Abstract
A number of hepatitis C virus (HCV) proteins, including NS5B, the RNA-dependent RNA polymerase, were detected in membrane fractions from Huh7 cells containing autonomously replicating HCV RNA replicons. These membrane fractions were used in a cell-free system for the analysis of HCV RNA replication. Initial characterization revealed a reaction in which the production of replicon RNA increased over time at temperatures ranging from 25 to 40°C. Heparin sensitivity and nucleotide starvation experiments suggested that de novo initiation was occurring in this system. Both Mn2+ and Mg2+ cations could be used in the reaction; however, concentrations of Mn2+ greater than 1 mM were inhibitory. Compounds shown to inhibit recombinant NS3 and NS5B activity in vitro were found to inhibit RNA synthesis in the cell-free system. This system should be useful for biochemical analysis of HCV RNA synthesis by a multisubunit membrane-associated replicase and for evaluating potential antiviral agents identified in biochemical or cell-based screens.
Hepatitis C virus (HCV) infects over 170 million people worldwide with ca. 70% failing to clear the virus. Chronic infection can lead to serious liver disease, including cirrhosis and hepatocellular carcinoma, making HCV infection the leading indication for liver transplantation in the United States (17). The current therapy, pegylated alpha interferon and ribavirin, is not effective in many patients; therefore, it is vital that more effective antiviral agents are identified.
HCV is a member of the family Flaviviridae and possesses a single-stranded, positive-sense RNA genome ca. 9.6 kb in length, encoding a single polyprotein (25). The genome is uncapped and an internal ribosome entry site (IRES) in the 5′-untranslated region (UTR) of the genome promotes translation initiation (42, 43). The 3′-UTR is 200 to 300 nucleotides (nt) in length consisting of a short variable region (ca. 40 nt), then a poly(U/UC) tract, and finally a conserved 98-nt sequence (21). The extreme 3′ end forms a stable 46-base stem-loop structure with the 3′-terminal U at the base of the stem in a predicted G-U base pair (7). The viral polyprotein is co- and posttranslationally cleaved by a combination of host and viral proteases to produce at least 10 individual proteins (25). The amino-terminal portion of the polyprotein comprises the structural proteins core or capsid protein (C), envelope protein 1 (E1) and E2, followed by p7, and the nonstructural proteins 2 (NS2), NS3, NS4A, NS4B, NS5A, and NS5B. Recent work demonstrated that NS3 to NS5B are sufficient for RNA replication (27). Specific functions have been defined for NS3, NS4A, and NS5B. NS3 is a bifunctional enzyme containing serine-protease activity within the N-terminal region, and C-terminal NTPase and/or helicase activities (13, 16, 23, 32). NS3 in conjunction with NS4A mediates cleavage at the 3-4A, 4A-4B, 4B-5A, and 5A-5B junctions (16, 25, 40). NS3 helicase activity is postulated to be required during RNA synthesis to unwind double-stranded regions of the template RNA, formed due to cis-derived secondary structures or annealing of plus- and minus-strand HCV RNA. NS5B is the catalytic subunit of the viral RNA replicase (4, 12, 26, 46), possessing the canonical GDD motif characteristic of RNA-dependent RNA polymerases (RdRp).
In order to develop antiviral agents, virus-specific processes are often targeted. One such process is replication of the RNA genome. Two systems are commonly used to assay HCV RNA synthesis. The first method relies on in vitro analyses of purified, recombinant NS5B. NS5B has been demonstrated to possess RNA synthetic activity (4, 12, 26, 29, 33, 46). However, although studies with purified polymerase provide insight into the enzymology of NS5B, they may not reflect authentic replication. For example, NS5B copies a wide range of template RNAs and DNA without preference, suggesting that NS5B is necessary but not sufficient for selective replication of the genome. Results are conflicting as to whether initiation of replication occurs by primer-dependent, copy back, or de novo RNA synthesis. In one case, purified NS5B could utilize full-length HCV genome RNA as a template (33). However, initiation of minus-strand synthesis occurred at the first non-base-paired nucleotide in the loop of the 3′-most stem-loop structure and not at or near the 3′ terminus, as one would intuitively predict (34). In terms of other components, one might speculate that NS3 is part of the replicase and that the associated helicase activity unwinds the double-stranded 3′-terminal bases facilitating initiation. Such a model is consistent with a recent report showing that NS5B requires a single-stranded 3′ terminus of the template for de novo initiation (15).
A second system in which HCV RNA replication can be analyzed is cell based and utilizes autonomously replicating HCV-derived RNAs (27). These replicon RNAs have the authentic HCV 5′- and 3′-UTRs. The HCV IRES drives the translation of a selectable marker such as neomycin resistance, and an internal encephalomyocarditis virus (EMCV) IRES directs translation of NS3 to NS5B (Fig. 1A). In vitro transcribed replicon RNAs are electroporated into the human hepatoma cell line Huh7 and placed under selection. The emergence of neomycin-resistant cells is indicative of RNA replication. Resistant cell colonies can be expanded, and RNA synthesis is observed by metabolic labeling. Recently, adaptive mutations that dramatically enhance the ability of HCV RNA replication to initiate in Huh7 cells have been identified in NS3, NS4B, NS5A, and NS5B (5, 14, 22). Optimized subgenomic and full-length HCV replicons (6, 14, 36), matched with highly permissive Huh7 sublines (6), now provide a means for genetic analysis of HCV RNA replication.
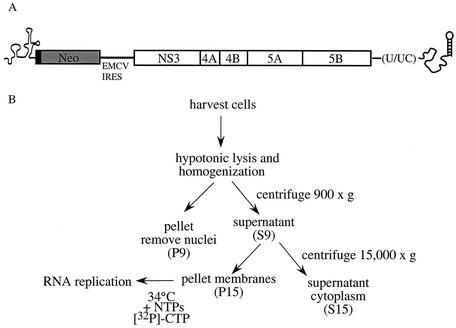
FIG. 1.
Replicon structure and fractionation scheme. (A) Schematic representation of the HCV subgenomic replicon RNA. The replicon contains the 5′- and 3′-UTRs of HCV and two cistrons encoding the first 12 amino acids of core fused to neomycin phosphotransferase (Neo) and NS3-NS5B. The HCV IRES located within the 5′-UTR and the EMCV IRES initiate translation of Neo and NS3-NS5B, respectively. (B) Procedure for isolation of membrane fractions from replicon-containing Huh7 cells. Cellular membranes were isolated by lysing cells in a hypotonic buffer, followed by two centrifugation steps. The cell lysate was centrifuged at 900 × g to pellet nuclei and unlysed cells (P9 fraction). The supernatant (S9) was further fractionated by centrifugation at 15,000 × g. The resulting supernatant (S15) and pellet (P15) were enriched for cytoplasm and cellular membranes, respectively.
Despite these advances, additional biochemical assays are needed for mechanistic studies. A cell-free system capable of authentic HCV RNA replication would permit the elucidation of the mechanism of viral replication, the components of the viral replicase, and the functions of various protein factors and RNA elements located within the genome. Since several of the HCV replicase proteins, including NS3-4A, NS4B, NS5A, and NS5B, are localized to the perinuclear membrane (8, 11, 18, 31, 45), we produced membrane fractions from cells harboring the subgenomic replicon. These fractions were shown to contain several HCV nonstructural proteins and RdRp activity. This system allows cell-free analysis of RNA replication by what is predicted to be a multicomponent HCV RNA replicase. In the present study we demonstrate the synthesis of HCV RNA in vitro, provide evidence that de novo initiation of RNA synthesis is occurring, characterize the general biochemical requirements for this reaction, and demonstrate the utility of this system for evaluating antiviral agents directed against HCV RNA replication.
MATERIALS AND METHODS
Cells.
Huh7 cells were grown at 37°C in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and nonessential amino acids. Huh7-clone A cells harboring autonomously replicating HCV replicon RNA were passaged in the same medium containing 1 mg of G418/ml.
Cellular fractionation.
To generate HCV replicon-containing membrane fractions (Fig. 1B), clone A cells were grown in 150-mm-diameter dishes to ca. 90% confluence (ca. 3 × 107 cells/dish). Cells were washed with 1× phosphate-buffered saline (PBS), detached by scraping, and harvested by low-speed centrifugation (900 × g). Cell pellets were resuspended in 1 ml of hypotonic buffer (10 mM Tris-HCl, pH 7.8; 10 mM NaCl), allowed to swell 15 min on ice, and disrupted with 50 strokes of a tight fitting pestle (0.020- to 0.056-mm clearance) in a Dounce homogenizer. Nuclei were removed by centrifugation (900 × g for 5 min at 4°C). Postnuclear homogenates were centrifuged at 15,000 × g for 20 min at 4°C. Pellets (P15) were resuspended in 100 μl of storage buffer (hypotonic buffer plus 15% glycerol) and stored at −80°C for no more than 90 days.
In vitro HCV RNA synthesis.
Standard reaction mixtures contained the following: 50 mM Tris-HCl (pH 7.8); 50 mM KCl; 5 mM MgCl2 (unless otherwise stated); 10 mM dithiothreitol; 10 μg of actinomycin D per ml; 5 mM creatine phosphate; 25 μg of creatine phosphokinase per ml; 1 mM concentrations of ATP, GTP, and UTP; 40 μM CTP; 1 mCi of [α-32P]CTP per ml; 800 U of RNasin per ml; 18 μl of P15 (material from ca. 5 × 106 cells); and H2O to a total volume of 50 μl. Reactions were incubated at 34°C for 60 min, after which 5 U of alkaline phosphatase (New England Biolabs) was added, and incubation continued for a further 20 min. Reactions were terminated by the addition of sodium dodecyl sulfate (SDS) to 2.5% and proteinase K to 100 μg/ml. RNA was isolated by phenol-chloroform extraction and ethanol precipitated. RNAs were denatured with glyoxal, separated by electrophoresis on 1% agarose-phosphate gel, and visualized by autoradiography.
Western blot analysis of cell fractionation.
To ensure equivalent loading of material onto the gel after each centrifugation, a sample of the material was centrifuged in parallel, separated, and brought to the same final volume with loading buffer. Samples were separated by SDS-polyacrylamide gel electrophoresis (8% polyacrylamide), transferred to nitrocellulose, and blocked with 5% milk in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, and 0.1% Tween 20 (pH 7.4). Blots were probed with anti-NS3 monoclonal antibody (1895 ViroStat; 1:100 dilution), anti-NS5A rabbit polyclonal antibody (SOD-NS5, 1:1,000 dilution; kindly provided by Shirley Wong, Chiron Corp.), and anti-NS5B monoclonal antibody (5B-3B1, 1:1,000 dilution; kindly provided by Darius Moradpour [31]). Proteins were detected by secondary antibody conjugated to horseradish peroxidase.
RNase H analysis.
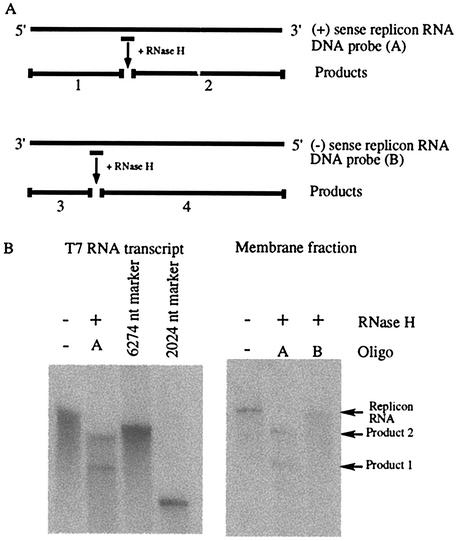
Ethanol-precipitated RNAs from the in vitro replication assay were further purified by using RNeasy kit (Qiagen) according to the manufacturer's protocol. Isolated RNA was ethanol precipitated an additional two times in the presence of 750 mM ammonium acetate. Precipitated RNA was washed with 70% ethanol and denatured for 2 min at 95°C in a buffer consisting of 80% formamide, 2 mM Tris-HCl (pH 7.5), and 0.2 mM EDTA. The denatured RNAs were diluted 10-fold in annealing buffer (20 mM Tris-HCl, pH 7.5; 100 mM KCl; 2.5 μg of tRNA) and annealed to 150 pmol of oligonucleotide by slow cooling from 80 to 30°C. Oligonucleotide A anneals to nt 3102 to 3121 of plus-sense replicon RNA (5′-GGCTGAAGTCGACTGTCTGG-3′), and oligonucleotide B corresponds to nt 2081 to 2100 of the plus-sense replicon RNA (5′-CCTTGACACCATGCACCTGC-3′) and is predicted to anneal to the minus-strand copy of the replicon RNA. An equal volume of digestion buffer (20 mM Tris-HCl, pH 7.5; 100 mM NaCl; 20 mM MgCl2; 20 mM dithiothreitol) was added to the reaction mixture, and digestion with 2 U of RNase H was performed at 37°C for 20 min. Products of digestion were isolated by phenol-chloroform extraction and ethanol precipitated. RNA was denatured with glyoxal, separated by electrophoresis, and visualized by autoradiography. Markers for the products of digestion of plus-strand RNA were generated by RNase H digestion of a labeled replicon RNA transcribed in vitro by using T7 RNA polymerase in the presence of oligonucleotide A. Additional markers were generated by runoff in vitro transcription of the replicon cDNA digested with BsrG1 (RNA product, 2,024 nt) or BglII (RNA product, 6,274 nt).
RESULTS
In vitro replication of HCV replicon RNA.
The replicon RNA (Fig. 1A) was derived from the HCV genotype 1b (Con1) replicon, I377/NS3-3′ (5, 27). The cDNA encoding this parental replicon RNA was originally generated by a PCR-based gene assembly procedure (5). The replicon RNA possesses the 5′-UTR of the HCV 1b genome and the first 12 codons of the C protein coding region fused in frame to the neomycin phosphotransferase II gene (Neo) that confers resistance to G418 upon expression. The Neo gene is followed by the EMCV IRES, which directs translation of the HCV replicase (NS3 to NS5B). The coding region is followed by the HCV 3′-UTR. In total, the replicon RNA is 7,989 nt long. The clonal cell line used in the present study, clone A, harbored a replicon RNA with two coding changes relative to the original I377/NS3-3′ sequence. These changes were a glutamine-to-arginine change in the NS3 protein (Q1112R; numbers refer to residues in the full-length Con1 polyprotein) and a serine-to-isoleucine change in the NS5A protein (S2204I). The S2204I change significantly increases G418 transduction efficiency and intracellular replicon RNA levels over those observed for the parental replicon (5).
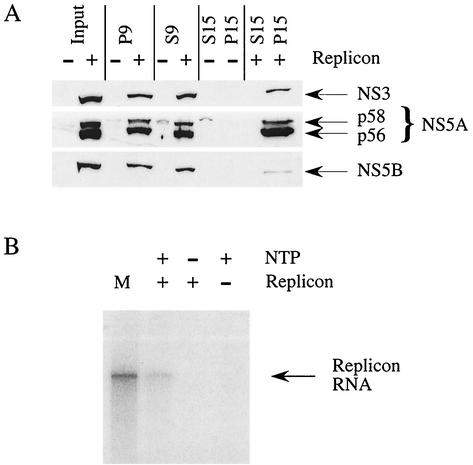
Replication of RNA viruses invariably occurs in association with cellular membranes, and crude membrane-associated replicase preparations have been successfully produced for several RNA viruses (3, 24, 35, 47). Not surprisingly, all HCV nonstructural proteins, including NS5B, associate with the perinuclear membrane (8, 18, 31, 45). Membrane fractions were prepared by hypotonic lysis and homogenization of clone A cells. Postnuclear homogenates were spun at 15,000 × g to pellet the membranes. Western blot analysis of this membrane fraction (P15) from HCV replicon-containing cells demonstrated an enrichment of NS3, NS5A, and NS5B compared to the supernatant (S15) (Fig. 2A). The P15 fractions enriched for HCV nonstructural proteins were assayed for replicase activity in the presence of a reaction mix, including [32P]CTP and actinomycin D. Since no additional RNA is added to the in vitro reaction mixture, the template corresponds to the endogenous replicon RNA. As shown in Fig. 2B, a radiolabeled RNA product was observed that comigrated with the originally transfected replicon RNA. Production of labeled RNA required the presence of cold nucleoside triphosphates (NTPs), indicating that the signal was not due to terminal addition of labeled nucleotide to full-length endogenous replicon RNA (Fig. 2B). Labeled RNA was not observed in reactions with membrane fractions from nontransfected Huh7 cells, indicating that the signal was dependent upon the presence of the replicon RNA (Fig. 2B). Membrane fractions from other replicon-containing cell lines have been tested with similar results (data not shown).
FIG. 2.
Locations of HCV proteins and replicase activity in the P15 fractions. (A) During the isolation, samples were prepared from each step and subjected to Western blot analysis with antisera directed against NS3, NS5A, and NS5B. NS5A appears as two bands (p56 and p58) for the basal and hyperphosphorylated forms. The majority of HCV proteins appear to be found in the P9 and P15 fractions. For comparison, equivalent amounts were loaded onto each lane. (B) Membrane fractions (P15) from replicon-containing cells (+) and Huh7 cells (−) were assayed for replicase activity. P15s were incubated with [32P]CTP in the absence (−) or presence (+) of a full complement of NTPs (ATP, GTP, and UTP). Denatured products were separated on a 1% agarose gel and visualized by autoradiography. Radiolabeled replicon RNA marker (lane M) was generated by in vitro transcription by T7 RNA polymerase of a linearized DNA template.
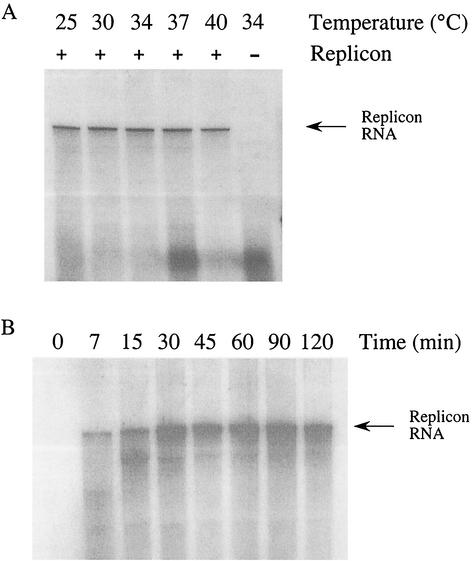
This in vitro RNA synthesis reaction was active over a temperature range of 25 to 40°C, synthesizing comparable amounts of RNA at the temperatures tested (Fig. 3A). A time course of the reaction demonstrated an increase in labeled replicon RNA up to ca. 45 min, after which the intensity of the signal remained the same (Fig. 3B). Smaller labeled products present at the 7 and 15 min time points disappeared with long incubation, suggesting that the smaller products represent replicon RNAs in the process of being synthesized.
FIG. 3.
Temperature range and time course of cell-free HCV RNA synthesis. Production of HCV RNA from P15 fractions under standard conditions (Materials and Methods) was assessed over a range of temperature (A) and time (B). RNA products were extracted, precipitated, and analyzed by denaturing agarose gel electrophoresis. (A) Reactions were incubated at 25, 30, 34, 37, and 40°C for 1 h and then terminated by the addition of SDS and proteinase K. P15 fractions from Huh7 cells lacking the replicon (−) were assayed at 34°C as a control for background RNA synthesis. (B) P15 fractions from replicon-containing cells were incubated at 34°C for 0, 7, 15, 30, 45, 60, 90, and 120 min under standard conditions. At each time point the reactions were stopped immediately by freezing in a dry ice bath prior to RNA extraction.
Divalent cation requirement for in vitro RNA replication.
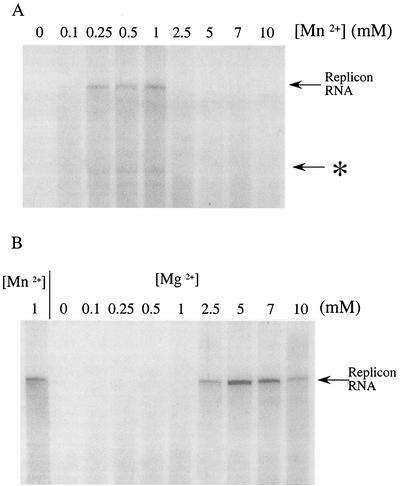
Previous studies utilizing purified NS5B to mediate RNA synthesis in vitro demonstrated that NTP incorporation was most efficient when Mn2+ was provided as the divalent cation (12, 29, 48). We tested the requirements of the replicase for Mn2+ and Mg2+ in the context of our in vitro replication assay. Production of full-length replicon RNA increased up to 5 mM Mg2+, whereas 1 mM was found to be optimal for Mn2+ (Fig. 4). Interestingly, manganese concentrations greater than 1 mM inhibited synthesis of full-length replicon RNA. Although nucleotide incorporation was not measured, these results suggest that higher concentrations of Mn2+ lead to a decrease in enzyme processivity, as demonstrated by the loss of production of full-length RNA. The production of smaller products in the presence of low concentrations of Mn2+ can be observed (Fig. 4A). This may further indicate a decrease in enzyme processivity in the presence of this ion. These same products were not observed in the presence of Mg2+. The addition of Mg2+ to reaction mixtures containing Mn2+ did not lead to an increase in full-length product (data not shown).
FIG. 4.
Divalent cation requirements for HCV RNA synthesis. In vitro RNA replication reactions were carried out in the presence of various concentrations (0.1 to 10 mM as indicated) of Mn2+ (A) or Mg2+ (B) at 34°C for 1 h. RNA products were extracted and analyzed by denaturing agarose gel electrophoresis (Materials and Methods), and the position of the full-length replicon RNA is indicated. (A) An asterisk denotes the position of an RNA that was smaller than the replicon that was formed with the addition of 0.25 to 1 mM Mn2+ to the reaction mixture. (B) Synthesis of RNA over the range of Mg2+ concentrations is compared to the same reaction done in the presence of the optimal Mn2+ concentration (1 mM).
Polarity of in vitro-synthesized RNA.
To determine the polarity of the RNA synthesized in these in vitro reactions, we subjected 32P-labeled RNA to RNase H digestion in the presence of oligonucleotides that anneal to either plus- or minus-sense replicon RNA (Fig. 5A). The results demonstrated that the vast majority of the RNA synthesized was of positive polarity (Fig. 5B). RNase H digestion in the presence of oligonucleotide A, complementary to plus-sense replicon RNA, yielded two labeled products of the predicted sizes (Fig. 5B). The production of two labeled products following digestion with RNase H in the presence of an oligonucleotide directed against plus-strand RNA provides further evidence that the signal observed in the in vitro reaction is not a consequence of terminal transferase activity. We were unable to detect products of digestion in the presence of oligonucleotide B targeted to minus-strand replicon RNA (Fig. 5B). Most positive-sense RNA viruses produce a negative-sense replication intermediate in smaller amounts than that of the genomic RNA; therefore, it is possible that minus-strand RNA was present but below the detection limit of this assay.
FIG. 5.
Polarity of HCV RNA products. RNase H digestion was employed as a means of determining the polarity of the RNA replication products. DNA oligonucleotides were designed to specifically hybridize to either plus-sense (+) or minus-sense (−) replicon RNA. (A) Diagram illustrating the locations of annealing oligonucleotide and predicted products after RNase H digestion. Oligonucleotide A should anneal to nt 3102 to 3121 of plus-strand replicon RNA; oligonucleotide B corresponds to nt 2081 to 2100 of the plus-sense replicon RNA and is predicted to anneal to minus-strand RNA. After hybridization of the DNA oligonucleotides, followed by RNase H digestion, the polarity of the RNA is determined by the production of products 1 and 2 for the plus (+) sense and products 3 and 4 for the minus (−) sense. (B) Products of RNase H digestion. RNase H digestions were performed on T7 transcribed RNA (left panel) and the RNA products of the cell-free reaction (right panel). Hybridizing DNA oligonucleotides used in the digestion are denoted by A and B. Two RNA markers of 6274 and 2024 nt were synthesized by in vitro transcription of a linearized DNA template. Oligo, oligonucleotide.
Evidence for de novo initiation of RNA synthesis.
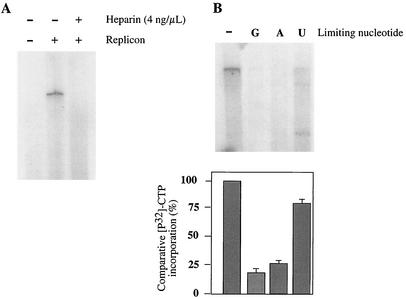
The template RNA was derived from cells that were actively replicating HCV replicon RNA. This led us to question whether the observed signal was a product of de novo initiation in vitro or elongation of RNAs whose synthesis had initiated in cells prior to membrane isolation. Due to the crude nature of the membrane fractions, we were unable to observe small products of de novo initiation directly (data not shown). We therefore employed well-established, indirect approaches for assessing whether de novo initiation might be occurring in the reactions. A characteristic of initiation of RNA synthesis is its sensitivity to heparin. Heparin, a polyanionic proteoglycan, inhibits the initiation of RNA synthesis for all RNA polymerases, including purified HCV NS5B (10, 30, 38, 41, 48). It is believed that the anionic heparin binds the polymerase prior to or during initiation, thus preventing RNA synthesis. However, a change in the stability of the ternary complex following or concurrent with the transition to elongation prevents inhibition by heparin. The addition of heparin (4 ng/μl) to the in vitro reaction led to a decrease in the production of labeled HCV RNA (Fig. 6A). This suggests that a significant fraction of the labeled full-length replicon RNA synthesized in this reaction is a consequence of de novo initiation. A longer exposure of the gel shown in Fig. 6A revealed a small amount of full-length product labeled in the presence of heparin (data not shown), which we assume is due to the continuation of RNA synthesis initiated in cells prior to membrane isolation.
FIG. 6.
Evidence for de novo initiation of RNA replication. (A) Heparin sensitivity of the in vitro replication reaction. Standard reactions were performed on P15s from Huh7 (− replicon) and clone A (+ replicon) cells in the presence (+) or absence (−) of 4 ng of heparin/μl. (B) Standard P15 reactions (represented by “−”) were performed with 1 mM ATP, GTP, or UTP; 40 μM CTP; and 1 mCi of [α-32P]CTP per ml. Nucleotide requirements for replicon RNA synthesis were tested by limiting the concentration (2 μM) of either GTP, ATP, or UTP (denoted as G, A, and U in the figure). Nucleotide incorporation was determined on the basis of denaturing agarose gel electrophoresis (top panel) or average trichloroacetic acid-precipitable counts from three independent experiments (bottom panel).
Additional evidence for de novo initiation was derived from experiments to determine the nucleotide requirements for the production of labeled HCV replicon RNA. In all RNA synthesis reactions mediated by RNA polymerases there is a requirement for a higher concentration of the initiating nucleotide than nucleotides required only for elongation. Studies with purified NS5B have demonstrated a requirement for high concentrations of GTP and ATP for de novo initiation of RNA synthesis when the templates used possess a 3′-terminal cytidylate or uridylate residue, respectively (19, 20, 29, 48). However, stimulation of NS5B-mediated de novo RNA synthesis by GTP, independent of the initiating nucleotide, has also been reported (20, 28). We performed reactions in which RNA was labeled with [32P]CTP and each of the other NTPs was limited (2 μM). The amount of full-length product decreased in each reaction where one of the NTPs was limiting; however, when ATP and GTP were limiting, there was significantly less nucleotide incorporation than when UTP was limiting (Fig. 6B). Uridylate and cytidylate are predicted to be the 3′ residues of the plus- and minus-strand replicon RNAs, respectively. The requirement for high concentrations of both ATP and GTP is consistent with de novo initiation of minus (ATP-dependent)- and plus (GTP-dependent)-strand RNA synthesis occurring in this system. However, it should also be noted that ATP and GTP concentration may also affect the helicase activity. The requirement for a high concentration of GTP may also be due to the general stimulatory effect of GTP on de novo initiation of RNA synthesis independent of the initiating nucleotide (20, 28; see also Discussion).
Inhibition of HCV replicon RNA synthesis in vitro by compounds directed against the viral polymerase and helicase.
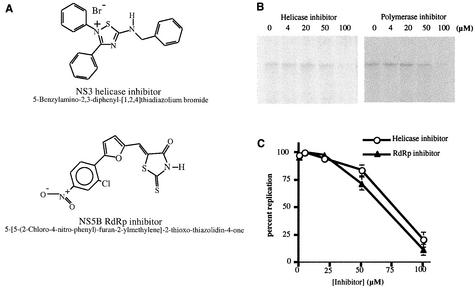
Membrane fractions from cells containing the HCV replicon RNA were incubated for 5 min at 25°C in the presence of candidate inhibitory compounds at concentrations from 10 to 250 μM. The compounds were dissolved and diluted in dimethyl sulfoxide plus equivalent volumes of the dilutions or in dimethyl sulfoxide alone, as a solvent control, and used in the experiments. After this preincubation period, the remaining components of the standard replication reaction were added (Cf inhibitor = 4 to 100 μM), and the reactions continued for 1 h at 34°C. We tested a number of compounds in this system (data not shown) and chose two to illustrate the efficacy of this system for antiviral evaluation. One compound, a 2,3,5-trisubstituted-1,2,4-thiadiazol-2-ium salt (Fig. 7A), has been demonstrated to inhibit helicase activity of purified NS3 in vitro (50% inhibitory concentration [IC50] = 25 μM; patent WO 00/24725 [Vertex]). This compound inhibits the ATPase activity of the purified NS3 protein and also the unwinding of double-stranded RNA. A second compound, a rhodanine derivative (Fig. 7A), inhibits the in vitro RNA synthetic activities of purified NS5B protein (IC50 = 30 μM; patent WO 00/10573 [ViroPharma]). As can be seen in Fig. 7B, when these compounds were titrated into the in vitro RNA replication reaction, RNA synthesis was inhibited at concentrations greater than 50 μM (Fig. 7B). The IC50 for the helicase inhibitor was approximately threefold higher in this assay than previously reported in assays with purified NS3 (ca. 80 μM compared to 25 μM). The same was true for the RdRp inhibitor (ca. 70 μM compared to 30 μM). The data shown in Fig. 7C are representative of three independent experiments.
FIG. 7.
Inhibition of HCV RNA synthesis by putative antiviral compounds. Various concentrations of NS3 and NS5B inhibitors were added to the membrane fractions and incubated at 25°C for 5 min prior to the addition of the other in vitro replication reaction components. Reactions and RNA product analysis were done by using standard conditions (Materials and Methods). (A) Chemical structures of the NS3 and NS5B inhibitory compounds. (B) P15 RNA synthesis reaction done in the presence of helicase inhibitor (left panel) or polymerase inhibitor (right panel). (C) The inhibitor titration experiments were performed in triplicate, quantitated by using a phosphorimager, and demonstrated graphically.
Since both of these compounds have been demonstrated to inhibit two HCV-specific catalytic processes, these data strongly support the conclusion that we are observing authentic HCV RNA replication in this in vitro system. Although it is obvious that the RdRp activity of NS5B is required for RNA replication, the role of the NS3-associated helicase activity in RNA replication is less clear. The fact that a compound directed against NS3 inhibits RNA synthesis suggests that the NS3 protein is an essential component of the viral RNA replication machinery.
DISCUSSION
We have developed a system in which the synthesis of HCV RNA is mediated by what we believe is the authentic replicase complex. Membrane fractions from Huh7 cells harboring HCV RNA replicons expressing NS3-5B were generated. These fractions were found to contain the endogenous replicon RNA and, when supplied with NTPs, were capable of replicating this RNA. Initial studies revealed that the majority of the RNA synthesized was positive-sense RNA. Also, RNA synthesis was sensitive to the presence of heparin and low concentrations of ATP and GTP, suggesting that de novo initiation of RNA synthesis may be occurring. We note, however, that neither heparin sensitivity nor differential sensitivity to low concentrations of ATP and GTP provides formal proof for de novo initiation. Recently, it has been reported that NS5B oligomerizes and that the monomers function in a cooperative fashion during RNA synthesis. This may cause the elongation reaction to be sensitive to heparin, although this has not been demonstrated (44). Further work is needed to explore this possibility.
The precise reason for the sensitivity of the reaction to low GTP concentrations remains unclear. Many reports in which the templates possessed a 3′-terminal cytidylate residue concluded that high concentrations of GTP were needed for initiation with a guanylate residue (19, 29, 48). Other reports, however, indicate that GTP has a stimulatory effect on initiation independent of the initiating nucleotide (20, 28). Recent structural analyses of the NS5B in complex with ribonucleotides revealed a GTP-binding site distant from the active site (9). While this may participate in regulating initiation of RNA synthesis, its importance needs to be explored by further mutational and biochemical studies.
The replication reaction was found to be stable for 2 h and active over a temperature range of 25 to 40°C. Interestingly, the replicase showed a difference in divalent cation preference to that of purified HCV RdRp. In our system the replicase required 5 mM Mg2+ for optimal synthesis of full-length replicon RNA. The presence of high concentrations of Mn2+ (>1 mM) inhibited full-length replicon synthesis. Smaller labeled products were also apparent in the presence of Mn2+, indicating a decrease in enzyme processivity. Previous studies demonstrated that the efficiency of nucleotide incorporation by NS5B increased up to a Mn2+ concentration of 7.5 to 10 mM at which point it leveled out (12, 48). However, the studies were performed by using short template RNAs no more than 100 nt in length, making it more difficult to observe differences in processivity. Ferrari et al. (12) did note that processivity of the HCV polymerase was low compared to other known polymerases, and it is well-known that processivity and fidelity of other nucleic acid polymerases are reduced by Mn2+ (1, 39).
The cell-free assay was used to screen a panel of compounds with reported inhibitory activity against HCV-encoded enzymes. A number of these compounds inhibited HCV RNA synthesis by the replicase at relatively high concentrations (Cf = 100 μM). These included a compound directed against the helicase/ATPase activity of the NS3 protein and another compound directed against the RdRp activity of NS5B. Both of these compounds inhibited RNA synthesis at high concentrations (Fig. 7B). This result not only demonstrates the use of this system for evaluating inhibitory compounds but also indicates that the RNA synthesis by the crude replicase requires both NS3 and NS5B, possibly as components of a multisubunit replicase. Compounds that target specific catalytic processes in single-subunit assays will be useful probes for studying replicase action and composition.
Further development will allow biochemical characterization of the process of HCV RNA replication, particularly the molecular composition of the active HCV replicase. Although we have presented indirect evidence that de novo initiation of RNA synthesis is occurring, efforts in the future will be directed toward exploring this possibility by more detailed product analysis. Certainly, the ability of the replicase to copy exogenous HCV RNA templates would greatly facilitate studies on initiation and on cis and trans replication elements. Unfortunately, we have thus far failed to detect replication of a template RNA added to the membrane fractions containing the replicase.
Another potential use of the cell-free system is to screen antiviral compounds directed against activities required for RNA synthesis. Further streamlining and scale-up will be required to use it as a primary screen; however, it could have immediate utility as a secondary screen for compounds identified as inhibitory in single-subunit or cell-based assays. It is interesting that the compounds used in the present study displayed a higher IC50 than they did in the enzymatic assays with purified recombinant protein in which they were initially identified. The reason(s) for this difference in inhibition is unclear, but it could be an issue of accessibility. If the protein against which a compound is targeted is in a complex with a number of other viral and cellular proteins, then access to the site of inhibitor binding may be limited, thus requiring a higher concentration of compound.
Given the effects of different divalent cations on the production of full-length RNA, it will be interesting to take this into account when inhibitory compounds are tested. Gliotoxin, a fungal metabolite, has been demonstrated to inhibit both purified HCV NS5B and poliovirus 3Dpol activity (12, 37). A second report showed that the inhibitory effect of gliotoxin on HCV NS5B was dependent on the divalent cation present in the reaction; effective inhibition occurred when Mn2+ was present but not when Mg2+ was used (2). Interestingly, when we tested gliotoxin in our system, no inhibition of full-length RNA synthesis occurred with either divalent cation (data not shown). This result again emphasizes possible differences between inhibitor action on a multisubunit replicase versus purified enzymatic components.
In conclusion, this crude cell-free system provides a valuable complement to single-subunit assays for biochemical dissection of HCV RNA replication and for evaluation of inhibitory compounds and determining their mode of action.
Acknowledgments
We thank Paul Olivo and Gary Franklin of Apath L.L.C. for helpful comments and technical advice and Darius Moradpour and Shirley Wong for providing HCV-specific antibodies.
This work was supported by Public Health Service grants AI24134 and CA57973 and the Greenberg Medical Research Institute. R.W.H. was supported by an Individual National Research Service Award from the Public Health Service (GM20451), and J.M. is a Merck Fellow of the Life Sciences Research Foundation.
REFERENCES
- 1.Adkins, S., S. S. Stawicki, G. Faurote, R. W. Siegel, and C. C. Kao. 1998. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4:455-470. [PMC free article] [PubMed] [Google Scholar]
- 2.Alaoui-Lsmaili, M. H., M. Hamel, L. L'Heureux, O. Nicolas, D. Bilimoria, P. Labonte, S. Mounir, and R. F. Rando. 2000. The hepatitis C virus NS5B RNA-dependent RNA polymerase activity and susceptibility to inhibitors is modulated by metal cations. J. Hum. Virol. 3:306-316. [PubMed] [Google Scholar]
- 3.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1988. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J. Virol. 62:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. DeFrancesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for the genetic study of subgenomic and genomic HCV replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 9.Bressanelli, S., L. Tomei, F. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamberlin, M. 1974. Selectivity of transcription. Annu. Rev. Biochem. 43:721-775. [DOI] [PubMed] [Google Scholar]
- 11.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallinari, P., D. Brennan, C. Nardi, M. Brunetti, L. Tomei, C. Steinkühler, and R. De Francesco. 1998. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 72:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. N. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 16.Hong, Z., E. Ferrari, J. Wright-Minogue, R. Chase, C. Risano, G. Seelig, C. G. Lee, and A. D. Kwong. 1996. Enzymatic characterization of hepatitis C virus NS3/4A complexes expressed in mammalian cells by using the herpes simplex virus amplicon system. J. Virol. 70:4261-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoofnagle, J. H. 1997. Hepatitis C: the clinical spectrum of disease. Hepatology 26:15S-20S. [DOI] [PubMed] [Google Scholar]
- 18.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 19.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashiwagi, T., K. Hara, M. Kohara, K. Kohara, J. Iwahashi, N. Hamada, H. Yoshino, and T. Toyoda. 2002. Kinetic analysis of C-terminally truncated RNA-dependent RNA polymerase of hepatitis C virus. Biochem. Biophys. Res. Commun. 290:1188-1194. [DOI] [PubMed] [Google Scholar]
- 21.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyono, K., M. Miyashiro, and I. Taguchi. 1998. Detection of hepatitis C virus helicase activity using the scintillation proximity assay system. Anal. Biochem. 257:120-126. [DOI] [PubMed] [Google Scholar]
- 24.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott/The Williams and Wilkins Co., Philadelphia, Pa.
- 26.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 29.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClure, W. R. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 54:171-204. [DOI] [PubMed] [Google Scholar]
- 31.Moradpour, D., E. Bieck, T. Hugle, W. Wels, J. Z. Wu, H. E. Blum, and R. Bartenschlager. 2002. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:593-601. [DOI] [PubMed] [Google Scholar]
- 32.Morgenstern, K. A., J. A. Landro, K. Hsiao, C. Lin, Y. Gu, M. S.-S. Su, and J. A. Thomson. 1997. Polynucleotide modulation of the protease, nucleoside triphosphatase, and helicase activities of a hepatitis C virus NS3-NS4A complex isolated from transfected COS cells. J. Virol. 71:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh, J.-W., T. Ito, and M. M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh, J.-W., G. T. Sheu, and M. M. C. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 35.Osman, T. A., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by template-dependent, membrane bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez, P. L., and L. Carrasco. 1992. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral polymerase 3Dpol. J. Virol. 66:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, J.-H., S. Adkins, G. Faurote, and C. C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligo ribonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 39.Tabor, S., and C. C. Richardson. 1989. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. USA 86:4076-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomei, L., C. Failla, E. Santolini, R. DeFrancesco, and N. La Monica. 1993. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 67:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 42.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, C., P. Sarnow, and A. Siddiqui. 1994. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 68:7301-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalm, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wölk, B., D. Sansonno, H.-G. Kräusslich, F. Dammacco, C. M. Rice, H. E.Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4a complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 47.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 48.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]