance liquid chromatography. Peptides 107HA (GGVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQGGGYPYDVPDYAGPG), 107FL (GGVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQGGGDYKDDDDY), and 178HA (YTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFGGGYPYDVPDYAGPG) were expressed in Escherichia coli and purified as previously reported (5) and are described below. Expression plasmids with a TrpLE fusion protein containing coding sequences for residues 552 to 595 (pTCLE-G2C) or 638 to 673 (pTCLE-178) of the LAI envelope gene product were gifts from Carl Wild (Panacos, Gaithersburg, Md.) and Terry Oas (Duke University, Durham, N.C.). Briefly, pTCLE-G2C and pTCLE-178 were modified by PCR to include sequences encoding GGG before the HA or FLAG epitopes at the C terminus of the envelope coding sequences. Expresssion plasmids were transformed into BL21(DE3) cells, induced with IPTG (isopropyl-β-d-thiogalactopyranoside), and lysed by using a French press. Inclusion bodies were pelleted, washed, and dissolved in 70% formic acid prior to cleavage with 50 mg of CNBr/ml. The dried products were dissolved in 6 M guanidine-HCl and separated by Sephadex G-50 (26 mm by 70 cm) gel filtration chromatography with 5% acetic acid. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analytic high-pressure liquid chromatography indicated that all of the peptides were >95% pure. All peptides were confirmed to have the expected molecular weight by using matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy and the expected inhibitory activity by using infectivity assays. A peptide (DLIAYLKQATKFRKDIAAKY), synthesized by combining T-cell epitopes from cytochrome c and sperm whale myoglobin (residues 1 to 11 and 12 to 20, respectively) and which has no inhibitory activity against HIV (data not shown), was obtained from Ira Berkower (Food and Drug Administration, Bethesda, Md.) and used as a negative control.
Reagents and cells.
Wild-type Env expression vector (pSM-HXB2), 293T cells, and 3T3 cells expressing human CD4 and chemokine receptors were provided by Dan Littman (New York University, New York). The Rev expression plasmid was provided by Tristram Parslow (University of California, San Francisco), and the furin expression plasmid was provided by Gary Thomas (Oregon Health Sciences University, Portland) and James Binley (Aaron Diamond AIDS Research Center, New York, N.Y.). sCD4 was a gift from Ray Sweet (SmithKline Beecham Pharmaceuticals, King of Prussia, Pa.). The T22 chemokine antagonist was purchased from BACHEM Bioscience (Philadelphia, Pa.). Phycoerythrin-conjugated anti-CXCR4 and allophycocyanin-conjugated anti-CD4 antibodies were purchased from (Pharmingen, San Diego, Calif.). Stromal cell-derived factor 1 (SDF-1) and RANTES chemokines were purchased from Peprotech, Inc. (Rocky Hill, N.J.). CHO cells stably expressing the HXB2 Env were previously described (23). Env mutants were created by oligonucleotide-directed mutagenesis as previously described (25) and verified to have the desired mutations by sequencing. 293T cells were cotransfected with Env and Rev expression plasmids and an additional furin expression plasmid as needed. At 48 h after transfection, cells were harvested for peptide coimmunoprecipitation as previously described (12).
Coimmunoprecipitation assay.
Briefly, 6 × 106 stable or transient Env-expressing cells were incubated with ca. 20 μg of peptides in the presence or absence of 3 μg of sCD4 or 6 × 106 target cells in 1 ml of complete medium for 90 min at 37°C. Cells were then washed three times to remove unbound peptide and then incubated with 15 μg of anti-HA antibody (12CA5; Roche, Indianapolis, Ind.) at room temperature for an additional hour. Cells were washed twice and then lysed with 1% Nonidet P-40. Clarified supernatants were immunoprecipitated with 25 μl of a 25% suspension of protein A-agarose (Invitrogen, Carlsbad, Calif.) overnight, and washed three times before separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with an anti-gp41 antibody (Chessie 8 [1]).
Chemokine treatment and flow cytometry.
A total of 2.3 × 107 3T3-CD4-X4 cells were incubated with 1 μg of SDF-1/ml, 1 μM T22, or 1 μg of RANTES/ml in complete medium at 37°C for 1 h and then distributed into tubes for coimmunoprecipitations or flow cytometry. For flow cytometry, 106 cells were stained with 20 μl of anti-CD4 antibodies, anti-CXCR4 antibodies, or isotype control antibodies for 30 min on ice. Cells were then washed and fixed with 2% paraformaldehyde prior to flow cytometry (Becton Dickinson LSR).
RESULTS
N-HR peptide binding to Env.
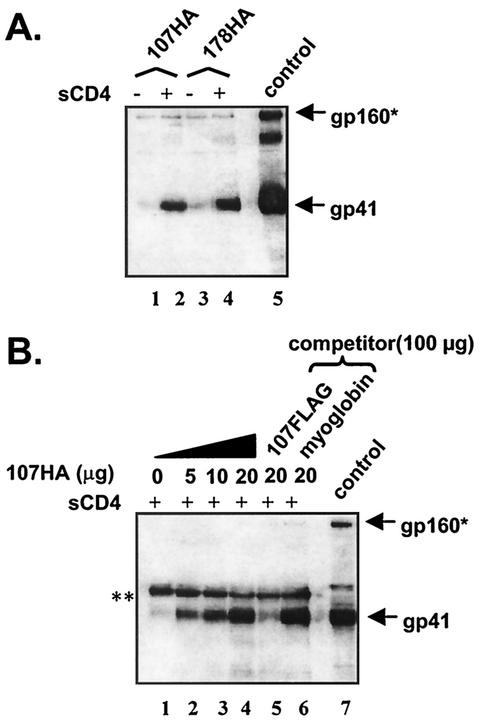
An epitope-tagged N-HR peptide (107HA) was incubated with Env-expressing cells (HXB2 strain) in the presence or absence of soluble CD4 (sCD4) at 37°C (Fig. 2A). In samples treated with sCD4, the 107HA coimmunoprecipitated gp41 (Fig. 2A, lane 2), similar to the C-HR peptide (178HA) (Fig. 2A, lane 4; see also reference 12). In some experiments, small amounts of gp41 were seen in samples incubated with 107HA or 178HA without exposure to CD4 (Fig. 2B, lane 1), but sCD4 consistently showed strong enhancement of coimmunoprecipitation of gp41 by both peptides in all experiments. 107HA binding to gp41 was dose dependent (Fig. 2B, lanes 1 to 4) and could be competed for by a fivefold excess of 107 peptide with a different epitope tag (107FLAG) but not by a fivefold excess of an irrelevant amphipathic peptide (myoglobin; Fig. 2B, lanes 5 and 6, respectively). These results demonstrate that the N-HR peptide binding to receptor-activated Env cells is specific and saturable. Similar findings were previously reported for a C-HR peptide (12).
FIG. 2.
Coimmunoprecipitation of gp41 by N-HR peptide (A) N-HR peptide binds receptor-activated gp41. HA-tagged peptides preferentially pull down receptor-activated gp41 (lanes 2 and 4). (B) Peptide binding is specific and saturable. N-HR peptide immunoprecipitates gp41 in a dose-dependent manner (lanes 1 to 4) and can be competed out by a fivefold excess of an untagged peptide (lane 5) but not by irrelevant peptide (lane 6). Gels shown are representative of at least three independent experiments. Control, surface immunoprecipition of Env-expressing cells with HIV+ immunoglobulin G (IgG); ✽, gp160 and/or a nonspecific bands; ✽✽, nonspecific (heavy-chain) band.
Receptor requirements for peptide binding.
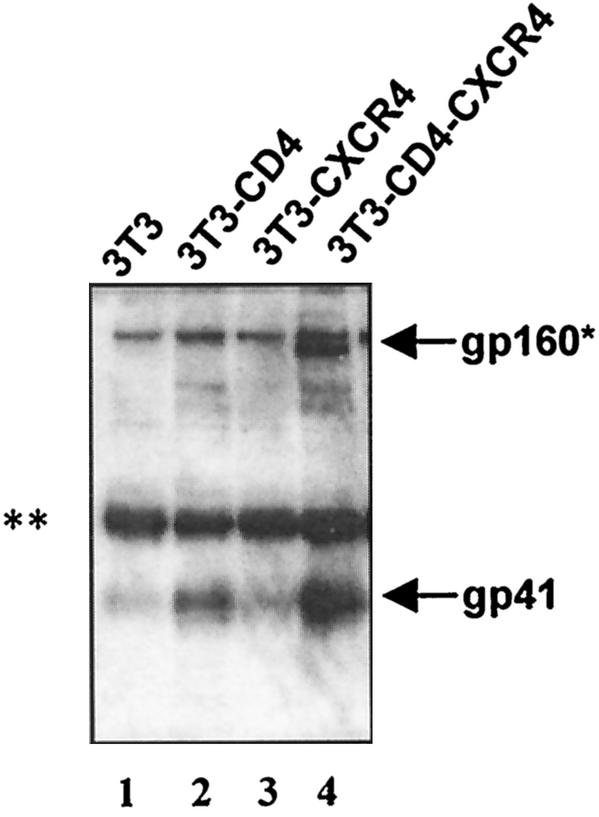
Experiments with sCD4 indicated that the CD4 receptor was sufficient for inducing conformational changes in Env to the prehairpin fusion intermediate that allow interactions with the N-HR and C-HR peptides. To more clearly define the receptor requirements for peptide binding, experiments were repeated with mouse 3T3 cells that express different combinations of the human CD4 and CXCR4 receptors on the cell surface (Fig. 3). Flow cytometry showed that all CD4-bearing cells expressed similar levels of CD4 (not shown). 3T3 target cells lacking human receptors or expressing only the CXCR4 receptor (Fig. 3, lanes 1 and 3, respectively) did not induce coimmunoprecipitation of gp41 by 107HA. However, 107HA coimmunoprecipitated gp41 in the presence of target cells expressing human CD4 only (Fig. 3, lane 2), which was further enhanced by the expression of both CD4 and CXCR4 receptors (Fig. 3, lane 4). Similar findings were reported for 178HA (12).
FIG. 3.
Receptor requirements for peptide binding to gp41. CD4 is sufficient, but chemokine receptor enhances peptide binding to Env cells. CD4 is sufficient (lane 2) and necessary (lane 3) for triggering conformational changes that allow the N-HR peptide to coimmunoprecipitate gp41, but the combination of CD4 and chemokine receptor is more efficient (lane 4). Lane 1 contains control cells without receptors. Gels shown are representative of at least three independent experiments. ✽, gp160 and/or a nonspecific bands; ✽✽, nonspecific (heavy-chain) band.
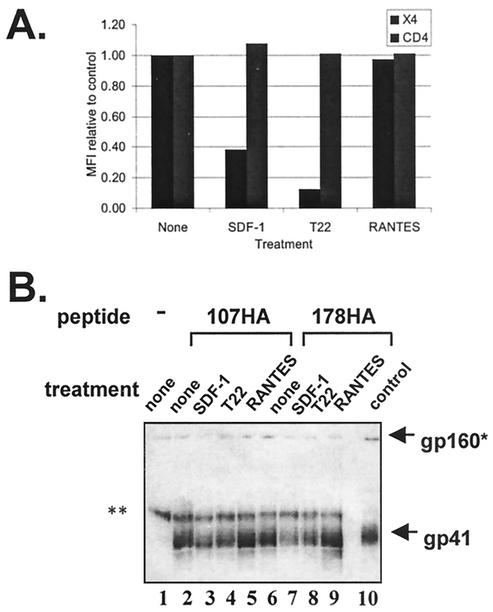
To confirm the role of the chemokine receptor in triggering conformational changes that allow peptide binding, further experiments were undertaken with 3T3 cells expressing both CD4 and CXCR4 that were pretreated with CXCR4 antagonists. Cells were incubated with SDF-1 or RANTES chemokines (specific for CXCR4 and CCR5 receptors, respectively) or T22, a peptide that was previously reported to block chemokine interactions with CXCR4 (19). Both SDF-1 and T22 blocked detection of CXCR4 at the cell surface, but RANTES had little effect on CXCR4 levels (Fig. 4A), a finding consistent with its specificity for CCR5. Pretreating cells with SDF-1 or T22 significantly inhibited 107HA and 178HA binding to gp41 (Fig. 4B, lanes 3 to 4 and 7 to 8), whereas pretreatment with RANTES did not (Fig. 4B, lanes 5 and 9). Our combined results indicate that CD4 is both sufficient and necessary for triggering conformational changes to the prehairpin intermediate in this Env, but the combination of CD4 and chemokine receptor further facilitates peptide binding.
FIG. 4.
Contributions of chemokine receptor in allowing peptide binding. (A) Flow cytometry of 3T3-CD4-CXCR4 cells treated with chemokine receptor inhibitors. SDF-1 and T22, but not RANTES, reduced detection of CXCR4 at the cell surface without reducing CD4 levels. The mean fluorescent intensity (MFI) was normalized to untreated control. (B) Chemokine receptor inhibitors diminish peptide binding to gp41. Pretreatment of CD4+ CXCR4+ 3T3 cells with SDF-1 (lanes 3 and 7) and peptide T22 (lanes 4 and 8) inhibits the ability of the peptides to coimmunoprecipitate gp41, but RANTES (lanes 5 and 9) does not. Control, surface immunoprecipition of Env-expressing cells with HIV+ IgG; X4, CXCR4; ✽, gp160 and/or a nonspecific bands; ✽✽, nonspecific (heavy-chain) band. Results shown in both panels are representative of at least three independent experiments.
Effects of Env mutations on peptide binding.
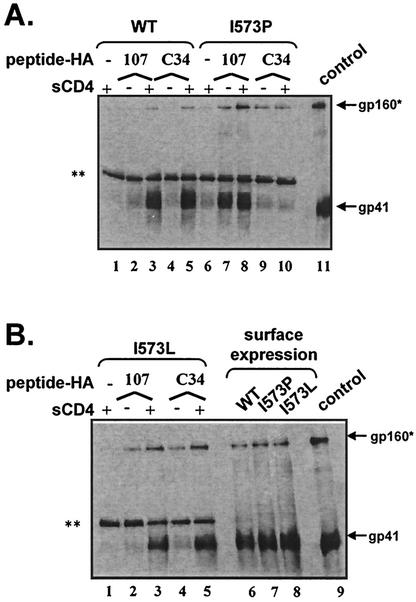
To investigate the binding sites for the peptides on gp41, we repeated peptide coimmunoprecipitation experiments with Envs that contain point mutations in the central region of the N-HR. An isoleucine-to-proline subsitution in position 573 (I573P) of the HXB2 Env was previously reported to abolish HIV infectivity, whereas an isoleucine-to-leucine (I573L) substitution preserves infectivity (10). When we reconstructed these mutations with our own expression vectors we found the same effects on Env function (data not shown), but the proline mutant had reduced precursor processing and cell surface expression compared to wild-type Env. To normalize Env processing and expression at the cell surface among mutant and wild-type Envs, cells were cotransfected with an expression plasmid containing furin along with the Env expression plasmid (Fig. 5B, lanes 6 to 8).
FIG. 5.
N-HR mutations in gp41 block C-HR but not N-HR peptide binding. (A) A nonconservative mutation (I573P) in the N-HR of gp41 blocks C-HR peptide binding (lanes 9 and 10) but not N-HR peptide binding (lanes 7 and 8). The I573P substitution also makes N-HR peptide binding CD4 independent. (B) A conservative mutation (I573L) in the N-HR of gp41 does not impair C-HR peptide binding (lanes 4 and 5) or N-HR peptide binding (lanes 2 and 3). Mutant and wild-type Envs are processed and expressed at the cell surface at comparable levels (lanes 6 to 8). The gels shown are representative of at least three independent experiments. Surface immunoprecipitation of Env-expressing cells used HIV+ IgG. ✽, gp160 and/or a nonspecific bands; ✽✽, nonspecific (heavy-chain) band.
Modeling of two overlapping C-HR peptides (C34HA and 178HA) by using atomic coordinates of the six-helix bundle suggested that the helix modeled by the C34HA peptide might be more directly affected by the mutation in the 573 position compared to the helix modeled by the 178HA peptide. Therefore, both C34HA and 178HA were tested for binding to the mutant Envs. We found that the proline (Fig. 5A, lane 10), but not the conservative leucine substitution (Fig. 5B, lane 5), greatly reduced the ability of both C34HA and 178HA (not shown) to coimmunoprecipitate gp41. These results strongly suggest that the C-HR peptides directly bind to the N-HR of gp41, as predicted from the structural studies of the six-helix bundle and a genetic study of a virus resistant to a C-HR peptide (DP-178) (20). In contrast, the I573P substitution did not impair the ability of 107HA to coimmunoprecipitate gp41 and surprisingly allowed this peptide to bind gp41 in a CD4-independent manner (Fig. 5A, lanes 7 and 8). Because 107HA binding to the I573P mutant was not measurably diminished compared to wild-type Env, these results suggest the peptide is binding to the C-HR of gp41 (pathway 2), at least with the 573 mutant. Additional mutants in the C-HR of gp41 were created to more directly assess peptide interactions with this region, but all nonconservative mutants tested were impaired in surface expression and/or precursor processing, despite extensive efforts to normalize expression to wild type. Consequently, appropriate C-HR mutants were not available to directly assess peptide binding to this region.
DISCUSSION
Our comparative studies of how N-HR and C-HR peptides bind gp41 shed light on the entry mechanism of Env and provide new information for drug and vaccine design. Using intact Env under fusogenic conditions, we show that both N-HR and C-HR peptides preferentially bind receptor-activated Env, rather than native Env. For both peptides, CD4 is sufficient for triggering conformational changes that expose the peptide-binding sites in the prehairpin intermediate, but the combination of CD4 and CXCR4 receptors is more efficient. These findings raise questions about the role of chemokine receptors in membrane fusion and the need for chemokine receptors to fully trigger conformational changes that lead to exposure of the fusion peptide or the six-helix bundle. In the case of the HXB2 Env, we show that CD4 is sufficient to expose the N-HR region, which is adjacent to the fusion peptide. CD4-induced triggering of Env to the six-helix bundle, in the absence of chemokine receptor, has also been previously reported (9, 14).
Nevertheless, the combination of CD4 and chemokine receptor enhances peptide binding to the prehairpin intermediate, but the reasons for this are not clear. The enhancement could reflect improved efficiency in triggering conformational changes when both receptors are present, and this level of efficiency may be required for membrane fusion. Alternatively, the chemokine receptors could promote membrane fusion by ensuring that the fusion peptide inserts into the target membrane rather than allowing it to insert into the viral membrane. In this scenario, enhanced peptide binding by the combination of CD4 and chemokine receptors might be explained by slower folding into the six-helix bundle when the fusion peptide is anchored into the target membrane, compared to folding when the fusion peptide inserts into the viral membrane during or after six-helix formation. It is also possible that Env association with the chemokine receptors promotes higher-order organization of Env-receptor complexes in specialized domains in the membrane, which may facilitate fusion-inducing conformational changes in multiple Envs.
Recent studies with HR peptides from the fusion protein of the simian virus 5 paramyxovirus show a mechanism of action similar to the HIV peptides (22). For both viruses, attachment of the viral fusion protein to target cells is required for peptide binding and inhibition of fusion. For simian virus 5, additional experiments involving temperature- and lipid-arrested fusion intermediates revealed that the N-HR peptide inhibited an earlier fusion intermediate than the C-HR peptide. To date, studies involving temperature shifts with the HIV N- and C-HR-peptides have not shown differences in binding to temporally distinct fusion intermediates (13, 18), but additional studies that focus on this issue are needed.
Experiments with the Env mutants further show that the peptides can bind to two regions in the prehairpin intermediate. A nonconservative mutation in the N-HR greatly reduced binding by two different C-HR peptides (178HA and C34HA), whereas a conservative mutation did not, indicating that the C-HR peptides most likely make direct contact with the N-HR in the hairpin intermediate. The I573P substitution probably disrupts local helical structure in the N-HR of gp41, and it is expected that C-HR peptide binding to the N-HR requires helical or coiled-coil structure in this region (2). Our peptide binding results are consistent with genetic changes in a virus resistant to the DP-178 C-HR peptide (20). Although only the C34HA peptide can be modeled to directly overlie the N-HR in the region of residue 573, the proline substitution in this position probably alters local structure of the N-HR region to affect nearby residues directly involved in binding to the 178HA peptide.
In contrast to the C-HR peptides, 107HA binding was not reduced by the same mutations in the N-HR of gp41, indicating that N-HR peptides corresponding to wild-type sequences of gp41 bind Env well even when they cannot bind the N-HR of gp41. As shown by the I573P mutant, our data strongly suggest that the N-HR peptide predominantly binds the C-HR of gp41 and mimics interactions in the six-helix bundle (pathway 2). In this model, the N-HR peptides probably bind the C-HR as an oligomer (2).
N-HR peptides with mutations that prevent binding to the C-HR have also been shown to inhibit HIV infection, presumably by binding to the N-HR of gp41 (2). Thus, we cannot rule out the possibility that N-HR peptides corresponding to wild-type sequences could also bind to the N-HR (pathways 1) in Envs with mutations in the C-HR or to either or both HR (pathways 1 and 2) of wild-type Env. If this can occur, then viruses may have to develop multiple mutations to resist inhibition by N-HR peptides. Finally, the I573P Env unexpectedly allowed the N-HR peptide to bind in a CD4-independent manner, implying that the N-HR region in native Env affects exposure of the C-HR. That the I573P Env was also inefficiently processed further indicates that this region of N-HR affects exposure or conformation of the cleavage site.
In summary, we show that peptides corresponding to wild-type sequences in the N- or C-HR of gp41 can trap the prehairpin fusion intermediate at two sites. The appeal of targeting these regions of gp41, which are exposed only transiently during the entry process, comes from the fact that these domains are among the most conserved regions of Env. Although the N-HR is better conserved than the C-HR, both HR peptides demonstrate potent inhibition against most HIV strains in vitro, and one C-HR peptide (T20) has already demonstrated efficacy in reducing viral load in vivo (16).
Acknowledgments
We thank Ira Berkower, Asier Sáez-Cirión, and Darón Freedburg (Food and Drug Administration, Bethesda, Md.) for critical reading of the manuscript. We also thank Carl Wild (Panacos Pharmaceuticals, Inc., Gaithersburg, Md.) and Terry Oas (Duke University) for the DP-107 and DP-178 expression plasmids and purification protocols.
This work was supported in part by the Intramural AIDS Targeted Antiviral Program at the National Institutes of Health, Bethesda, Md. (C.D.W.).
REFERENCES
- 1.Abacioglu, Y., T. Fouts, J. Laman, E. Claassen, S. Pincus, J. Moore, C. Roby, R. Kamin-Lewis, and G. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371-381. [DOI] [PubMed] [Google Scholar]
- 2.Bewley, C. A., J. M. Louis, R. Ghirlando, and G. M. Clore. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. [DOI] [PubMed]
- 3.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caffrey, M., J. Kaufman, S. Stahl, P. Wingfield, A. M. Gronenborn, and G. M. Clore. 1999. Monomer-trimer equilibrium of the ectodomain of SIV gp41: insight into the mechanism of peptide inhibition of HIV infection. Protein Sci. 8:1904-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderone, T. L., R. D. Stevens, and T. G. Oas. 1996. High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J. Mol. Biol. 262:407-412. [DOI] [PubMed] [Google Scholar]
- 6.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational changes of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 9.de Rosny, E., V. Vassell, P. T. Wingfield, C. T. Wild, and C. D. Weiss. 2001. Peptides corresponding to the heptad repeat motifs in the transmembrane protein (gp41) of human immunodeficiency virus tyep 1 elicit antibodies to receptor-activated conformatinos of the envelope glycoprotein. J. Virol. 75:8859-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubay, J. W., S. J. Roberts, B. Brody, and E. Hunter. 1992. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J. Virol. 66:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 13.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 16.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 17.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 18.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami, T., T. Nakajima, Y. Koyanagi, K. Tachibana, N. Fujii, H. Tamamura, N. Yoshida, M. Waki, A. Matsumoto, O. Yoshie, T. Kishimoto, N. Yamamoto, and T. Nagasawa. 1997. A small molecule CXCR4 inhibitor that blocks T-cell line-tropic HIV-1 infection. J. Exp. Med. 186:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 22.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss, C. D., and J. M. White. 1993. Characterization of stable Chinese hamster ovary cells expressing wild-type, secreted, and glycosylphosphatidylinositol-anchored human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 67:7060-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 25.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled coil domain of the human immunodeficiency virus type 1 (HIV-1) gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wild, C., J. W. Dubay, T. Greenwell, T. J. Baird, T. G. Oas, C. McDanal, E. Hunter, and T. Matthews. 1994. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc. Natl. Acad. Sci. USA 91:12676-12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 angstrom resolution. J. Struct. Biol. 126:131-144. [DOI] [PubMed] [Google Scholar]