Abstract
Saposin B is a small, nonenzymatic glycosphingolipid activator protein required for the breakdown of cerebroside sulfates (sulfatides) within the lysosome. The protein can extract target lipids from membranes, forming soluble protein-lipid complexes that are recognized by arylsulfatase A. The crystal structure of human saposin B reveals an unusual shell-like dimer consisting of a monolayer of α-helices enclosing a large hydrophobic cavity. Although the secondary structure of saposin B is similar to that of the known monomeric members of the saposin-like superfamily, the helices are repacked into a different tertiary arrangement to form the homodimer. A comparison of the two forms of the saposin B dimer suggests that extraction of target lipids from membranes involves a conformational change that facilitates access to the inner cavity.
The saposins are a group of four structurally related activator proteins that function in conjunction with hydrolase enzymes for the stepwise breakdown of glycosphingolipids within the lysosome (1, 2). These proteins act by modifying the local environment of target lipids, and “activate” the breakdown of their substrates by presenting them in a form in which the lipid scissile bonds are accessible to the active sites of the specific enzymes. Presumably, the hydrolases cannot form a catalytic complex with the glycosphingolipids when these are in unmodified membrane bilayers.
Each of the four saposins activates one or more lipid exohydrolases. For example, saposin B (also known as the cerebroside sulfate activator, or CS-Act) facilitates the hydrolysis of the sulfate group from cerebroside sulfate by arylsulfatase A, resulting in the formation of galactosylceramide (3). This glycolipid is then catabolized to ceramide by β-galactosylceramidase in a reaction activated by saposin C. There seems to be more than one mechanism of activation by the saposins, and this may further depend on the particular target lipid. Thus, saposin B is able to extract and solubilize cerebroside sulfates from membranes, allowing arylsulfatase A to act on the small, diffusible protein-lipid complexes. Saposin B may also have a physiological role in activating the hydrolysis of the ganglioside GM1 to GM2 by lysosomal β-galactosidase, and in this case, the activator may act by modifying the local lipid structure at the membrane surface to allow catalysis to proceed (4).
The saposins belong to a large and diverse family of small, cysteine-rich proteins that share a common ability to interact with membranes but act in a wide variety of functions. Other members of the “saposin-like” superfamily of proteins include the lung surfactant-associated protein B (SP-B), the tumorolytic protein NK-lysin, granulysin, the pore-forming amoebapores, and the membrane-targeting domain of some enzymes (5, 6). The saposin-like proteins are ≈80 residues in length and have a characteristic pattern of conserved cysteines (Fig. 1A; refs. 7 and 8). To date, the three-dimensional modeling of the saposins and other members of the saposin-like family (9–11) has relied on the NMR structure of the monomeric NK-lysin, which is a relatively compact monomer made up from five α-helices (12). This fold has also been seen in the vacuolar-targeting domain of prophytepsin (13) and the antimicrobial peptide bacteriocin AS-48 (14); however, these three structures provide little insight as to how the small saposin B dimer can solubilize relatively large lipid ligands. Here, in the 2.2-Å crystal structure of saposin B, we find conserved secondary structure with the previously determined monomeric structures but striking differences at the tertiary and quaternary level. The unexpected dimer structure has important implications for the understanding of the mechanism of lipid binding in this family of proteins and illustrates a hitherto unknown mechanism of adaptation of function within a fold superfamily.
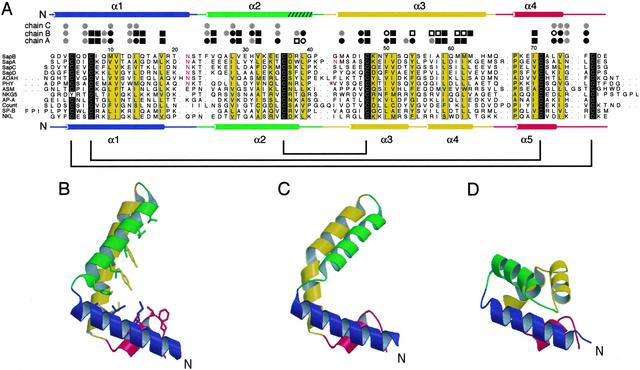
Figure 1.
(A) Multiple alignment of representative saposin-like proteins from eight different functional families (6). Sequence numbering is according to saposin B. The sequence alignment was generated with clustalw and includes manual adjustments. Identical amino acids are shaded black; conserved positions are shaded yellow. Experimentally determined N-glycosylation sites are indicated with red lettering. Human saposin B (SapB) is aligned with human saposins A, C, and D (SapA, SapC, and SapD), acyloxyacyl hydrolase (AOAH), plant phytepsin (PHY), acid sphingomyelinase (ASM), granulysin (NKG5), pore-forming amoebapore A (AP-A), Countinin (Count), surfactant-associated protein B (SP-B), and porcine NK-lysin (NKL). Leading and trailing dots indicate cases where the saposin motif is a sub-sequence of a larger protein. Note that in the phytepsin “swaposin” domain, the sequence block VVSQ… TFDG precedes the block ADPM… NRLP, as marked by the red asterisk (32). The secondary structures of saposin B chain A (Top) and NK-lysin (Bottom) are shown as cylinders representing helices, and the cysteines that are linked by disulfide bonds are connected with brackets. A turn of 310 helix at the end of helix α2 in saposin B is indicated by hatching. The saposin B residues are assigned to the following environment classes: residues contributing >20 Å2 to the dimer interface (circles); residues facing the inner cavity but not part of the dimer interface (squares); residues with side chains lining the inner hydrophobic cavity (black symbols); residues with side chain lining the opening to the cavity (white symbols). The inner cavity is formed only in the AB dimer. (B–D) Ribbon diagrams of chains A and B of saposin B and of the NK-lysin monomer, respectively. The side chains from chain A that are exposed to the inner cavity are indicated in stick representation.
Methods
Expression and Crystallization.
The expression, purification, and crystallization of the selenomethionine-substituted protein has been described (15). Briefly, human saposin B was expressed in Escherichia coli strain AD494(DE3) and purified with heat treatment, followed by anion exchange, size-exclusion, and hydrophobic interaction chromatographies. Crystals belonging to space group P3121 with unit cell dimensions of a = 72.14 Å and c = 94.37 Å were grown by vapor diffusion by mixing equal amounts of protein solution and reservoir buffer (16% polyethylene glycol 3350/0.2 M magnesium acetate/0.1 M sodium cacodylate, pH 5.8) and equilibrating against 1.0 ml of reservoir solution. The I46C and M10C mutants were generated with the QuikChange site-directed mutagenesis kit (Stratagene), and protein purification was carried out as with the native protein. MS was carried out as described (15).
Crystallographic Data Collection and Structure Determination.
All diffraction data were collected at 100 K. Because of evidence of partial selenium oxidation during expression and crystallization, crystals were soaked for 30 sec in a cryoprotectant solution of mother liquor containing 15% glycerol and 0.1% H2O2 in an attempt to convert all of the selenium atoms to a uniform oxidization state (16). Multiwavelength anomalous diffraction (MAD) data were collected at two wavelengths (λ1 = 0.9789 Å, λ2 = 0.9793 Å) at beamline F2 at Cornell High Energy Synchrotron Source (CHESS), Cornell University, but a third dataset at an energy remote from the Se absorption edge could not be collected because of the formation of ice on the crystal surface. Data processing and reduction were done with MOSFLM and SCALA (17). A partial selenium substructure was determined with SnB (18) and refined with SHARP (www.globalphasing.com), producing a phase set with a figure of merit of 0.595 on acentric reflections. Density modification with DM (17) resulted in an interpretable map, and an atomic model was traced with the program O (19). Refinement and addition of water molecules and ligands were done with the program package CNS (20). The final model has 96.3% of the residues in the most favored regions of the Ramachandran plot. The crystallographic statistics are presented in Table 1. Molecular graphics were prepared withMOLSCRIPT (21) and PYMOL (www.pymol.org).
Table 1.
Data collection and refinement statistics
| Data collection | ||
| Wavelength, Å | 0.9789 | 0.9793 |
| Resolution, Å | 37–2.2 | 37–2.2 |
| Unique reflections | 14,653 | 14,644 |
| Redundancy | 17.1 | 10.5 |
| Completeness, % (all data) | 98.3 (98.3)* | 98.3 (98.3) |
| Completeness, % (data with I/σ(I) > 3) | 84.2 (49.6) | 80.3 (41.0) |
| 〈I/σ(I)〉 | 8.5 (2.5) | 9.0 (2.6) |
| Rmerge, % | 6.5 (29.3) | 5.9 (28.9) |
| Refinement | ||
| Resolution, Å | 20–2.2 | |
| Number of reflections (F/σ(F) > 0) | 14,613 | |
| Rcryst, % | 22.2 (23.5) | |
| Rfree,5%, % | 26.2 (29.1) | |
| rmsd bonds, Å | 0.0077 | |
| rmsd angles, ° | 1.20 | |
| Number of non-hydrogen atoms/〈B〉, Å2 | ||
| Protein | 1822/44.5 | |
| Water | 90/46.7 | |
| PE ligand | 46/87.6 |
All values in parentheses are for the high-resolution shell (2.20–2.34 Å).
Activity Assay.
Measurements of the activation of cerebroside sulfate hydrolysis by saposin B were carried out as described (22) with two exceptions: (i) [14C]stearic-1 cerebroside sulfate (American Radiolabeled Chemicals, St. Louis) was used in place of the rat brain [35S]cerebroside sulfate; and (ii) the final determination of activity was based on the amount of [14C]galactosylceramide in the organic phase rather than the inorganic sulfate in the aqueous phase. Briefly, 10 μg of [14C]cerebroside sulfate (1,000 cpm/μg) were added to a solution containing 2.5 units of arylsulfatase A and varying amounts of saposin B, and the reaction mixtures were incubated at 37°C for 2 h. The lipids were extracted, separated by thin layer chromatography, and quantitated by liquid scintillation counting of the scraped bands.
Results and Discussion
Conformational Differences Between Monomers.
The crystallographic asymmetric unit contains three independent peptide chains (referred to as chains A, B, and C) that form two distinct homodimers. The first dimer is formed by chains A and B (dimer AB) and has a pronounced asymmetry. The second dimer (dimer CC′) is formed by chain C and a mate related by a crystallographic twofold axis and is thus symmetric. Each chain consists of four amphipathic α-helices arranged in a long hairpin that is bent into a simple V-shape (Fig. 1 B and C). The convex outer surface of the monomer is hydrophilic, whereas the concave inner surface of the “V” is lined with the side chains from hydrophobic residues. As with other members of the saposin-like family, the fold is stabilized by two disulfide bridges connecting α1 and α4 near the N and C termini of the chains and by one bridge between α2 and α3 near the hairpin turn. As a result, there are tight pairwise α-helical interactions over the length of the monomer.
Chains A and C are structurally similar and superimpose with an rms deviation (rmsd) of 0.90 Å over the Cα atoms of residues 2–78. However, chain B is bent into a significantly sharper “V,” and can be reasonably superimposed with chains A and C at either the base (residues 2–20 and 56–78, rmsd = 1.0 Å) or hairpin (residues 26–54 rmsd = 1.2 Å) regions. The key structural differences in the B chain relative to the A and C chains are (i) a reorganization of the first loop and partial unfolding at the start of helix α2, and (ii) a highly localized kink in α3 at residue Tyr-54 (φ, ψ = −100.4°, +10.2°). In chains A and C, helix α3 is only slightly curved and does not have the sharp kink that is obvious in the B chain.
Comparison with NK-Lysin.
NK-lysin (12) and the other saposin-like proteins (13, 14) show a similar overall fold consisting of five amphipathic α-helices wound into a closed monomer with a small hydrophobic core. A comparison of the saposin B chains with the NK-lysin structure reveals similarities in the secondary structure and in the lateral interactions between the disulfide-bonded helices, but the more open saposin B monomer conformation leads to a significantly different tertiary organization (Fig. 1). Remarkably, the differences are largely caused by structural changes at the same two points of variability that occur within the set of saposin B chains, except that they are more pronounced. First, a sharper turn in loop 1 of NK-lysin results in direct contacts between helices α1 and α2. Second, the sharp break between NK-lysin helices α3 and α4 places these two helices at roughly right angles to each other. This kink in NK-lysin occurs at Phe-52 (φ, ψ = −140°, +40°) and corresponds directly to the smaller kink in the α3 helix of chain B of saposin B. The same break in the equivalent helix is also seen in the saposin-like insert of prophytepsin (13). In the monomeric saposin-like proteins, these two features result in direct contacts between the hairpin and base region of the chains, generating a closed monomer that buries the core residues and precludes the saposin B-type dimerization (see below).
Dimer Structure.
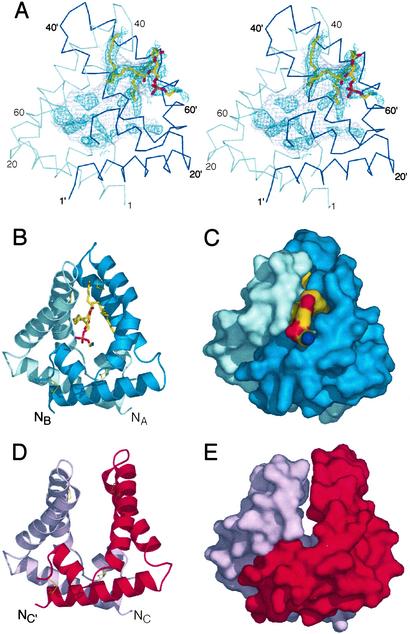
Saposin B dimers are formed by clasping together two V-shaped monomers. In the AB dimer, the two chains interlock to form a shell-like monolayer of α-helices, with a long and extended horseshoe-shaped interface that buries ≈1,260 Å2 of surface area, or 22% of the monomer surfaces. However, most of the hydrophobic residues that line the concave surface of the monomers remain exposed to a large interior cavity, and the protein does not have a packed hydrophobic core (Fig. 2). The volume of the internal cavity is ≈900 Å3, and as a result of the asymmetry in this dimer, there is a single 13 × 6 Å opening to the cavity at the subunit interface. It has long been suspected that the hydrophobic pocket of saposin-B is formed at the dimer interface (23).
Figure 2.
The saposin dimer encloses a large hydrophobic cavity. (A) Stereo view of the AB dimer. The Cα trace of the A chain is in light blue, and the B chain is in dark blue. Residues from the B chain are labeled with primes. The surface of the inner cavity is in gray mesh, and the positive Fo-Fc density calculated before the inclusion of the ligand is indicated in dark blue mesh (contoured at 1.5 σ). The final refined phosphatidylethanolamine fragment is in stick representation. (B and C) Ribbon and solvent-accessible surface diagrams of the AB dimer, with coloring as in A. The lipid headgroup is exposed to solvent, and the acyl chains are contained within the cavity. Disulfide bonds are indicated with thin yellow lines. (D and E) Ribbon and surface representations of the symmetric CC′ dimer with the chains in pink and red. There are tight interchain contacts at the base, but no contacts between the hairpin regions and no ordered ligands within the solvent-exposed core.
Bound Phospholipid.
During the refinement of the structure, difference electron density maps revealed regions of elongated electron density within the AB dimer, which we have modeled as a partially ordered phospholipid (Fig. 2A). Support for the presence of copurifying lipid in our saposin B preparations was obtained from electrospray ionization MS. In a water/acetonitrile/formic acid solvent, positive ions were observed at m/z values of 744.8, 718.8, 716.8, 690.5, and 662.5. These values were within 0.01% of the expected masses for singly protonated forms of phosphatidylethanolamine (PE) with C18:1/C18:1, C18:0/C16:1, C18:1/C16:1, C16:0/C16:1, and C14:0/C16:1 acyl chains, respectively. A selection from these ions was analyzed further by tandem MS (MS/MS) at a variety of collision gas concentrations, and in all cases, the expected fragmentation pattern for the assigned phospholipids was observed (24). Additional confirmation was obtained by MS/MS on negative ions measured in a water/acetonitrile/triethylamine solvent. The copurification of E. coli phospholipids has been seen with other heterologously expressed lipid-binding proteins (25, 26).
Based on the MS results, we have modeled a partially ordered fragment of PE into the difference electron density with the lipid headgroup exposed at the opening to the cavity and the two acyl chains sequestered within the cavity (Fig. 2A). We see evidence for additional lipid conformers in both the headgroup and the acyl chain regions, but we have not modeled this disorder. Because of the uncertainties in the specific features of the lipid conformation, the current data does not justify a detailed analysis of the protein–lipid interactions in this complex. Additional electron density is also seen deeper within the encapsulated interior, but these features cannot be modeled with confidence. Saposin B is known to have broad capabilities for lipid solubilization (3, 27). Although a phospholipid is not the target substrate for the activation reaction, PE and cerebroside sulfate have similar relative binding affinities in vitro (27), and because both lipids are present in lysosomal membranes, it is likely that complexes of saposin B with PE occur in vivo.
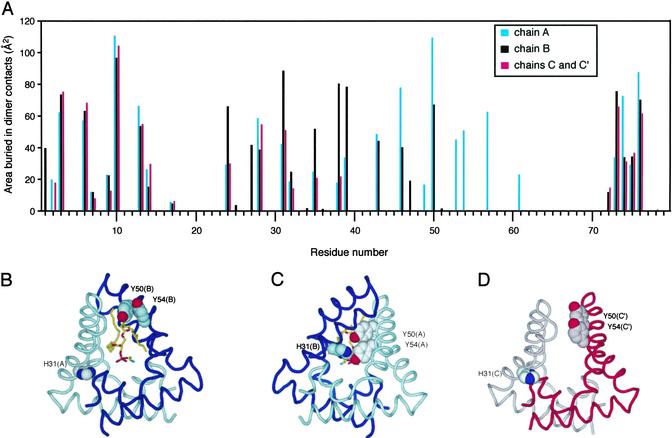
Intersubunit Contacts.
The dimerization surface can be subdivided into contacts between the base regions and contacts between the hairpins (Fig. 3). At the base, there is a tight anti-parallel coiled-coil interaction between the two N-terminal helices and a close packing of each C terminus with helix α2 of the opposite monomer. In the CC′ dimer, these are essentially the only interchain contacts, and they result in the burial of 773 Å2 of surface area. A second set of interactions occurs in the AB dimer between the hairpin turn of chain B to helix α3 of the A chain. These contacts bury an additional 486 Å2 of surface area, and because this surface is continuous with the interface at the base region, this seals off much of the interior cavity.
Figure 3.
(A) Surface burial in the dimer for each of the residues in the three crystallographically independent chains. The open “front” (B) and sealed “back” (C) of the AB dimer are shown. The lipid headgroup protrudes from the opening to the cavity in the front, whereas contacts among His-31(B), Y50(A), and Y54(A) seal the cavity on the back. (D) The relative orientations of residues H31(C), Y50(C′), and Y54(C′) (and the equivalent residues related by the twofold symmetry, not shown in this view) in the CC′ dimer are similar to those found near the opening to the AB dimer.
The asymmetry in the AB dimer results in significant differences in the interresidue contacts across the two chains. The pairs of tyrosines at positions 50 and 54 are aligned on one face of helix α3. In chain B, these residues participate in the pore opening on one side of the dimer, whereas in the A chain, they form part of the closed surface at the “back” of the cavity. His-31 from the opposite chain seals the back of the cavity by forming a hydrogen bond with the phenolic oxygen of Tyr-54, and the close approach of these three aromatic residues is made possible by the tilting in of the hairpin region of chain B. The opening of the cavity is mostly lined with hydrophobic residues (Fig. 1A), with the exception of Tyr-50 and Tyr-54 from chain B and Arg-38 of chain A. This arginine may play a role in the binding of the sulfate group of cerebroside sulfate, but in general, the structural basis for lipid recognition and specificity of the activation reaction by saposin B remains unclear.
The CC′ dimer is significantly more open and there are no interchain contacts between the hairpins. The hydrophobic cavity is, therefore, more exposed to the bulk solvent in this dimer, and the relative orientation of residues H31, Y50, and Y54 are similar to those seen on the pore opening side of the AB complex. Very weak residual electron density is observed within the cavity of the CC′ dimer, and we refer to this dimer as the open or unliganded dimer.
Dimer Flexibility Is Important for Function.
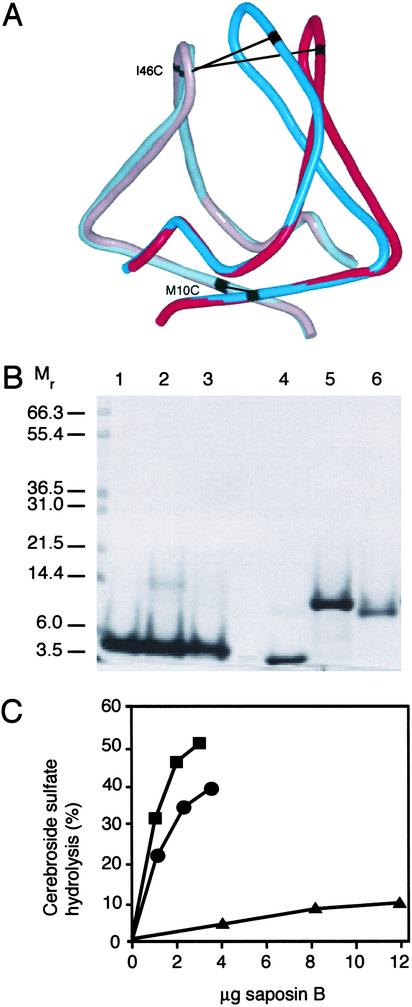
To verify the functional relevance of the dimer and the observed conformational differences between the AB and CC′ dimers, we introduced single cysteine residues at positions near the molecular twofold axis for the formation of interchain disulfide bonds (Fig. 4). Mutant M10C is at the base of the dimer in a region where there is little difference between the two dimers (Cβ-Cβ distances = 5.1 and 5.3 Å in AB and CC′, respectively), whereas mutation I46C is near the hairpin turn where the differences are much larger (Cβ-Cβ distances = 6.3 and 15.6 Å in AB and CC′, respectively). The formation of DTT-reducible dimers was confirmed by SDS/PAGE (Fig. 4B) and by MS (data not shown). The wild-type and both mutant proteins were competent for cerebroside sulfate binding, as measured by isoelectric focusing and fluorescence spectroscopy after incubation of the protein with lipid for 24 h at 37°C (data not shown). However, the rate of stimulation of the cerebroside sulfate hydrolysis reaction by arylsulfatase A was severely compromised in the I46C mutant but not in the M10C mutant (Fig. 4C). Thus, a monomeric species is not required for the function of saposin B, because an obligate dimer (M10C) is able to function as an activator. However, flexibility in the hairpin region is important for the activity (I46C), and we suggest that structural changes in the hairpin region are important in the process of extracting lipids from the membranes. Because the I46C crosslinked saposin B is competent for sulfatide binding and retains a small but measurable level of activity, we propose that this form of the protein uses other less kinetically favored routes for the formation of the lipid complexes.
Figure 4.
(A) Superposition of the AB and CC′ dimers, shown in tube representation. Chains A (light blue) and C (pink) superimpose well, as do the base of chains B (dark blue) and C′ (red). The kink in the third helix of chain B is evident in the overlay with the C′ chain. Residues Met-10 (base) and Ile-46 (hairpin) are near the molecular (pseudo)-symmetry axes and are indicated in black. Equivalent residues in the dimers are connected by black lines. (B) SDS/PAGE of reduced and nonreduced saposin B with and without the additional cysteines. Wild-type saposin B is in lanes 1 and 4, the M10C mutant is in lanes 2 and 5, and I46C is in lanes 3 and 6. DTT was added to lanes 1–3 only. (C) Activation of cerebroside sulfate hydrolysis by arylsulfatase A in the presence of wild-type recombinant saposin B (squares), M10C (circles), and I46C (triangles) crosslinked dimers. Less than 2% hydrolysis of the input sulfatide was observed in the absence of activator under the assay conditions.
Flexibility at the opening to a hydrophobic cavity is a feature that is suspected to be important for the recognition and extraction of lipids from membranes in many other lipid-transport proteins (26, 28–30). In particular, the GM2 activator protein (GM2-AP) is functionally related to the saposins and activates the hydrolysis of the ganglioside GM2 to GM3 by β-hexosaminidase A. However, this activator is a 22-kDa monomer that adopts a β-cup topology that has no structural similarity to saposin B (26). At a global level, however, the GM2-AP is related to the saposin-B dimer in that it has similar overall dimensions and contains a large accessible hydrophobic cavity with an opening lined with a flexible loop. Despite the fact that saposin B is an all α-helical homodimer and that GM2-AP is a monomer consisting mainly of β-structure, saposin B and GM2-AP may share a similar molecular mechanism for glycosphingolipid activation, as supported by the crosslinking data presented above.
Insights into Mechanism.
The existence of the large hydrophobic cavity, the encapsulated phospholipid, and the conformational variability seen between the chains all support a lipid solubilization mechanism for saposin B. These features are consistent with the observation that saposin B can act as a lipid-transport protein (31). We propose that the open form of the saposin B dimer (or conformers similar to it) can interact directly with membrane surfaces, and promote a reorganization of the lipid alkyl chains by virtue of the partly accessible hydrophobic surfaces of the dimer. With some lipids, and perhaps with some of the saposins, these stable structures at the membrane surface may suffice to activate hydrolysis reactions. In the case of cerebroside sulfate, saposin B can effect the full extraction of the lipid, and we propose that this step involves the conformational changes that we have seen at the two hinge points in the B chain of the AB dimer. The high exposure of the cerebroside sulfate headgroup in a soluble saposin B-lipid complex would allow for the formation of a productive enzyme-substrate complex with arylsulfatase A.
Acknowledgments
We thank Lu Chen, Christian Engel, and Farid Ahmad for helpful discussions. This work was supported by Canadian Institutes for Health Research Grant MOP-37967 (to G.G.P.). The work at University of California, Los Angeles was supported by National Institutes of Health Grant NS31271 (to A.L.F.) and the W. M. Keck Foundation. The Cornell High Energy Synchrotron Source (CHESS) is supported by National Science Foundation Award DMR 97-13424, and the Macromolecular Diffraction at the CHESS facility is supported by National Institutes of Health Award RR-01646, through its National Center for Research Resources.
Abbreviations
- PE
phosphatidylethanolamine
- rmsd
rms deviation
- MS/MS
tandem MS
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1N69).
References
- 1.Sandhoff K, Kolter T. Trends Cell Biol. 1996;6:98–103. doi: 10.1016/0962-8924(96)80999-8. [DOI] [PubMed] [Google Scholar]
- 2.Vaccaro A M, Salvioli R, Tatti M, Ciaffoni F. Neurochem Res. 1999;24:307–314. doi: 10.1023/a:1022530508763. [DOI] [PubMed] [Google Scholar]
- 3.Fluharty A L. Trends Glycosci Glycotechnol. 1995;7:167–189. [Google Scholar]
- 4.Wilkening G, Linke T, Uhlhorn-Dierks G, Sandhoff K. J Biol Chem. 2000;275:35814–35819. doi: 10.1074/jbc.M006568200. [DOI] [PubMed] [Google Scholar]
- 5.Munford R S, Sheppard P O, O'Hara P J. J Lipid Res. 1995;36:1653–1663. [PubMed] [Google Scholar]
- 6.Zhai Y, Saier M H., Jr Biochim Biophys Acta. 2000;1469:87–99. doi: 10.1016/s0304-4157(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 7.Vaccaro A M, Salvioli R, Barca A, Tatti M, Ciaffoni F, Maras B, Siciliano R, Zappacosta F, Amoresano A, Pucci P. J Biol Chem. 1995;270:9953–9960. doi: 10.1074/jbc.270.17.9953. [DOI] [PubMed] [Google Scholar]
- 8.Waring A J, Chen Y, Faull K F, Stevens R, Sherman M A, Fluharty A L. Mol Genet Metab. 1998;63:14–25. doi: 10.1006/mgme.1997.2646. [DOI] [PubMed] [Google Scholar]
- 9.Bruhn H, Leippe M. Biol Chem. 1999;380:1001–1007. doi: 10.1515/BC.1999.124. [DOI] [PubMed] [Google Scholar]
- 10.Whitelegge J P, Penn B, To T, Johnson J, Waring A, Sherman M, Stevens R L, Fluharty C B, Faull K F, Fluharty A L. Protein Sci. 2000;9:1618–1630. doi: 10.1110/ps.9.9.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaltash S, Palmblad M, Curstedt T, Johansson J, Persson B. Biochim Biophys Acta. 2000;1466:179–186. doi: 10.1016/s0005-2736(00)00199-1. [DOI] [PubMed] [Google Scholar]
- 12.Liepinsh E, Andersson M, Ruysschaert J M, Otting G. Nat Struct Biol. 1997;4:793–795. doi: 10.1038/nsb1097-793. [DOI] [PubMed] [Google Scholar]
- 13.Kervinen J, Tobin G J, Costa J, Waugh D S, Wlodawer A, Zdanov A. EMBO J. 1999;18:3947–3955. doi: 10.1093/emboj/18.14.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez C, Langdon G M, Bruix M, Galvez A, Valdivia E, Maqueda M, Rico M. Proc Natl Acad Sci USA. 2000;97:11221–11226. doi: 10.1073/pnas.210301097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn V E, Faull K F, Whitelegge J P, Higginson J, Fluharty A L, Privé G G. Protein Expression Purif. 2003;27:186–193. doi: 10.1016/s1046-5928(02)00597-1. [DOI] [PubMed] [Google Scholar]
- 16.Sharff A J, Koronakis E, Luisi B, Koronakis V. Acta Crystallogr D. 2000;56:785–788. doi: 10.1107/s090744490000370x. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 18.Smith G D, Nagar B, Rini J M, Hauptman H A, Blessing R H. Acta Crystallogr D. 1998;54:799–804. doi: 10.1107/s0907444997018805. [DOI] [PubMed] [Google Scholar]
- 19.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Fluharty A L, Katona Z, Meek W E, Frei K, Fowler A V. Biochem Med Metab Biol. 1992;47:66–85. doi: 10.1016/0885-4505(92)90009-n. [DOI] [PubMed] [Google Scholar]
- 23.Wynn C H. Biochem J. 1986;240:921–924. doi: 10.1042/bj2400921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy R C. Mass Spectrometry of Phospholipids: Tables of Molecular and Product Ions. Denver, CO: Illuminat; 2002. [Google Scholar]
- 25.Beamer L J, Carroll S F, Eisenberg D. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 26.Wright C S, Li S C, Rastinejad F. J Mol Biol. 2000;304:411–422. doi: 10.1006/jmbi.2000.4225. [DOI] [PubMed] [Google Scholar]
- 27.Fluharty C B, Johnson J, Whitelegge J, Faull K F, Fluharty A L. J Neurosci Res. 2001;63:82–89. doi: 10.1002/1097-4547(20010101)63:1<82::AID-JNR10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 28.Anderson T A, Levitt D G, Banaszak L J. Structure (London) 1998;6:895–909. doi: 10.1016/s0969-2126(98)00091-4. [DOI] [PubMed] [Google Scholar]
- 29.Schouten A, Agianian B, Westerman J, Kroon J, Wirtz K W, Gros P. EMBO J. 2002;21:2117–2121. doi: 10.1093/emboj/21.9.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roderick S L, Chan W W, Agate D S, Olsen L R, Vetting M W, Rajashankar K R, Cohen D E. Nat Struct Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 31.Vogel A, Schwarzmann G, Sandhoff K. Eur J Biochem. 1991;200:591–597. doi: 10.1111/j.1432-1033.1991.tb16222.x. [DOI] [PubMed] [Google Scholar]
- 32.Ponting C P, Russell R B. Trends Biochem Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]