Abstract
Formation of human immunodeficiency virus type 1 (HIV-1) particles takes place at the plasma membrane of cells and is directed by the Pr55Gag polyprotein. A functional assembly domain (the M domain) within the N-terminal portion of Pr55Gag mediates the interaction of Gag with cellular membranes. However, the determinants that provide specificity for assembly on the plasma membrane, as opposed to intracellular membranes, have not been identified. Recently, it was reported that Pr55Gag interacts with lipid raft microdomains of the plasma membrane. We sought to identify the domains within Pr55Gag that contribute to lipid raft association of Gag. Here we demonstrate that the I domain is required for interaction with detergent-resistant membrane fractions (DRMs). Mutation of key I-domain residues or loss of myristylation abrogated the association of Gag with DRMs. Thus, the I domain and the M domain combine to mediate Gag-lipid raft interactions as defined by these biochemical criteria. However, Gag protein complexes defined by flotation studies were much denser than classical lipid rafts, failed to incorporate classical lipid raft marker proteins, and were not disrupted by cholesterol extraction. Large sheets of Gag protein were identified in DRM fractions upon examination by electron microscopy. These results indicate that HIV-1 Pr55Gag forms detergent-resistant complexes at the cellular periphery that are distinct from lipid raft microdomains.
Human immunodeficiency virus (HIV) particles assemble at the plasma membranes of infected cells. The HIV type 1 (HIV-1) Pr55Gag polyprotein is the primary mediator of particle assembly. Pr55Gag forms the core of developing virions, which are first visible by electron microscopy studies as electron-dense patches underlying the plasma membrane (11). The pathway taken by Gag to reach the plasma membrane is unknown but is clearly distinct from the secretory pathway through which the HIV envelope glycoprotein complex travels. Pr55Gag localizes to discrete, punctate regions underlying the plasma membranes of cells (14, 31). This finding suggests that cellular factors present at the periphery of cells contribute to the particle assembly site, perhaps through providing a specific microenvironment in which Gag molecules can multimerize and begin the budding process. Defining the specific cellular microenvironment where particles assemble may allow the assembly process to be dissected and allow for the design of specific inhibitors of the assembly process.
Recently, several reports have indicated that Gag is indeed targeted to lipid raft microdomains of the plasma membrane (20, 23, 24). Lipid rafts are conceived as spatially differentiated microdomains that are enriched in cholesterol and glycosphingolipids (13, 34). By providing an environment that selectively concentrates some proteins and excludes others, rafts provide compartmentalization for important cellular processes, such as sorting of plasma membrane proteins and cellular signaling processes (9, 16, 19, 37). Lipid rafts are enriched in glycosylphosphatidylinositol (GPI)-anchored proteins, Src family kinases, and some transmembrane proteins (6, 34). Lipid rafts and raft components are generally identified by isolation of detergent-resistant membranes (DRMs) using Triton X-100 (TX-100) extraction at low temperatures and equilibrium flotation centrifugation (5, 34). Viruses such as measles virus and influenza virus utilize lipid rafts in the particle assembly process, and some viral structural proteins such as influenza virus hemagglutinin (HA) fractionate in a manner identical to that of classical raft marker proteins (21, 33). Thus, there is precedent for pathogenic viruses to utilize lipid raft microdomains as an exit pathway from the cell.
If Gag does indeed associate with rafts, then this finding should provide important clues to the determinants of specific viral assembly at the plasma membrane. It is well-known that myristic acid, together with determinants within the N-terminal portion of the HIV-1 matrix (MA) protein region of Gag, form a functional assembly domain that mediates Gag-membrane interaction (the M domain). Ono and Freed (24) and Lindwasser and Resh (20) recently demonstrated that additional determinants within the C-terminal portion of Gag are also required for flotation with lipid rafts. The I domain is a functional assembly domain located within the nucleocapsid (NC) region that also plays a role in plasma membrane targeting of HIV Gag proteins (30, 31). In this study, we sought to define the contribution of the HIV-1 I domain to Gag protein association with lipid raft microdomains. We found that the I domain and the M domain are both required for lipid raft association using biochemical flotation criteria similar to those used by previous investigators. Discrete mutations that eliminate I-domain function were found to eliminate the putative Gag-raft interaction. However, we noted that the density of detergent-resistant Gag complexes differed drastically from that of cellular membranes, a finding similar to that of Lindwasser and Resh (20). Furthermore, lipid raft marker proteins did not segregate with detergent-resistant Gag complexes but rather could be widely separated by density from Gag. Classical interventions such as cholesterol depletion that disrupt raft architecture failed to alter Gag DRCs. Finally, we demonstrated large complexes of Gag in detergent-resistant, buoyant fractions by electron microscopy. We conclude that Gag forms distinct detergent-resistant protein complexes at the periphery of cells that are distinct from lipid raft microdomains.
MATERIALS AND METHODS
Plasmids, cells, and transfection.
The plasmids used for expression of Pr55Gag and Gag-green fluorescent protein (GFP) fusion proteins that define the HIV-1 I domain have been previously described (30, 31). The vaccinia virus/T7 polymerase hybrid expression system was employed as previously described for production mutant Gag and Gag-GFP fusion proteins. For this expression system, 10 μg of expression vector DNA was transfected into BSC-40 cells using an in-house lipofection reagent 30 min following infection with recombinant vaccinia virus vTF7-3. Transfections with proviral DNA and raft protein controls were performed using the calcium phosphate transfection method in 293T cells. pNLPR- is a proviral expression plasmid that contains a triple alanine substitution in the protease active site. This plasmid was created in Chris Aiken's laboratory by inserting a BssHII-EcoRI fragment from R9.PR- (40) into pNL4-3. pPLAP is an expression plasmid also constructed in the Aiken laboratory for expression of placental alkaline phosphatase (PLAP) in mammalian cells. It was generated by first cloning the NotI-EcoRI fragment from HX-B/PLAP (7) into the intermediary cloning vector pMECA (35) and then removing the resulting XhoI-EcoRI fragment into the cytomegalovirus promoter-based expression plasmid pCMX/PL-1 (3). pFyn-GFP is an expression plasmid for Fyn bearing a C-terminal GFP tag and was obtained from Mark Philips (8) through Anne Kenworthy's laboratory. LYFPGT46 (26)was obtained from Kai Simons through Anne Kenworthy's laboratory. LYFPGT46 is a nonraft protein that contains the CD46 cytoplasmic tail, the transmembrane domain of the low-density lipid receptor, and yellow fluorescent protein (YFP) as the ectodomain. pCDNA/GagOpt is an expression plasmid bearing a codon-optimized Gag sequence obtained by PCR cloning of Gag from pVRC3900 (15) into the HindIII and BamHI sites of pCDNA3.1. Oligonucleotides for cloning of Gag for this plasmid included the forward oligonucleotide AACAAGCTTGCCACCATGGGCGCCCGCGCCAGC and reverse oligonucleotide CGGGATCCTTATTGTGACGAGGGGTC .
BSC-40 cells were used for most studies employing the vaccinia virus/T7 expression system and were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum and antibiotics at 37°C in 5% CO2. Expression experiments employing proviral plasmids and codon-optimized Gag expression were performed in 293T cells maintained in identical culture medium. Experiments in which both Gag and raft marker expression were required were performed in 293T cells.
Cellular fractionation and equilibrium flotation centrifugation.
Cells were harvested for analysis 15 to 16 h posttransfection when the vaccinia virus/T7 expression system was used or 48 to 72 h posttransfection following expression plasmid transfection in 293T cells. Typically, three 10-cm2-diameter dishes of near-confluent BSC-40 or 293T cells were included in each experimental arm. Cells were washed and then allowed to swell in hypotonic buffer (10 mM Tris-Cl [pH 8.0] plus protease inhibitors) for 15 min on ice. Cells were then broken by gentle Dounce homogenization. After the buffer was adjusted to 0.1 M NaCl, nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min. Supernatants containing cytosolic and membrane components were then adjusted to 50% iodixanol from a stock solution of 60% iodixanol. Forty percent and 10% solutions of iodixanol were prepared using a sodium chloride diluent according to the manufacturer's recommendations (Nycomed Pharma, Oslo, Norway) and layered on top of the 50% iodixanol step. The preparation was then centrifuged in a Sorvall RP55A rotor at 55,000 rpm for 2 h at 4°C, following which fractions were collected from the top of the gradient. Gradient fractions were then analyzed directly by fluorometry or further processed for immunoprecipitation or Western blotting where indicated. For gradients where collection of larger amounts of Gag and control were desirable, similar gradients formed in 13-ml SW41 centrifuge tubes were utilized. Specifically, 50%/40%/30%/10% iodixanol gradients were formed in SW41 centrifuge tubes using 2.5 ml of 50% iodixanol, 4 ml of 40% iodixanol, 4 ml of 30% iodixanol, and 2.5 ml of 10% iodixanol. Flotations in the Beckman SW41 rotor were performed at 35,000 rpm for 16 h at 4°C. Equal fractions were harvested from the top of the gradient for analysis.
Detection of Gag-GFP and controls by fluorometry.
Gag-GFP proteins in cellular fractions were detected by fluorescence spectrophotometry as previously described (30, 31). A standard curve of recombinant enhanced GFP (Clontech) was utilized with each experiment to determine the linear range and limits of detection. Samples were quantified on a microplate fluorometer (Fluostar; BMG LabTechnologies) using an excitation wavelength of 450 nm and an emission wavelength of 510 nm. For iodixanol gradient fractions, a control gradient from cells lacking any GFP expression was included in the analysis as a background control, and values were subtracted from those of test samples.
Detection of Gag protein and control proteins by nonfluorometric means.
Gag protein was detected in some experiments by radiolabeling with [35S]cysteine-methionine and immunoprecipitation with pooled HIV patient sera. Gag protein in cells expressing NL4-3PR- was labeled for 2 h in cysteine- and methionine-deficient media supplemented with 200 μCi of [35S]cysteine-methionine per ml. PLAP activity in fractions was detected using a chemiluminescent substrate for alkaline phosphatase (AP substrate; Lumigen, Southfield, Mich.); luminescence was quantitated in a TopCount plate-based detection system (Packard Instruments).
For additional separation of raft components and Gag protein, a two-phase separation procedure was used. First, DRM fractions were prepared as described above using 50%/40%/10% iodixanol gradients. The fluorescence of each fraction was determined by fluorometry, and peak fluorescing fractions (at the 40%/10% iodixanol interface) were pooled. This sample was then loaded on top of a preformed 20 to 75% linear sucrose density gradient and subjected to centrifugation at 35,000 rpm for 16 h at 4°C.
Cholesterol extraction.
293T cells were labeled overnight by incubation with 50 μCi of [3H]cholesterol per ml. After the cells were washed, fresh medium containing 10 mM methyl-β-cyclodextrin (MβCD) was added, and the cells were incubated for 1 h at 37°C. Portions of cellular and supernatant samples were collected for scintillation counting. The remaining cells were washed and then subjected to lysis, flotation centrifugation, and analysis of fluorescence as described above.
Electron microscopy.
Cells were prepared and processed for electron microscopy as previously described (30). For microscopic examination of lipid raft components, postnuclear supernatants incorporating Gag-GFP protein were extracted with 1% TX-100 and separated by flotation centrifugation, and the fractions demonstrating peak fluorescence activity (40%/10% iodixanol interface) were harvested. Control lipid raft preparations were prepared from cells in the absence of Gag-GFP expression in an identical manner, and the 40%/10% iodixanol interface was identified by refractometry and harvested. These fractions were then pelleted in 1.5-ml microcentrifuge tubes at 100,000 × g for 30 min. Following fixation and sectioning, analysis was performed on a Philips CM-12 electron microscope equipped with a high-resolution charge-coupled device camera. For immunoelectron microscopic analysis, pellets were embedded using LR white or Eponate as described previously (38, 39). In brief, thin sections (100 nm thick) were etched with freshly made 5% H2O2 and incubated with anti-MA rabbit antiserum (36). The sections were allowed to react first with secondary antibodies of sheep anti-mouse immunoglobulin G conjugated with biotin after neutralization in equal volumes of 2% bovine serum albumin and then with streptavidin complexed with gold particles of 10 nm in diameter (Amersham Pharmacia Biotech, Piscataway, N.J.). All the incubation procedures were performed at room temperature in a humidified chamber in a laminar flow hood. The thin sections were then stained with uranyl acetate and lead citrate and examined under a JEM-1230 electron microscope.
RESULTS
Cosedimentation of Gag and raft marker proteins on flotation gradients.
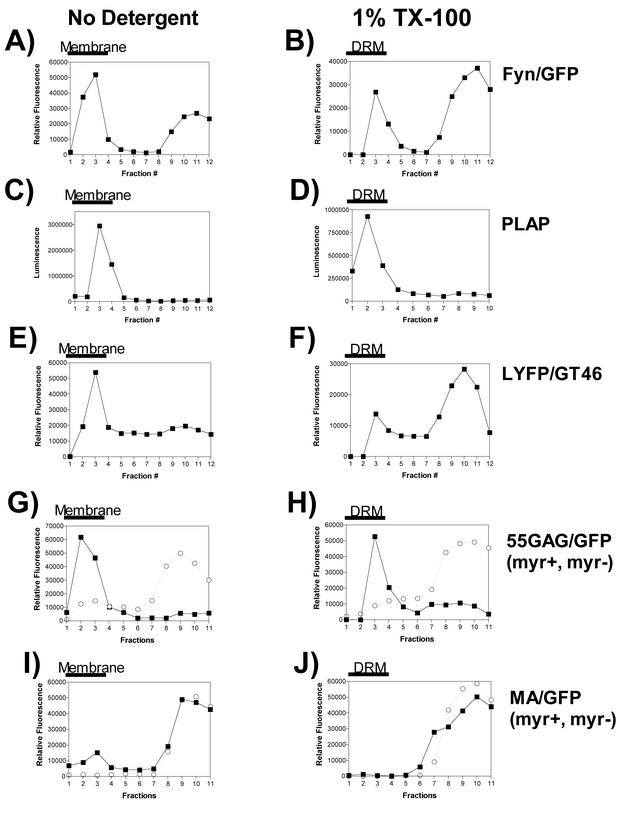
HIV-1 Gag has been reported to associate with lipid rafts. To define the assembly domains within Pr55Gag that are required for Gag-raft interactions, we developed an equilibrium flotation centrifugation assay for Gag and raft markers. Gag and raft marker proteins bearing a fluorescent tag (GFP or YFP) or enzymatic activity (PLAP) were employed in this analysis to facilitate quantitative analysis of the protein content in each fraction. To identify lipid raft-associated proteins, the postnuclear supernatants were divided into two equal portions prior to formation of iodixanol step gradients, and one portion was normalized to 1% TX-100. All reactions were performed on ice, and the centrifugation step was maintained at 4°C to maintain lipid raft structure. Fyn-GFP is a raft-associated marker that has been validated in several previous studies (8). Fyn-GFP was found in both membrane-associated and cytosolic (bottom) fractions of the gradient (Fig. 1A). Following TX-100 extraction, a significant peak of Fyn-GFP remained in the membrane (designated DRM) (Fig. 1B). This result is consistent with the known raft association of Fyn. PLAP, a GPI-linked protein, was examined in an identical manner on iodixanol flotation gradients and quantitated by a luminescence assay for alkaline phosphatase activity. PLAP was found exclusively in the membrane fractions (Fig. 1C) and remained completely associated with DRMs (Fig. 1D). Next, we examined an artificial transmembrane protein (LYFPGT46) that is excluded from lipid rafts. As expected, LYFPGT46 was dissociated from membrane fractions upon TX-100 treatment (Fig. 1E and F). The characteristic raft and nonraft behavior of these marker proteins on iodixanol flotation gradients allowed us to ask if Pr55Gag is associated with the DRM fraction. 55GAG/GFP was found in membranes and DRMs, in a manner identical to that of PLAP (Fig. 1G and H). In contrast, MA/GFP was inefficiently associated with membranes in the absence of TX-100 extraction (Fig. 1I) and was absent in the DRM fraction (Fig. 1J). These results indicate that Gag and DRMs cosediment on 50%/40%/10% iodixanol flotation gradients, a result that is consistent with the reported interaction of Gag with lipid rafts. In addition, determinants within Pr55Gag but outside of MA are required for DRM association.
FIG. 1.
Analysis of raft markers and detergent-resistant Gag complexes on iodixanol flotation gradients. Cells were transfected with the expression construct indicated at the right side of each panel and processed for flotation centrifugation on 50%/40%/10% iodixanol gradients in the absence of detergent (left panels) or following TX-100 extraction (right panels). The thick black bar over the top four fractions of each gradient indicates the location of membrane or DRM following centrifugation. In panels G to J, the requirements for myristylation and the I domain were examined by incorporation of myristyl-acceptor glycine substitutions in four Gag-GFP constructs. In these panels, results with both myristylated (myr+) (filled squares) and nonmyristylated (myr−) (open circles) Gag proteins are shown.
Myristylation is required for membrane and DRM association.
The M domain of Pr55Gag is essential for membrane association and therefore should also be required for the association of Gag with lipid rafts. To directly test whether the M domain and the I domain are both required for raft association, we performed equilibrium flotation analysis with myristylation-defective and -competent versions of Gag constructs with and without the I domain. As stated above, 55GAG/GFP was predominantly membrane associated and remained associated with DRMs following TX-100 extraction (Fig. 1G and H). Myristylation-deficient 55GAG/GFP was predominantly cytosolic in both untreated and detergent-treated samples (Fig. 1G and H). MA/GFP was inefficiently membrane associated and absent from DRMs (Fig. 1I and J). The myristylation-deficient version of MA/GFP was completely absent from cellular membranes and from DRMs (Fig. 1I and J). These data indicate that the M domain is required, but not sufficient, for Gag association with DRMs. The ability of 55GAG/GFP, but not MA/GFP, to associate with DRMs suggests that functional assembly domains outside MA are required for Gag-DRM association.
Mapping the determinants within Pr55Gag required for DRM association.
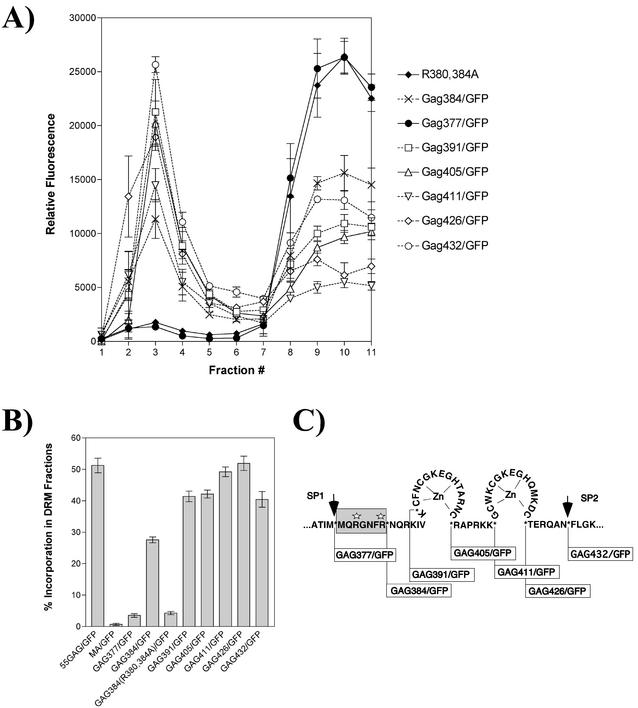
We hypothesized that the I domain of Pr55Gag was required for lipid raft interactions. The I domain is a determinant of retroviral particle density that has been mapped to positively charged residues within the NC region of Pr55Gag (30, 31). To test this hypothesis, equilibrium flotation centrifugation was performed on cell lysates expressing a well-characterized series of Gag-GFP fusion proteins previously used to map the I domain (30). The fusion junctions between NC sequences and GFP for each construct are shown in Fig. 2C. The Gag-GFP fusion construct expressing MA, CA, and SP1 fused to GFP did not associate with DRMs (Gag377/GFP [Fig. 2A]). In contrast, a similar construct that included just the first seven residues of NC was significantly associated with DRMs (Gag384/GFP [Fig. 2A]). The function of the N-terminal I domain has previously been demonstrated to require two arginine residues within the first seven residues of NC (R380 R384). Substitution of these two arginine residues with alanine resulted in a loss of DRM association (R380, 384A [Fig. 2A]). All of the more C-terminal fusions were significantly associated with DRMs, consistent with the presence of intact I domains (Fig. 2A). These results indicate that the I domain plays an essential role in cosedimentation of Gag with DRMs. The results are consistent with the report of Ono and Freed (24) that sequences in NC contribute to raft association and extend these findings to directly implicate the I domain. To compare quantitatively the percentages of DRM-associated Gag protein, flotation analysis was performed for each construct three times, and the mean percentage of Gag protein was determined (calculated as GFP fluorescence in gradient fractions 1 to 4 divided by total fluorescence in all gradient fractions). As shown in Fig. 2B, the N-terminal I domain (represented by GAG384/GFP) conferred significant DRM association, while addition of sequences within the NC region of Gag enhanced the efficiency of DRM association. Although achieved in this case via flotation analysis, these data are remarkably similar to data previously reported for pelleting of Gag DRCs (30). Together, these data establish that the I domain is required for the association of Gag with DRMs. Because the I domain has been proposed to play a key role in Gag-Gag multimerization (30, 31), these results suggest that multimerization of Gag is required for DRM association.
FIG. 2.
Analysis of the role of the I domain in Gag protein flotation with DRMs. A panel of Gag-GFP fusion constructs that contain nested C-terminal fusion sites spanning the NC region of Gag was examined for association with DRM fractions. Postnuclear extracts treated with TX-100 were examined by flotation centrifugation using 50%/40%/10% iodixanol step gradients. Quantitation of Gag-GFP protein in each fraction was performed by microplate fluorometry. (A) Fluorescence values for Gag-GFP fusion constructs. The top of the gradient is at the left. (B) Gag protein content in fractions 1 to 4 was designated the DRM fraction, and the percentage of Gag in DRMs was calculated as the fluorescence in fractions 1 to 4 divided by total fluorescence in all fractions. Experiments were performed three times, and results are presented as the means ± standard deviations. (C) Map of I-domain constructs utilized in these experiments. The N-terminal (minimal) I domain (shaded box) and the two arginines that were substituted with alanine in GAG384(R380,384A) (white stars) are shown. Protease cleavage sites (arrows) and fusion breakpoints to which GFP was fused (asterisks) are indicated.
Independent segregation of Gag protein and lipid raft marker proteins.
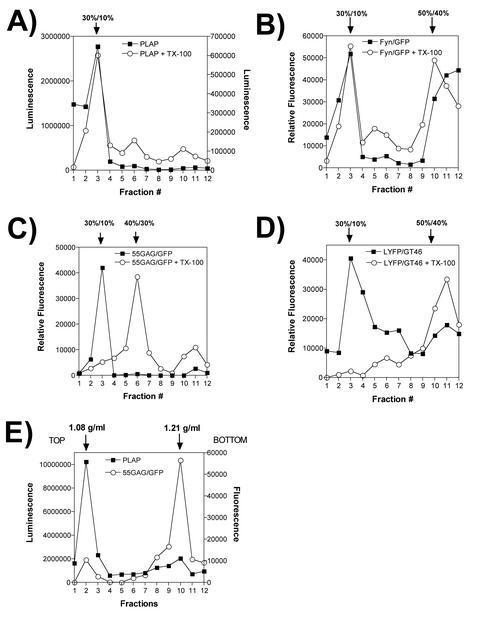
The experiments described above were performed with the hypothesis that the detergent-resistant Gag proteins identified by equilibrium flotation centrifugation represent Gag interacting with lipid raft microdomains of the plasma membrane. If this is true, then our experiments establish that the I domain and the M domain contribute to interactions with lipid rafts. However, we considered the possibility that Gag may form DRCs that do not represent Gag-lipid raft interactions. To test this hypothesis, we performed equilibrium centrifugation of Gag and raft markers under altered gradient conditions. Raft components are known to float to a 30%/10% iodixanol interface, as established in studies with influenza virus HA and a number of classical raft markers (32). We therefore added a 30% iodixanol step to our analysis (making a 50%/40%/30%/10% gradient). PLAP behaved in a manner consistent with a lipid raft-associated protein, remaining in the 30%/10% iodixanol interface following TX-100 extraction (Fig. 3A). Fyn/GFP remained localized to this interface in a similar manner (Fig. 3B). Surprisingly, Gag-GFP shifted almost completely to the 40%/30% iodixanol interface following TX-100 extraction (Fig. 3C). This experiment was repeated on three occasions with identical results. As a further control for the integrity of this equilibrium flotation procedure, we employed the nonraft artificial plasma membrane protein LYFPGT46. As expected for a nonraft component, detergent extraction solubilized the protein and prevented it from floating out of the 50% gradient fraction (Fig. 3D). These results suggested that the density of detergent-resistant Gag complexes was inconsistent with membrane density but that Gag complexes did have a buoyant density lighter than that of 40% iodixanol.
FIG. 3.
Independent segregation of Gag DRCs and lipid raft markers. Proteins were expressed and cells were processed in an identical manner to the proteins and cells shown in Fig. 1 to 3, and postnuclear supernatants were subjected to equilibrium flotation on 50%/40%/30%/10% iodixanol gradients. Flotation results obtained from samples extracted with TX-100 (open circles) and results obtained without detergent treatment (closed squares) are shown. All processing was performed on ice or at 4°C. (A) PLAP activity was assessed using a chemiluminescence assay for alkaline phosphatase activity. (B) Fyn-GFP quantitation by fluorometry. (C) 55GAG/GFP fluorescence profile before and after TX-100 extraction. (D) LYFPGT46, a nonraft transmembrane marker protein, was quantitated by fluorometry in the absence of detergent or following TX-100 treatment. (E) Separation of raft markers and Gag-GFP was performed on 20 to 75% sucrose gradients. The density of peak fractions was measured by refractometry and is indicated above the graph.
To precisely define the buoyant density of Gag-GFP complexes, we performed equilibrium flotation centrifugation on a 50%/40%/10% iodixanol gradient as before, collected the fluorescence peak at the 40%/10% iodixanol interface, and loaded this sample on the top of a 20 to 75% linear sucrose gradient for equilibrium density centrifugation. In the same sample, we measured PLAP as a raft marker. PLAP remained near the top of the sucrose density gradient following centrifugation, at a density of 1.08 g/ml (Fig. 3E). This result is consistent with the known density of the plasma membrane and of lipid rafts (6). However, the Gag complexes were found to be of a much higher density (1.21 g/ml [Fig. 3E]). This density is significantly higher than those of membrane components and is close to that of HIV cores.
Insensitivity of Gag complexes to cholesterol extraction.
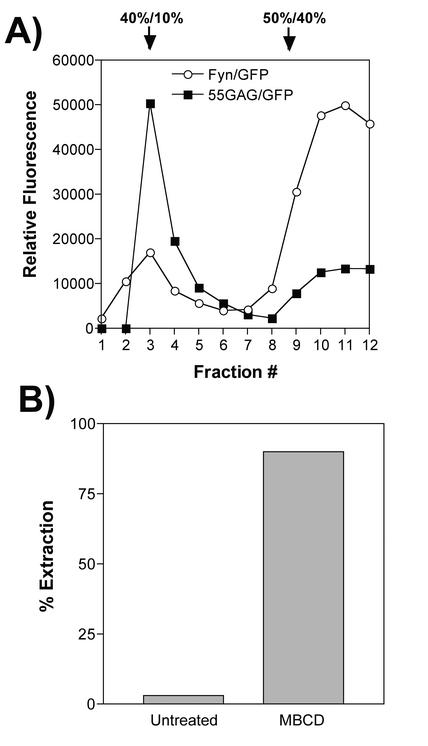
Lipid rafts are disrupted upon extraction of cholesterol by MβCD (6). To further test the hypothesis that Gag associates with lipid rafts, we labeled cells expressing Gag-GFP or Fyn-GFP with [3H]cholesterol. Cholesterol was then extracted from the plasma membranes of the cells with 10 mM MβCD prior to lysis and flotation centrifugation analysis. The amounts of radiolabeled cholesterol in the cells and supernatants were quantitated, and the efficiency of extraction was calculated. Figure 4B demonstrates that the cholesterol extraction process was efficient. Following extraction, cells were lysed, a postnuclear supernatant was prepared, and this supernatant was normalized to 1% TX-100. Upon equilibrium flotation analysis (50%/40%/10%iodixanol gradients), the flotation characteristics of Gag complexes were unchanged (Fig. 4A). In contrast, Fyn-GFP was largely dissociated from the top fractions of the gradient (Fig. 4A). These data indicate that disruption of lipid rafts by cholesterol extraction does not alter the buoyant density of detergent-resistant Gag complexes.
FIG. 4.
Extraction of cholesterol and Gag-raft flotation. (A) 55GAG/GFP and Fyn-GFP were examined via equilibrium flotation centrifugation following extraction of cholesterol from cellular membranes with 10 mM MβCD. These experiments were performed simultaneously but in separate centrifuge tubes to facilitate detection of each GFP fusion protein. (B) Efficiency of cholesterol extraction was monitored by measuring [3H]cholesterol in supernatants versus cells of treated and untreated cells. The percent extraction was calculated as total counts per minute in supernatant following MβCD treatment divided by the total counts in cells plus supernatant.
Detergent-resistant Gag complexes form rapidly following Gag expression.
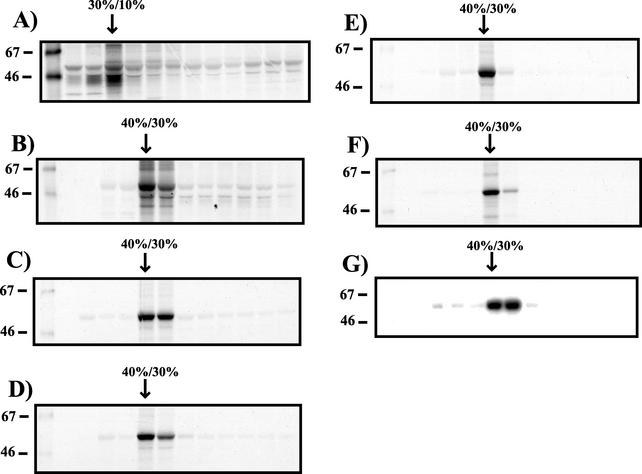
Gag-GFP fusion constructs were used in all of the flotation experiments described above. To determine if these results were applicable to Gag proteins in the absence of the GFP marker, we expressed Gag from a proviral construct containing an inactivating mutation in protease (pNLPR-). Pr55Gag floated to the 30%/10% iodixanol interface when no detergent was included in the experiment (Fig. 5A). Following TX-100 extraction, Pr55Gag shifted to the 40%/30% iodixanol interface (Fig. 5B), in agreement with our findings using Gag-GFP fusion proteins (Fig. 3C). We next considered the possibility that the dense, detergent-resistant Gag fraction could represent an aberrant pathway for Gag that is present only after Gag is overexpressed in cells. If so, we would expect to see a lighter, detergent-resistant Gag fraction representing Gag-lipid raft association at earlier time points following expression. Cells expressing Gag were pulsed for 2 h with [35S]cysteine-methionine and examined by equilibrium flotation on 50%/40%/30%/10% iodixanol gradients following a 1-, 2-, 4-, and 6-h chase. Following the 1-h chase, the major Gag protein fraction floated to the 40%/30% iodixanol interface (Fig. 5C). All subsequent time points revealed similar results, with some diminution in the total detergent-resistant Gag protein present in the cell following the 6-h chase (Fig. 5D to F). These results indicate that the dense Gag complexes are formed quickly within the cell and represent the major detergent-resistant Gag complex present either at steady state (Fig. 5B) or during early stages of particle formation (Fig. 5C to F). These results do not rule out the possibility that at even earlier time points, another population of detergent-stable Gag that may resemble lipid raft-associated complexes exists.
FIG. 5.
Equilibrium flotation analysis of Gag expression from proviral DNA, and pulse-chase analysis of Gag DRM association. (A) 293T cells were transfected with pNLPR- and labeled 48 h later with [35S]cysteine-methionine. Cells were harvested and processed for equilibrium flotation analysis on 50%/40%/10% iodixanol gradients in the absence of detergent. Each fraction was immunoprecipitated with HIV patient sera. The position of the top (30%/10%) iodixanol interface is indicated. (B) Cells transfected with pNLPR- were labeled and harvested for gradient analysis as for panel A but were treated with 1% TX-100 on ice prior to centrifugation. (C) 293T cells transfected with pCDNA/GagOpt were labeled for 2 h with [35S]cysteine-methionine, washed, and harvested following a 1-h chase period in the absence of radioactive amino acids. Gradient separation and analysis were performed as described for panel B. (D) Analysis of detergent-resistant Gag complexes from pCDNA/GagOpt following a 2-h cold chase. (E) Analysis of detergent-resistant Gag complexes following a 4-h cold chase. (F) Analysis of detergent-resistant Gag complexes following a 6-h cold chase. (G) Immature viral cores prepared from supernatants of cells expressing pNLPR- by TX-100 extraction were analyzed on 50%/40%/30%/10% iodixanol gradients. The positions of molecular size markers (in kilodaltons) are shown to the left of the blots.
A possible explanation for the buoyant, detergent-resistant Gag seen in these experiments is the formation of core-like Gag complexes in cells. We next compared the behavior of immature viral cores to that of the cellular complexes. Immature particles were harvested from the supernatant of cells expressing NLPR-, filtered, and purified by centrifugation through a 20% sucrose cushion. Following resuspension, immature cores were prepared by treating the particles with 1% TX-100. Upon examination by equilibrium flotation analysis, immature viral cores behaved in a manner identical to that of cellular detergent-resistant Gag complexes (Fig. 5G). These results suggest that a core-like multimer of Gag accounts for the buoyant density of cellular detergent-resistant Gag complexes.
Electron microscopic analysis of detergent-resistant Gag complexes.
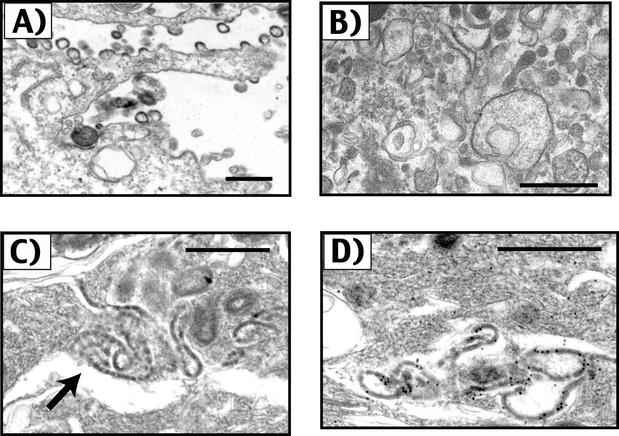
Gag protein cores are distinguishable upon electron microscopy by the electron-dense Gag shell and by their spherical morphology. Electron microscopic analysis was performed to determine if cores or core-like structures could be identified in DRM fractions prepared by flotation centrifugation. To facilitate the collection of Gag complexes in this analysis, we utilized Gag-GFP fusion proteins. After identification of the peak GFP-containing fractions from an equilibrium flotation gradient by fluorometry, the fractions were pelleted at 100,000 × g and prepared for transmission electron microscopic analysis. Examination of intact cells expressing 55GAG/GFP prior to harvesting revealed numerous budding 55GAG/GFP particles as previously described (Fig. 6A). The detergent-resistant, buoyant pellets from control detergent-resistant membrane (raft) preparations contained largely amorphous material and vesicles (Fig. 6B). A careful search for core-like structures from control preparations was negative. The appearance of DRMs from cells in which Gag was expressed were largely similar. However, scattered within the amorphous material in DRMs from Gag-expressing cells were collections of electron-dense material that formed sheets that in cross-section resembled the layer of Gag in an immature viral core (Fig. 6C). Although these sheet-like structures were easily found in scanning multiple sections, no intact viral cores were identified. To determine if these collections were indeed formed by Gag protein, immunogold labeling was employed using an anti-MA antisera as the primary antibody. This analysis indicated that the electron-dense, sheet-like structures represented Gag protein within the detergent-resistant, buoyant gradient fractions (Fig. 6D). Immunogold staining of multiple control preparations from cells not expressing Gag were negative (data not shown). We conclude that the major detergent-resistant species of Gag identified by gradient analysis is neither an intact core nor a Gag-lipid raft complex but rather represents Gag multimers that have formed large sheets following extraction of the plasma membrane with TX-100.
FIG. 6.
Electron microscopic analysis of detergent-resistant Gag complexes. (A) Gag-GFP virus-like particles are shown budding from the plasma membrane of BSC-40 cells. Magnification, ×22,000. (B) Amorphous material and vesicles were found in control DRM pellets lacking Gag expression. Magnification, ×20,000. (C) Sheet-like collections of Gag-GFP protein within the DRM pellet. Magnification, ×42,916. (D) Gag-GFP collections identified by immunogold staining using an anti-MA antiserum as the primary antibody. Magnification, ×50,000. Bars = 500 nm.
DISCUSSION
The role of plasma membrane microdomains in the virus assembly processes is an area of active investigation. It is well-established that some viruses, such as influenza virus and measles virus, interact with rafts during the budding process (2, 17, 21, 27, 33). Rafts may provide a platform for assembly, allowing selective enrichment of viral proteins in discrete locations on the plasma membrane and thus providing a focal nidus for the assembly process. Much of the evidence for the association of viral proteins with rafts is derived from biochemical separations in which viral structural proteins float with DRMs to a low-density position following equilibrium centrifugation. In the case of influenza virus, both HA (33) and neuraminidase (NA) (18) have been found to associate with DRMs. The behavior of HA on biochemical separations of cellular membranes resembles that of classical raft-associated proteins such as the GPI-anchored protein PLAP (33). For example, HA associates with a low-density membrane fraction following extraction with TX-100 in the cold, and the interaction of HA and DRMs is lost when cholesterol is extracted from the membrane. Localization of HA and NA allows recruitment of M1 protein to the lipid raft assembly site through interactions with the HA (and perhaps NA) cytoplasmic tail (2). M1 is essential for virus assembly and budding (12), indicating that influenza virus glycoproteins recruit internal structural proteins to rafts to drive the particle assembly process. More recently, this paradigm of viruses using lipid rafts as platforms for assembly has been extended to include Semliki Forest virus (1), Ebola and Marburg viruses (4), and HIV-1 (20, 22, 24, 29).
Rafts have been shown to play a role in HIV budding through several lines of evidence. Nguyen and Hildreth found that HIV particles selectively incorporate raft-associated proteins (22). These investigators also showed that some HIV proteins, specifically MA and transmembrane protein (TM), were associated with DRMs in infected Jurkat cells. A role for Gag in selectively interacting with lipid rafts was suggested by these studies. The interaction of Gag with lipid rafts was more intensively studied by Lindwasser and Resh (20). In this study, a significant proportion of Gag protein was found to separate from cytosolic Gag in a flotation gradient following TX-100 extraction in the cold. Importantly, multimerization was implicated in the formation of an atypical raft-like structure termed a “barge.” Both myristylation and the more C-terminal regions of Gag were required for the formation of barges in their study. It was postulated from these data that Gag targets lipid rafts and subsequently alters the density of the rafts through multimerization of Gag, forming a barge. Similar results implicating the involvement of the I domain of HIV and the M domain in raft association were reported by Ono and Freed (24). Colocalization of Gag and raft marker proteins on sucrose flotation gradients was demonstrated in their study, suggesting that a direct interaction of Gag with lipid rafts could be established by gradient flotation analysis.
This study was initiated to further define the domains within Gag that mediate Gag-lipid raft interactions. Specifically, we wished to define the role of the I domain in Gag-lipid raft interactions through the use of Gag-GFP fusion constructs that had previously been characterized for I-domain function by particle density and membrane localization experiments (30). We modeled our separation techniques on those from the influenza virus literature (32) and chose several well-characterized raft markers as controls. We first confirmed the results described by others that under some gradient conditions, Gag will float together with lipid raft markers. In our gradients, both raft markers and Gag DRCs float to an interface between 40% and 10% iodixanol. This finding is analogous to the 65%/10% sucrose interface utilized by Ono and Freed (24) to show coflotation of Gag with raft markers. Using this criteria, we were able to precisely map the C-terminal domain required for raft association to the I domain. Most convincing was the fact that a mutant construct previously shown to be deficient in I-domain function through substitution of two arginine residues with alanine [GAG384(R380,384A)] was absent from DRMs, while the nonsubstituted construct (GAG384/GFP) was found together with DRMs. Similar to previous reports, the M domain was required but not sufficient for the association of Gag with DRMs. Although our mapping studies were unambiguous, we felt that the gradient conditions may have introduced some bias into the conclusions, since a wide range of densities can be encompassed between the 40% and 10% iodixanol gradient steps. We suggest that both the 50%/40%/10% iodixanol flotation method reported here and the 65%/10% sucrose method used by others (24) group components of widely varying densities into a single interface, such that conclusions regarding the significance of the cosedimentation are questionable. We addressed this by altering the gradient conditions to separate membranes from denser cellular components on a 50%/40%/30%/10% iodixanol gradient as described below.
It has been clearly demonstrated that raft membranes are low-density complexes. Brown and Rose measured the density of membranes containing PLAP to be 1.08 g/ml (6). When we altered the gradient conditions to include an additional density layer, Gag DRCs separated dramatically from light membrane fractions. The measured density of Gag complexes in our study (1.21 g/ml) is quite distinct from that of membranes and quite different from those of classical raft markers. We were gratified to find that raft markers such as PLAP were found in our experiments at 1.08 g/ml, precisely the density previously demonstrated by Brown and Rose (6). Although the identification of Gag complexes that are denser than classical raft results is similar to the findings of Lindwasser and Resh (20), we did not find evidence suggesting that these denser complexes represent a unique form of lipid raft. Specifically, lipid raft markers were absent from the dense complexes of Gag, even when the lipid raft markers (Fyn/GFP and PLAP) were overexpressed. Additional evidence comes from the fact that cholesterol extraction failed to alter the density of the Gag complexes. In the absence of these classical criteria (low density, disruption with cholesterol extraction, and cosedimentation with raft markers), we conclude that Gag DRCs do not represent complexes of Gag with lipid rafts.
The electron microscopy data shown in Fig. 6 indicate that Gag proteins form large, sheet-like structures following the extraction of cellular membranes with nonionic detergents. Because Gag proteins were present in discrete locations prior to extraction (Fig. 6A), it is likely that the large sheets of Gag are somewhat artificial by-products of detergent treatment. We suggest that the following is occurring: electron-lucent Gag multimers first form on cellular membranes in a process requiring both the M domain and the I domain. As the multimers enlarge, they become electron-dense at specific sites of particle budding. Although a membrane is required for formation of large Gag complexes, we suggest that it is not the raft components of the membrane that determine the ultimate density of the complex but rather the array of Gag proteins itself. In this way, Gag DRCs resemble in density a completed immature viral core. Raft proteins are predicted to be neither specifically included nor excluded from the developing Gag core by this process. However, upon extraction of most of the lipid components of the membrane, Gag multimers are forced together and become complexed in a large sheet of protein (as illustrated in Fig. 6C and D). This would explain the relatively high density of the Gag DRCs and provide an explanation for the lack of effect of cholesterol extraction. The I domain is required for Gag-Gag multimerization, so no DRCs would form in the absence of the I domain. We note that some lipids may remain complexed with the dense Gag multimers, although cholesterol itself appears not to be necessary on the basis of the results presented in Fig. 4.
Our results do not exclude a potential role for lipid rafts in the HIV assembly process. Other viral proteins such as gp41 may target Gag proteins more specifically to lipid rafts, as suggested by a recent report (29). It is also possible that Gag briefly interacts with lipid rafts before forming the denser complexes detected upon TX-100 extraction that segregate independently from lipid raft markers. Rafts have been reported to be quite heterogeneous in nature (28), so our choice of raft markers in this study may not reflect the characteristics of all known subtypes of rafts. However, our results indicate that the biochemical evidence for a direct Gag-raft interaction is weak. We and others have previously reported that Gag targets distinct, punctate locations on the plasma membrane (14, 25, 30). These punctate sites of Gag represent the predominant form of Gag at the plasma membrane, and their presence correlates with the detergent-resistant Gag complexes that are found in cells following biochemical analysis of cellular lysates (30). It seems likely that factors other than lipid rafts account for this punctate membrane localization. While these collections of Gag are easily seen by confocal microscopy, rafts are not distinguishable by microscopic analysis in discrete locations unless raft components are first cross-linked by antibodies (13). If Gag complexes act as raft “patching” agents acting from the cytoplasmic leaflet of the membrane, then we would expect raft components such as PLAP and Fyn to be concentrated in Gag DRCs. This was not observed in this study. Consistent with this, our preliminary results suggest that raft marker proteins bearing fluorescent tags do not concentrate in areas of Gag puncta but remain diffusely distributed in the plasma membrane (data not shown).
Further research is needed to define the cellular factors that determine the specificity of plasma membrane localization of HIV-1 Pr55Gag. An intriguing area for future research is the interaction of Gag with components of the vacuolar protein sorting pathway, as recently illustrated by the characterization of TSG101 as a binding partner for the PTAP domain within p6 (10). Perhaps in the future, specific components of the endosomal machinery or the peripheral actin cytoskeleton will be identified that are active in directing Gag to specific locations on the plasma membrane.
Acknowledgments
This work was funded in part by grant R01 AI40338. A.D. was supported by NIH training grant T32 CA09385.
We thank Chris Aiken and Donald Wyma for providing the pPLAP expression plasmid and pNLPR- and for helpful advice as this project progressed. We thank Kai Simons for LYFPGT46 and Mark Philips for Fyn-GFP, Anne Kenworthy for help in the use of these marker plasmids, and Gary Nabel for pVRC3900. Some of the electron microscopic analysis was performed with the assistance of the Research Electron Microscopy Core Laboratory of the Vanderbilt-Ingram Cancer Center Cell Imaging Shared Resource.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antman, K. H., and D. M. Livingston. 1980. Intracellular neutralization of SV40 tumor antigens following microinjection of specific antibody. Cell 19:627-635. [DOI] [PubMed] [Google Scholar]
- 4.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B. K., M. B. Feinberg, and D. Baltimore. 1997. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J. Virol. 71:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 9.Dhanvantari, S., and Y. P. Loh. 2000. Lipid raft association of carboxypeptidase E is necessary for its function as a regulated secretory pathway sorting receptor. J. Biol. Chem. 275:29887-29893. [DOI] [PubMed] [Google Scholar]
- 10.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 11.Gelderblom, H. R. 1991. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS 5:617-637. [PubMed] [Google Scholar]
- 12.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, Y., W. P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikonen, E. 2001. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13:470-477. [DOI] [PubMed] [Google Scholar]
- 17.Keller, P., and K. Simons. 1998. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 140:1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundu, A., R. T. Avalos, C. M. Sanderson, and D. P. Nayak. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 70:6508-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafont, F., and K. Simons. 2001. Raft-partitioning of the ubiquitin ligases Cbl and Nedd4 upon IgE-triggered cell signaling. Proc. Natl. Acad. Sci. USA 98:3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen, D. H., and D. Taub. 2002. CXCR4 function requires membrane cholesterol: implications for HIV infection. J. Immunol. 168:4121-4126. [DOI] [PubMed] [Google Scholar]
- 24.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pralle, A., P. Keller, E. L. Florin, K. Simons, and J. K. Horber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puertollano, R., J. A. Martinez-Menarguez, A. Batista, J. Ballesta, and M. A. Alonso. 2001. An intact dilysine-like motif in the carboxyl terminus of MAL is required for normal apical transport of the influenza virus hemagglutinin cargo protein in epithelial Madin-Darby canine kidney cells. Mol. Biol. Cell 12:1869-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roper, K., D. Corbeil, and W. B. Huttner. 2000. Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2:582-592. [DOI] [PubMed] [Google Scholar]
- 29.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 33.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 35.Thomson, J. M., and W. A. Parrott. 1998. pMECA: a cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. BioTechniques 24:922-924, 926, 928. [DOI] [PubMed] [Google Scholar]
- 36.Varthakavi, V., P. J. Browning, and P. Spearman. 1999. Human immunodeficiency virus replication in a primary effusion lymphoma cell line stimulates lytic-phase replication of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:10329-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verkade, P., and K. Simons. 1997. Lipid microdomains and membrane trafficking in mammalian cells. Histochem. Cell Biol. 108:211-220. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J. J., Y. Lu, and L. Ratner. 1994. Particle assembly and Vpr expression in human immunodeficiency virus type 1-infected cells demonstrated by immunoelectron microscopy. J. Gen. Virol. 75:2607-2614. [DOI] [PubMed] [Google Scholar]
- 39.Wang, J. J., S. Sandefur, P. Spearman, C. T. Chiou, P. H. Chiang, and L. Ratner. 2001. Tracking the assembly pathway of human immunodeficiency virus type 1 Gag deletion mutants by immunogold labeling. Appl. Immunohistochem. Mol. Morphol. 9:371-379. [DOI] [PubMed] [Google Scholar]
- 40.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55Gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]