Abstract
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Studies of HCV replication and pathogenesis have so far been hampered by the lack of an efficient tissue culture system for propagating HCV in vitro. Although HCV is primarily a hepatotropic virus, an increasing body of evidence suggests that HCV also replicates in extrahepatic tissues in natural infection. In this study, we established a B-cell line (SB) from an HCV-infected non-Hodgkin's B-cell lymphoma. HCV RNA and proteins were detectable by RNase protection assay and immunoblotting. The cell line continuously produces infectious HCV virions in culture. The virus particles produced from the culture had a buoyant density of 1.13 to 1.15 g/ml in sucrose and could infect primary human hepatocytes, peripheral blood mononuclear cells (PBMCs), and an established B-cell line (Raji cells) in vitro. The virus from SB cells belongs to genotype 2b. Single-stranded conformational polymorphism and sequence analysis of the viral RNA quasispecies indicated that the virus present in SB cells most likely originated from the patient's spleen and had an HCV RNA quasispecies pattern distinct from that in the serum. The virus production from the infected primary hepatocytes showed cyclic variations. In addition, we have succeeded in establishing several Epstein-Barr virus-immortalized B-cell lines from PBMCs of HCV-positive patients. Two of these cell lines are positive for HCV RNA as detected by reverse transcriptase PCR and for the nonstructural protein NS3 by immunofluorescence staining. These observations unequivocally establish that HCV infects B cells in vivo and in vitro. HCV-infected cell lines show significantly enhanced apoptosis. These B-cell lines provide a reproducible cell culture system for studying the complete replication cycle and biology of HCV infections.
Hepatitis C virus (HCV) has been the major etiological agent of posttransfusion non-A, non-B hepatitis and currently afflicts >100 million people worldwide. Acute HCV infection is usually subclinical without obvious symptoms. About 15 to 20% of patients can mount a successful immune response to clear the virus in the acute phase; however, 80 to 85% of patients become chronic carriers, and these patients are at high risk of developing liver cirrhosis and/or hepatocellular carcinoma.
Besides causing liver pathology, HCV infection is frequently associated with mixed cryoglobulinemia, non-Hodgkin's B-cell lymphoma, and Sjögren's syndrome, all of which involve B-cell proliferation (8, 10, 27, 37, 49; P. Pioltelli, G. Zehender, G. Minti, A. Monteverde, and M. Galli, Letter, Lancet 347:624-625, 1996), suggesting that HCV may infect B cells or affect B-cell functions in natural infection. Negative-strand HCV RNA has been detected by reverse transcriptase (RT) PCR in the peripheral lymphocytes, bone marrow, lymph nodes, and central nervous system of some HCV patients (23, 30, 34). Analysis of positive-strand HCV RNA sequences and quasispecies patterns suggested that HCV RNAs in these cells are different from those in the serum (22). However, the possibility that HCV replicates in extrahepatic cells remains controversial because of the lack of isolation and characterization of viruses from the infected cells. Further, the use of RT-PCR for detection of viral RNA in these studies could not rigorously rule out possible contamination by the virus from the serum. Several laboratories have also shown that HCV can infect B-cell (30), T-cell (18, 32, 39), and hepatoma cell (14, 41) lines in culture, but the infection is usually transient and inefficient. Nevertheless, these studies suggested that B or T cells could support HCV replication, albeit inefficiently, at least in vitro.
The molecular cloning of the HCV genome has made possible the delineation of the gene functions and the potential mechanism of pathogenesis of this virus. Recently, establishment of self-replicating HCV subgenomic (2, 25) and genomic (13, 33) replicons in Huh-7 cells has also provided an important new tool for the study of HCV replication mechanisms. However, these systems do not allow the study of viral infection or assembly or virus particle production (33). Furthermore, the HCV replicon cannot replicate in cell lines other than Huh-7. The utility of the HCV replicon system for studying the pathogenesis of HCV or the biology of the complete HCV life cycle is thus limited.
In this study, we established three in vivo HCV-infected B-cell lines directly from chronically HCV-infected patients. At least one of these B-cell lines (SB) persistently produces infectious virions. HCV produced from the HCV-infected B cells could establish secondary infection in primary human hepatocytes and lymphocytes in vitro. The establishment of B-cell lines infected with HCV in vivo provides unequivocal evidence that HCV infects B cells during the course of natural infection. Using these cell lines, we demonstrated that HCV infection causes cytopathic effects in B cells. These cell systems will be useful for studying the biology of the complete replication cycle of viral infection.
MATERIALS AND METHODS
Cell culture and virus infection. (i) Establishment of SB cells.
SB cells were isolated from the spleen of an HCV-infected patient with type II mixed cryoglobulinemia and monocytoid B-cell lymphoma. The spleen was surgically removed as part of clinical patient care. The mononuclear cells were isolated by pressing the spleen tissue through stainless steel mesh and purified by centrifugation through 30% Percoll solution. The cells were maintained in standard RPMI 1640 medium with 20% fetal bovine serum (FBS) without any supplement. Every 5 days, the cells were sedimented by natural gravity for 30 min at 37°C. Half of the medium was removed and replaced with an equal volume of fresh medium.
(ii) Establishment of B-cell lines from PBMCs of HCV-infected patients.
Peripheral blood mononuclear cells (PBMCs) were isolated from HCV-infected individuals by Ficoll-Paque centrifugation (Amersham Pharmacia Biotech, Wikströms, Sweden). Ten million mononuclear cells were suspended in 2.5 ml of RPMI 1640 containing 20% FBS and incubated with 2.5 ml of fresh Epstein-Barr virus (EBV) preparation produced from exponentially growing B95-8 cells (43). Two hours later, 5 ml of RPMI 1640 containing 20% FBS and 1 μg of cyclosporine (Sigma, St. Louis, Mo.)/ml was added. The EBV-immortalized B cells were observed after 2 to 3 weeks. Then, the cells were passaged by partial medium change every 3 to 4 days as described above.
(iii) In vitro infection of Raji cells and PBMCs.
To establish HCV infection of Raji cells, 40 ml of SB cell culture supernatant was collected, filtered through a 0.45-μm-pore-size filter, and concentrated to ∼5 ml by ultrafiltration using Centripuls YM-100 (Millipore Corp.-Amicom Bioseparations, Bedford, Mass.). One milliliter of the concentrated supernatant was incubated with 1 × 106 to 5 × 106 Raji cells in 3 to 5 ml of RPMI 1640 containing 20% FBS. After 5 days, the culture supernatant was removed and washed three times with prewarmed PBS. The cells were maintained at 37°C in RPMI 1640 containing 20% FBS, and the media were partially changed every 3 to 4 days as described above.
To establish HCV infection of B cells from normal PBMCs, the PBMCs from a healthy individual were isolated by Ficoll-Paque centrifugation. To serve as a control, half of the concentrated SB culture supernatant (5 ml) was spread on a 60-mm-diameter plate and placed under a germicidal UV lamp at a distance of 50 cm for 2 to 5 min to inactivate the virus (26). The untreated or UV-irradiated SB supernatant (2.5 ml) was then added, together with 2.5 ml of fresh EBV preparation, to 107 PBMCs. After 2 hours of incubation, 5 ml of RPMI 1640 containing 20% FBS and 1 μg of cyclosporine/ml was added. After 5 days, the cells were washed three times with prewarmed PBS, and fresh medium containing cyclosporine was added. One month later, the surviving cells were transferred to RPMI 1640 containing 20% FBS and maintained in culture. The cell line was designated JT.
(iv) Single-cell cloning.
To determine the percentage of infected cells containing HCV RNA, the cells were cultured by single-cell limiting dilution in a 96-well U-bottom plate. After 2 to 3 weeks, the plate was duplicated and cultured for another week. The duplicated plate was used for nested RT-PCR to detect HCV RNA in each clone.
(v) HCV infection of primary human hepatocytes.
Primary human hepatocytes were obtained from the Liver Center of the Keck School of Medicine, University of Southern California. Three million cells were seeded on a 60-mm-diameter collagen-coated plate. Six hours later, nonadherent cells were removed and the adherent cells were washed twice with culture medium and grown in serum-free culture medium as described previously (42). The concentrated SB culture supernatant was then added to the hepatocytes, and the medium was changed daily to maintain the viability of the hepatocytes. Hepatocyte culture supernatant was collected at various times and used for HCV RNA detection by RT-PCR.
Immunofluorescence staining.
B cells in suspension were cytospun to slides, fixed with 4% paraformaldehyde, and permeabilized with 0.4% Triton X-100. The cells were then stained with either a polyclonal rabbit anti-core antibody (1:200) or a monoclonal anti-NS3 antibody (1:50) (Novocastra Laboratories Ltd., Newcastle upon Tyne, United Kingdom). The bound antibody was detected with a fluorescein isothiocyanate-conjugated anti-rabbit antibody or anti-mouse antibody. Positive cells were observed under a confocal or immunofluorescence microscope.
Detection of intracellular HCV NS3 protein expression by flow cytometry.
Half a million SB cells or 1 million Raji cells were suspended in 100 μl of phosphate-buffered saline (PBS)-0.1% saponin-1 mM CaCl2-1 mM MgSO4-0.05% NaN3-1% bovine serum albumin-10 mM HEPES (PBS-S)-5% nonfat dry milk solution and incubated at room temperature for 1 h with gentle rocking. Monoclonal anti-NS3 antibody (1:25; Novocastra Laboratories, Ltd.) was added to the cell suspension and incubated at room temperature for 1 h. As an isotype control, 1 μg of purified mouse immunoglobulin G2b (IgG2b) monoclonal immunoglobulin isotype standard (BD Biosciences)/ml was added to the cell suspension. After incubation for 1 h, the cells were washed three times with PBS-S buffer. Then, the cells were stained with 1 μg of fluorescein-conjugated goat anti-mouse IgG2b antibody (Southern Biotechnology Associates, Inc.)/ml and incubated at room temperature for 30 min. After being washed, the cells were fixed with 1% paraformaldehyde and analyzed by flow cytometry.
RPA.
Total RNA was extracted using the acid-guanidinium isothiocyanate-phenol-chloroform method (5). One hundred micrograms of total cellular RNA was used for the RNase protection assay (RPA) according to the published method (24) with slight modifications. The antisense (AS) and sense (S) probes used in RPA were cloned by RT-PCR from the 5′ untranslated region (UTR) of HCV RNA in SB cells into the pBluescript SK vector (Stratagene, La Jolla, Calif.). The 32P-labeled AS or S riboprobe was generated by in vitro transcription of the plasmid with T7 or T3 RNA polymerase (Ambion, Austin, Tex.). For detecting negative-strand HCV RNA, total cellular RNA was pretreated with RNase A (0.5 μg/ml; Ambion) to remove excess positive-strand RNA, extracted with phenol-chloroform to remove RNase A, and precipitated. For detecting positive-strand HCV RNA, total RNA was processed by the same procedure as for negative-strand RNA except that the RNA was not pretreated with RNase A. The precipitated RNA was suspended in 30 μl of hybridization buffer {40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 6.7), 400 mM NaCl, 1 mM EDTA, 80% formamide} containing 2 × 105 cpm of 32P-labeled cRNA probe. Hybridization was carried out at 58°C for 12 to 15 h. After hybridization, the RNA was digested with 10 μg of RNase A/ml and 0.5 μg of RNase T1/ml in 10 mM Tris-HCl (pH 7.5)-5 mM EDTA-300 mM NaCl. The RNA was then resolved on a 6% denaturing polyacrylamide gel.
Detection of positive- and negative-strand HCV RNA by RT-PCR.
Total RNA was extracted by acid-guanidinium thiocyanate-phenol-chloroform from cells or serum. For positive-strand HCV RNA, reverse transcription was performed using primer HCV-282r (5′-CACTCGCAAGCACCCTATCAG-3′) (12); the cDNA was then amplified by PCR using the primers 5′-TTCACGCAGAAAGCGTCTAGCCAT-3′ and 5′-TCGTCCTGGCAATTCCGGTGTACT-3′ for 40 cycles (94°C for 1 min, 58°C for 1.5 min, and 72°C for 1.5 min). For negative-strand-specific RT-PCR, 5 μg of total RNA was mixed with a sense primer, 5′-CGCGCGACTAGGAAGACTTC-3′ (23), and reverse transcribed at 65°C for 30 min using C. therm polymerase (Roche Molecular Biochemicals, Mannheim, Germany). One-fifth of the cDNA was then amplified using AmpliTaq DNA polymerase (Perkin-Elmer Corp.-Applied Biosystems, Foster City, Calif.) in the presence of 0.5 M betaine plus the sense primer and an antisense primer, 5′-ATAGAGAAAGAGCAACCAGG-3′ (23), for 35 cycles, each at 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. One-tenth of the PCR product was analyzed by agarose gel electrophoresis and Southern blot hybridization with a 32P-labeled probe, 5′-GCCGACCTCATGGGGTACAT-3′, which is internal to the PCR primers. Synthetic HCV negative- and positive-strand RNAs were generated by in vitro transcription and used for determining the sensitivity and specificity of the reaction.
Single-stranded conformational polymorphism (SSCP) analysis.
SSCP analysis of hypervariable region 1 (HVR1; amino acids 384 to 414 from the N terminus of the HCV polyprotein) was performed as previously described (6). In brief, to amplify the region flanking HVR1, a nested hot-start RT-PCR was performed with the primers described by Weiner et al. (X14, X18, X4, and X19) (45). One microliter of the PCR product was diluted in SSCP loading buffer (95% formamide, 20 mM EDTA [pH 8.0], 0.05% bromophenol blue, and 0.05% xylene cyanol) to a final concentration of 50 ng/μl. Five microliters of the diluted PCR product was preheated at 95°C for 5 min, chilled on ice, loaded onto a 10% polyacrylamide gel, and electrophoresed at 10 mA for 5 h. The temperature of the electrophoresis buffer was kept at 20°C. Bands on the gel were detected by silver staining.
Quantitation of HCV RNA by real-time RT-PCR.
HCV RNA in the cell culture supernatant was quantified by real-time RT-PCR using the TaqMan Chemistry System (Perkin Elmer Corp.-Applied Biosystems). Total RNA was extracted from the cells or concentrated culture supernatant by the acid-guanidinium thiocyanate-phenol-chloroform method (5). In vitro-transcribed full-length HCV RNA was used as a standard. Real-time RT-PCR was performed using a sense primer (5′-TGCGGAACCGGTGAGTACA-3′), an antisense primer (5′-CTTAAGGTTTAGGATTCGTGCTCAT-3′), TaqMan probe [5′-6-FAM d(TTGGGTTGCGAACGGCCTTGTGGTAC)BHQ-1-3′], and the TaqMan Gold RT-PCR kit (Applied Biosystems) with the ABI Prism 7900 sequence detector system. After an initial reverse-transcription reaction at 48°C for 30 min followed by inactivation of RT and activation of AmpliTaq Gold DNA polymerase at 95°C for 10 min, PCR amplification was performed for 55 cycles (denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min). Following amplification, real-time data acquisition and analysis were performed using SDS 2.0 software (Perkin-Elmer Corp.-Applied Biosystems).
Immunoblotting.
The virus particles in the SB cell culture supernatant (from 180 ml) were pelleted by ultracentrifugation with a 20% sucrose cushion. The pellet and 50 μg of total cell lysate were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Polyclonal rabbit anti-HCV core (1:1,000) or monoclonal anti-HCV NS3 antibody (1:50) was used for immunoblotting.
Quantitation of core protein in cell culture supernatant.
The presence of HCV core proteins in culture supernatant of SB cells or control cell lines was investigated using a commercially available enzyme immunoassay (Ortho trak-C; Ortho Clinical Diagnostics, Raritan, N.J.). Briefly, 100 μl of 10- to 20-fold-concentrated culture supernatant was mixed with 50 μl of the sample pretreatment buffer and incubated at 56°C for 30 min in Eppendorf tubes. After cooling down, 100 μl of the sample diluent and 100 μl of the treated sample were added to the detection plate, and the plate was incubated for 1 h at room temperature with shaking. After being washed, 200 μl of anti-core horseradish peroxidase conjugate was added to each well, and the plate was incubated at room temperature for 30 min. The reaction was developed with the addition of o-phenylenediamine · 2HCl. The amount of HCV core protein was determined by interpolation of the optical density of the sample against a standard curve.
HCV RNA was quantified in the culture supernatant of SB cells by the HCV Amplicor Monitor assay (version 2.0) (Roche Molecular Systems, Pleasanton, Calif.). Amounts of HCV RNA in the culture supernatants of control cell lines were determined by a nested-RT-PCR method.
Equilibrium sucrose gradient ultracentrifugation.
One milliliter of the 10-fold-concentrated SB culture supernatant was layered on top of 3.5 ml of a discontinuous density gradient of sucrose prepared from 0.7 ml each of 60, 50, 40, 30, and 10% (wt-vol) sucrose in NTE buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA) and centrifuged at 36,000 rpm in a Beckman SW55 rotor for 16 h at 4°C. Fractions (350 μl each) were collected from the bottom of the tube. The density of each fraction was determined using the Abbe Mark II Plus Refractometer model 10494. HCV RNA from each fraction was quantitated by TaqMan real-time RT-PCR as described above.
Apoptosis assay.
The terminal deoxyribonucleotidyltransferase (TdT)-mediated dUTP-biotin nick end-labeling (TUNEL) assay was performed using a TdT-FragEL or Fluorescein-FragEL DNA fragmentation detection kit (Oncogen, Darmstadt, Germany) according to the manufacturer's instruction with slight modification. For costaining of HCV NS3 protein and apoptotic cells, the cells were first stained for apoptosis; after the final wash, monoclonal anti-NS3 antibody was added and the cells were incubated at 37°C for 30 min. After the cells were washed with PBS three times, rhodamine-labeled goat anti-mouse antibody was added and the cells were incubated for 30 min. Finally, the cells were mounted with Fluorescein-FragEL mounting medium (Oncogen) containing DAPI (4′,6′-diamidino-2-phenylindole). The annexin V-binding assay was performed using the annexin V-fluorescein isothiocyanate apoptosis detection kit (Oncogen).
RESULTS
Establishment of SB cell line.
Because of the strong association of HCV infection with non-Hodgkin's B-cell lymphoma (8, 37, 49) and the various kinds of circumstantial evidence implicating the presence of HCV RNA in PBMCs of HCV-infected patients, we attempted to establish B-cell tumor cell lines from HCV-infected patients in the hope that some B-cell lines may carry persistent infection of HCV. Such cell lines would be useful for studying the biology of HCV infection, and their existence would unequivocally establish that HCV can infect lymphoid cells in vivo. We started with an HCV-infected patient with type II mixed cryoglobulinemia and monocytoid B-cell lymphoma. The mononuclear cells were cultured, and a permanent cell line (named SB cells) was successfully established. These cells have been maintained in culture for >15 months, with a doubling time of ∼4.3 days. The cells were subsequently shown to be a monoclonal B-cell line with a rearranged IgM gene but without IgG class switching (data not shown). The cells were determined to be devoid of EBV DNA.
SB cells are persistently infected by HCV.
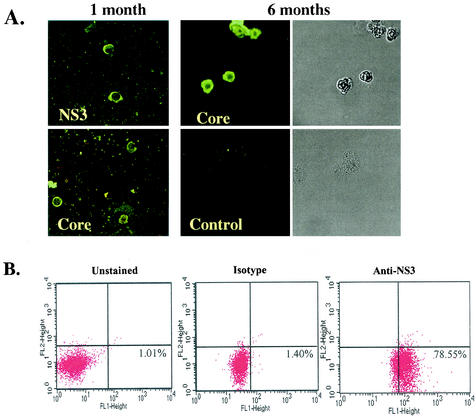
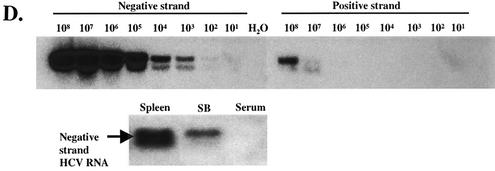
To determine whether these cells were infected with HCV, we performed immunofluorescence staining of HCV proteins in SB cells (Fig. 1A). At 1 month of culture, HCV core protein and the nonstructural protein NS3 could be detected in some of the cells. At 6 months, cytoplasmic staining of core protein was detected in almost all the cells, in contrast to the lack of staining in control Raji cells, an established EBV+ B lymphoblastoid cell line (LCL). Approximately 77% of SB cells stained positive for HCV NS3 protein as detected by flow cytometry (Fig. 1B). We also used an RPA to detect HCV RNA (Fig. 1C). We were able to detect ∼1 ng of HCV positive-strand RNA in 100 μg of total cellular RNA. However, HCV negative-strand RNA could not be detected by RPA in the same amount of SB cell RNA. Therefore, a negative-strand-specific RT-PCR, followed by Southern blotting, was employed. This method could distinguish the negative-strand from the positive-strand HCV RNA by 106-fold (Fig. 1D, top). The negative-strand HCV RNA could be seen in both the original spleen tissue and SB cells even after 1 year in culture, but not in the patient's serum (Fig. 1D, bottom). Quantitation of the negative-strand-RNA signal suggests the detection of <1 copy of HCV negative-strand RNA per SB cell. This low copy number could be due to the instability of HCV negative-strand RNA and/or the low sensitivity of the negative-strand-specific RT-PCR (a sacrifice for the high specificity). These data together indicate that HCV replicates in both the spleen and SB cells even after a prolonged period.
FIG. 1.
SB cells are persistently infected by HCV. (A) Detection of HCV protein expression by immunofluorescence staining in SB cells at different time points of culture. Uninfected Raji cells were used as a control. Phase contrast images of SB cells (upper) and uninfected Raji cells (lower) are shown on the right. (B) Detection of intracellular NS3 protein expression in SB cells by flow cytometry. Unstained SB cells served as the “no-antibody” control; isotype control antibody served as the background staining control. (C) Detection of positive-strand HCV RNA in SB cells by RPA. The expected size of the protected band from HCV RNA in SB cells is 123 bp. Different amounts of in vitro-transcribed sense and antisense HCV RNA (IVT-S and IVT-AS) were used for quantitation. The AS probe was used for detecting positive-strand RNA, and the S probe was used for detecting negative-strand RNA. Because of the presence of vector sequences in both the probes and the in vitro-transcribed RNA, the protected sizes for IVT-S and IVT-AS are 215 and 233 bp, respectively. HCV(+), SB RNA hybridized to the AS probe; HCV(-), same RNA hybridized to the S probe. (D) Detection of negative-strand HCV RNA by strand-specific RT-PCR and Southern blotting. The top blot shows the validation of the strand specificity of the assay. Serial dilutions of both positive- and negative-strand in vitro-transcribed HCV RNAs from 108 molecules to 1 molecule were subjected to negative-strand-specific RT-PCR using the sense primer for reverse transcription. The bottom blot shows that negative-strand HCV RNA was detected in the spleen and SB cells but not in the patient's serum.
Characterization of HCV RNA quasispecies in SB cells.
To identify the HCV RNA species in SB cells, we first cloned and sequenced the 5′ UTR and NS5B region of HCV RNA. Based on a sequence homology search, the HCV RNA from SB cells was determined to belong to genotype 2b (data not shown).
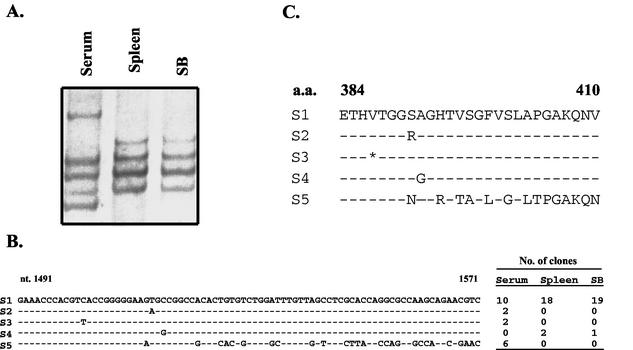
To understand the origin of HCV in SB cells, we performed RT-PCR of the HVR1 and analyzed the HCV quasispecies by the SSCP method (6). Figure 2A shows that the HCV quasispecies pattern in SB cells is very similar to that in the spleen but different from that in the serum of the same patient. These regions were cloned and sequenced. The results showed that HCV RNA in the serum consists of at least four different species, three of which are unique to the serum HCV. In contrast, those in the spleen and SB cells consist of a predominant RNA species plus a single minor RNA species (S4) which was not detected in the serum. This result indicates that HCV RNA in SB cells is most likely derived from the spleen, which may have harbored a distinct HCV quasispecies population. The HCV quasispecies is more heterogeneous in the serum, which may reflect the HCV species in the liver. However, we were unable to obtain liver biopsy tissue to confirm this interpretation. This finding nevertheless indicates that the origin of HCV in SB cells is the spleen but not the patient's serum and establishes that HCV replicates in B cells during natural infection.
FIG. 2.
HCV quasispecies of the SB cell. (A) SSCP analysis of HCV quasispecies from the patient. (B) Nucleotide sequences of the HVR (nucleotides [nt] 1491 to 1571) of HCV RNA from SB cells (1 month old) and spleen and serum of the same patient. HVR1 was amplified by nested RT-PCR, and the PCR products were cloned into the pCRII-TOPO vector (Invitrogen). Twenty independent clones from each sample were sequenced from both ends. (C) Amino acid sequences of the HVR (amino acids [a.a.] 384 to 410) deduced from the sequences in panel B. The dashes represent nucleotides or amino acids identical to those of S1. The asterisk represents a silent mutation.
Establishment of EBV-immortalized B-lymphoid cell lines from HCV-infected patients.
The successful establishment of SB cells clearly indicates that B cells are infected with HCV in vivo. However, since SB cells were derived from a B-cell lymphoma, we were not certain whether HCV infection of B cells occurs only in tumor cells or is a general phenomenon associated with HCV infection. Previous studies addressing this question relied on RT-PCR detection of viral RNA in PBMCs (22, 30). To unequivocally resolve the issue, we attempted to establish B-cell lines by EBV immortalization of PMBCs from HCV-infected patients without B-cell lymphoma. We succeeded in establishing several EBV-immortalized B-lymphoid cell lines from four different HCV-infected individuals. Two of these cell lines were positive for HCV RNA by RT-PCR and for NS3 protein by immunofluorescence staining (Fig. 3). The HCV core sequences from both cell lines were cloned and sequenced to identify the genotype of the HCV RNA from the two cell lines. HCV RNAs from both cell lines are genotype 1b (data not shown). HCV RNA remained positive after 3 months of culture in these cell lines. Since immunofluorescence studies of the cell lines showed only a small proportion of cells to be positive for the HCV NS3 protein (Fig. 3), we performed single-cell cloning of these cells by limiting dilution; each clone was then grown into 106 cells and used for RT-PCR analysis of HCV RNA. About 12.5% of 011A cells and ∼7.5% of 016A cells were positive for HCV RNA (data not shown). Together, these data provide further evidence that HCV infects B cells in vivo.
FIG. 3.
Establishment of B-cell lines from PBMCs of HCV-infected patients by EBV immortalization. Shown are indirect immunofluorescence staining of HCV NS3 protein on EBV-immortalized B cells from patients 011A (A) and 016A (B) after 2 months in culture (top) and detection of HCV RNA by RT-PCR after 1 month in culture (bottom). The 5′ UTR was used for RT-PCR. PCR without an RT reaction was used as a negative control for cDNA contamination.
SB cells produce HCV particles.
We next examined whether any of these cell lines produced HCV particles. Among the cell lines, we found that only SB cells consistently release a significant amount of HCV RNA into the supernatant. The positive-strand HCV RNA was detected by RT-PCR in the SB cell culture supernatant (Fig. 4A, left). Real-time RT-PCR analysis showed that the amount of HCV RNA released from SB cell culture steadily increased to ∼28,000 copies per ml during 5 days of culture (Fig. 4A, right), indicating that the virus is continuously released. Furthermore, immunoblotting of the concentrated culture medium detected HCV core protein (Fig. 4B). Only a trace amount of the core protein could be detected in the cell lysate, probably because the amount of cellular lysate that could be loaded onto the gel was limited (Fig. 4B, bottom). In contrast, the NS3 protein was detected in the SB cell lysate but not in the culture supernatant (Fig. 4B, top).
FIG. 4.
SB cells continuously produce HCV particles. (A) On the left is shown the detection of HCV RNA by RT-PCR from SB cells and culture supernatant. Uninfected Raji cells were used as a negative control. On the right is shown the quantitation by real-time RT-PCR of HCV RNA in the SB cell culture supernatant during 5 days of culture after medium change. One hundred million SB cells were cultured in 10 ml of medium, and 1 ml was harvested for RT-PCR every day. (B) Western blotting of HCV NS3 (top) and core protein (bottom) in the cell lysate (Cells) and culture supernatant (Sup.). The asterisks indicate HCV core or NS3 protein. (C) Equilibrium sucrose gradient sedimentation of the SB cell culture supernatant. The HCV RNA in each fraction was measured by real-time RT-PCR.
The HCV core protein in the SB culture supernatant was quantified by the Ortho trak-C assay, designed to quantify total HCV core protein in samples of serum or plasma. As controls, we included seven supernatant samples from overgrown (>15 × 106 cells/ml) EBV-transformed LCLs established from HIV+ HCV+ patients. These cell lines have been in culture for a mean time of 6 months and were negative for HCV RNA by nested RT-PCR. In previous experiments, we noticed that the Ortho trak-C assay exhibited some background when supernatants from overgrown tissue cultures were used, but minimal or no background was observed when PBMC lysates or liver homogenates were used (G. Picchio, unpublished observations). Table 1 summarizes the results obtained from two independent experiments. After subtracting the mean background level obtained with the control LCL, we determined that the 10-fold-concentrated SB culture supernatant contained 20.4 (experiment 1) and 21.2 (experiment 2) pg of core protein/ml (mean concentration, 20.8 pg/ml). The concentration of HCV RNA in the SB supernatant was 203,903 IU/ml. The HCV RNA/core protein ratio (mean, 9,803) found in the supernatant of SB cells is closely similar to the one reported in serum samples of HCV-infected individuals (mean, 8,000) (3).
TABLE 1.
Core protein concentrations in tissue culture supernatants
| Cell line supernatanta | Concn (pg/ml)
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| LCL1 | 5.8 | 5.4 |
| LCL2 | 4.3 | 4.2 |
| LCL3 | 4.0 | 3.8 |
| LCL4 | 7.3 | 7.1 |
| LCL5 | 4.7 | 4.5 |
| LCL6 | 3.7 | 3.7 |
| LCL7 | 3.9 | 4.0 |
| LCL (mean background) | 4.8 | 4.6 |
| SB | 25.4 | 27.1 |
| SB (corrected) | 20.6 | 22.5 |
| RPMI + 10% FBSb | 0.5 | 0.1 |
LCL1 to 7 are independent EBV-transformed LCLs established from HIV+ HCV+ patients.
Below lower limit of detection.
The buoyant density of the HCV particles from SB culture supernatant in sucrose was 1.13 to 1.15 g/ml (Fig. 4C). A second small peak at fraction 5 with a density of 1.2 g/ml was occasionally observed, which may represent the nucleocapsid of HCV, as previously described (17). Further characterization of HCV virions from SB culture supernatant is in progress. Together, these data indicate that SB cells produce HCV particles.
HCV from SB cells could infect primary human hepatocytes and other B cells.
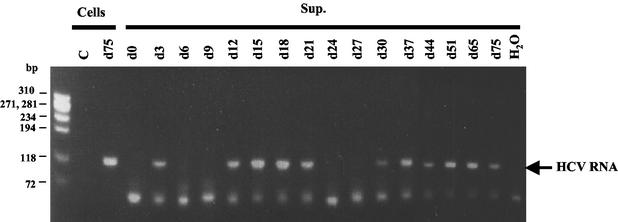
To determine whether the virus particles produced by this cell line were infectious, the SB cell culture supernatant was used to infect primary human hepatocytes. After inoculation (day zero), the medium was changed every day and harvested for RT-PCR analysis. On day 3, positive-strand HCV RNA was detected in the supernatant, which likely represents the virus remaining from the initial inoculum (Fig. 5). On day 6 and day 9, no viral RNA was detected in the supernatant. From day 12 to day 21, viral RNA was again detected in the supernatant. HCV RNA was undetectable on day 24 and day 27, but it came back on day 30 and lasted up to day 75, shortly after which the cells died. In this culture, we observed the appearance of some cell foci of hepatocytes (data not shown). The fluctuation of HCV RNA in hepatocyte culture supernatant was reproducible in several primary human hepatocyte cultures derived from different sources, though the length of the culture period varied. This result indicates that SB cells produce infectious viral particles which can infect primary human hepatocytes. The reason for the fluctuation in HCV RNA production is still unclear, but it reflects typical HCV infections in patients and chimpanzees (20, 38, 40).
FIG. 5.
HCV produced from SB cells can infect primary human hepatocytes. Virus produced from primary human hepatocytes infected with SB cell culture supernatant (Sup.) was analyzed by RT-PCR of HCV RNA. Uninfected primary human hepatocytes (Cells) were used as a control (C). Cellular RNA on day 75 (d75) was used as the positive control.
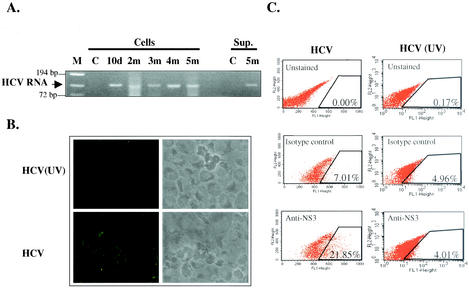
Since the HCV in the SB cell line appears to be derived from the spleen, we next tested whether the virus produced was also infectious for other B cells. We inoculated an established B LCL (Raji cells) with the concentrated SB cell culture supernatant. HCV positive-strand RNA was detectable in both the cell lysate and culture supernatant for up to 5 months (Fig. 6A). HCV NS3 expression could be detected as early as 10 days postinfection (Fig. 6B) and remained positive for up to 5 months in culture; however, the number of positive cells gradually decreased (data not shown). As a control, separate Raji cells were infected with the UV-irradiated culture supernatant from the SB cells. No NS3 protein staining was detected (Fig. 6B). The percentage of NS3-positive Raji cells was determined by flow cytometry to be ∼15% (after subtracting the background staining [isotype control]) (Fig. 6C). However, as in natural HCV infection, the detection of HCV protein expression by immunofluorescence staining is not as sensitive as the detection of HCV RNA by RT-PCR. Therefore, we further determined the percentage of HCV-infected Raji cells by single-cell cloning and detection of HCV RNA in each individual clone by RT-PCR. The results showed that ∼80% of HCV-infected Raji cells contained HCV RNA (Fig. 6D).
FIG. 6.
Infection of Raji cells by HCV from SB cells. (A) Continuous detection of HCV RNA in HCV-infected Raji cells. Raji cells were inoculated with SB culture supernatant (Sup.). Cells or culture supernatant were harvested at various times (d, day; m, month) and used for RT-PCR analysis. C, uninfected Raji cells; M, molecular size marker. (B) Detection of HCV NS3 protein by indirect immunofluorescence staining in HCV-infected Raji cells. Raji cells infected with UV-irradiated SB supernatant [HCV (UV)] were used as a control. (C) Detection of intracellular NS3 protein expression in HCV-infected Raji cells (left) and Raji cells infected with UV-irradiated HCV (right) by flow cytometry. (D) Single-cell cloning of HCV-infected Raji cells. HCV-infected Raji cells were diluted into single cells and grown in a 96-well U-bottom plate. After 2 to 3 weeks, 81 clones were obtained and analyzed by RT-PCR to detect HCV RNA. Sixty-four clones were positive for HCV RNA. A through G designate different groups of cell clones processed for RT-PCR.
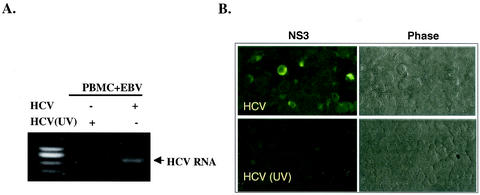
We also used the SB culture supernatant to infect normal PBMCs in primary culture. HCV RNA was detected by RT-PCR in the cells for up to 6 weeks (data not shown), but the PBMCs eventually died. To produce immortalized cell lines, we coinfected normal PBMCs with the SB culture supernatant and EBV. An immortalized B-cell line, named JT cells, was established and found to be positive for HCV RNA as detected by RT-PCR (Fig. 7A) and for NS3 protein by immunofluorescence staining for >3 months in culture (Fig. 7B). As a control for HCV infection and viral replication, PBMCs inoculated with EBV and UV-irradiated SB culture supernatant did not show any HCV RNA or proteins. These data suggest that the virus produced from SB cells can establish persistent infection in other B cells in vitro.
FIG. 7.
In vitro Infection of normal PBMCs with HCV from SB cells. (A) Fresh PBMCs were coinfected with EBV and SB supernatant (HCV) or UV-irradiated SB supernatant [HCV (UV)]. Cells and culture media from 1-month-old culture were harvested, and HCV RNA was detected by RT-PCR. +, present; −, absent. (B) The same cells described in the legend to panel A were used for detection of HCV NS3 protein by immunofluorescence staining 3 months postinfection. Phase, phase-contrast image.
Enhanced apoptosis in HCV-infected B-cell lines.
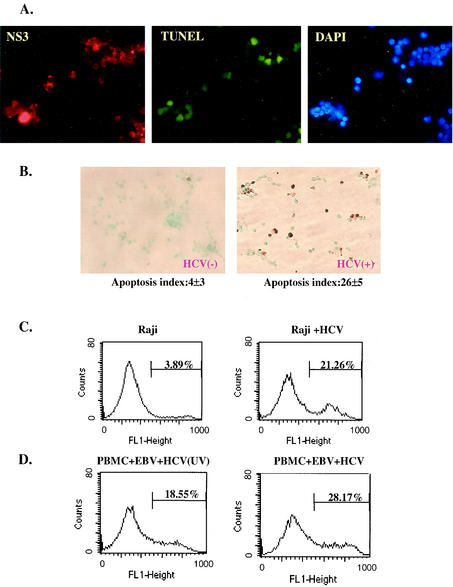
We noted that SB cells had a very low growth rate with a high percentage of apoptotic cells, suggesting a possible cytopathic effect of HCV infection. Trypan blue exclusion assay indeed showed a high rate of cell death in this cell line (data not shown). To determine if the apoptotic cells were positive for HCV, we stained the apoptotic cells with TdT-dependent fluorescein-dUTP followed by anti-NS3 antibody staining for HCV protein expression. Approximately 30% of the SB cells were apoptotic (Fig. 8A), as shown by green fluorescence; all of them also stained positive for HCV NS3 protein.
FIG. 8.
HCV infection causes apoptosis in HCV-infected B cells. (A) SB cells were costained with anti-HCV NS3 antibody, fluorescence-labeled deoxynucleoside triphosphate for TUNEL assay, and DAPI for visualization of all cells. Most of the TUNEL-positive cells were also positive for NS3. (B) Detection of apoptotic cells by TUNEL assay in uninfected Raji cells (left) and HCV-infected Raji cells (right). Apoptotic cells were stained brown. The percentage of apoptotic cells was determined as the average of four fields under the microscope. This experiment was repeated five times. The range of percentages of apoptosis was 1 to 5% for Raji cells and 21 to 29% for HCV-infected Raji cells. (C) Analysis of apoptotic cells by fluorescence-activated cell sorter in HCV-infected (Raji + HCV) and uninfected Raji cells by annexin V-binding assay. (D) Analysis of apoptotic-cell populations in PBMCs coinfected with EBV and SB culture supernatant (HCV) or UV-irradiated SB supernatant [HCV (UV)]. The assays were done on the cells 2 months postinfection.
To establish that B-cell apoptosis is associated with HCV infection, we performed the TUNEL assay on the HCV-infected and uninfected Raji cells (Fig. 8B). The uninfected Raji cells demonstrated 1 to 5% apoptotic cells. In contrast, 21 to 29% of the persistently HCV-infected Raji cells were apoptotic. We also performed an early-apoptosis assay, the annexin V-binding assay; the results showed that 21% of the persistently HCV-infected Raji cells were apoptotic compared to 4% of the uninfected Raji cells (Fig. 8C). This figure is in rough agreement with the results of the TUNEL assay. We also performed a similar study of the immortalized PBMCs infected in vitro with HCV and EBV or with EBV and UV-irradiated HCV. The cells were analyzed by the annexin V-binding assay 30 days after virus infection, when only the immortalized cells survived. The HCV-infected PBMCs showed a significantly higher apoptotic rate than the cells infected with the UV-inactivated virus (Fig. 8D). This result clearly indicates that the higher apoptotic rate of HCV-infected cells was not caused by the components of the culture medium but was associated with HCV infection. The reason for the relatively high apoptotic level of EBV-immortalized HCV− PBMCs compared to the uninfected Raji cells is not clear. Since no inflammatory cells are present in this in vitro culture system, these combined findings indicate that HCV infection and replication cause direct cytopathic effects on B cells.
DISCUSSION
In this report, we have provided unequivocal evidence showing that HCV can infect and replicate in B cells both in vivo and in vitro. HCV replication in B cells in vivo has been a controversial issue which had been addressed by several different reports (21, 22, 28, 47). Previous studies have detected viral RNA (positive and negative strands) in B cells from PBMCs and spleens of some, but not all, HCV patients. However, the virus has never been directly demonstrated or isolated. In our studies, we also found that the sera of some HCV-infected patients were positive for HCV RNA but their PBMCs were negative (data not shown). Our findings here that the 011A, 016A, and SB cell lines, which were established from HCV-infected patients, were positive for HCV proteins and RNA after many months in culture confirmed that B cells indeed were infected by HCV in vivo. In particular, SB cells continue to produce infectious HCV particles in culture. The reason why PBMCs or B cells harbored HCV in some patients but not others is not clear, but it could be related to the virus load, the stage of HCV infection, or the viral genetic sequence. It is possible that the high rate of HCV genetic variation during virus replication may generate and select certain viral genomes to replicate or persist in certain reservoirs. Thus, HCV may evolve into either hepatotropic or lymphotropic strains. Also, the genetic backgrounds of different patients, such as expression of HCV receptors or coreceptors on different cell types in certain patients, may also determine whether lymphoid cells are infected or not. In the two B-cell lines established by immortalization of PBMCs from HCV-infected patients, only 7.5 to 12.5% of the cells were infected. In contrast, at least 77% of SB cells were infected with HCV. Since this cell line is monoclonal (unpublished observation), it is conceivable that HCV-infected B cells were transformed and amplified. In B-cell lines established by in vitro infection with the SB cell culture supernatant, ∼40 (JT) to 80% (Raji) of cells were infected. A possible reason for the high efficiency of B-cell infection in vitro is the absence of T cells in our tissue culture system. These findings indicate the high infectivity of the SB culture supernatant.
Using these cultured cell lines, we have demonstrated that HCV infection causes apoptosis. HCV has previously been regarded as noncytocidal, and HCV pathogenesis has been thought to be mainly due to the immunopathogenic mechanisms (4). The apoptotic effect of HCV infection, at least in B cells, is particularly surprising, in view of the fact that HCV can establish persistent infection in these cell lines. These findings suggest that HCV may also stimulate cell growth to counter the apoptosis. Several viral proteins, such as core or NS5a, may either sensitize cells to apoptotic signals or stimulate cell proliferation to counter apoptosis, probably depending on cellular conditions (9, 11, 16, 29, 35, 36, 46). In our preliminary data, we found that SB cells produce IgM antibody, which recognizes NS3 and presumably IgG-NS3 complex in the body (15). These antigen-antibody interactions may provide the proliferation signal to the virus-infected cells, thus overcoming the HCV-induced apoptosis. Furthermore, PBMCs from patients with chronic hepatitis C associated with type II mixed cryoglobulinemia have been reported to have high frequencies of bcl-2 rearrangements (19, 48), which may induce an antiapoptotic effect. Our preliminary findings suggest that these HCV-infected B cells have increased levels of somatic hypermutation in immunoglobulin genes and some oncogenes (unpublished observation), which may also contribute to the survival of the cells. Raji cells carry an EBV genome that encodes proteins, such as EBNA and LMP-1, which may stimulate cell proliferation (7). Thus, various viral and cellular factors may combine to cause apoptosis and/or stimulation of cellular proliferation.
In our in vitro B-cell infection system, SB culture supernatant could reproducibly infect both established B-cell lines and primary PBMCs. Up to 80% of cultured cells can be infected in vitro. The viral RNA and protein persisted in the infected cells for an extended period in culture. The infectivity of SB culture supernatant appears to be higher than most of the reported in vitro infection using HCV patients' sera, even though the viral RNA titer in SB culture supernatant is lower than those in some patient's sera (usually in the range of 106 copies per ml). It is possible that patients' sera contain antibodies or other inhibitors that may interfere with HCV infection in vitro. It is interesting that HCV particles released in the SB culture supernatant had a buoyant density of 1.13 to 1.15 g/ml, which is considerably higher than most of the reported buoyant densities of HCV particles from the patients' sera (as low as 1.06 g/ml [1, 17, 31, 44]), most likely because of the binding of serum lipids or lipoproteins to the virus particles. Indeed, the buoyant densities of HCV particles from the sera showed significant heterogeneity, depending on the types of lipoproteins associated with the virus (1). These lipids may affect the infectivity of the virus. The HCV released from SB cells is likely free from lipid association and thus gives more accurate buoyant density and higher infectivity. It is also possible that HCV from SB cells may have acquired certain mutations to adapt to replication in vitro, particularly in B cells. However, the virus produced from SB cells is infectious for both primary hepatocytes and B cells. Thus, it is not clear whether the putative growth advantage of SB virus is its adaptation to in vitro infection in general or to B cells specifically. The cloning and sequencing of the entire viral genome is in progress.
In summary, we have established a cell line persistently infected with HCV. This cell line supports the complete replication cycle of HCV and produces infectious viral particles. The persistent state of HCV infection in the cell line mimics the chronic infection in natural HCV infections. Furthermore, the virus particles produced from the cell line are infectious for other B cells, as well as primary human hepatocytes, thus allowing reproducible studies of viral infections. The cell line will be useful for evaluating antiviral agents that target the entire HCV life cycle from virus entry to virus production. It will also enable studies of the biology of HCV infection in culture.
Acknowledgments
This work was partially supported by grants AI 40038 and R03AI45873 from the National Institutes of Health.
The primary hepatocytes were provided by the Cell Culture Core of the Liver Center of the Keck School of Medicine, University of Southern California (supported by NIH grant P30DK48522).
REFERENCES
- 1.Andre, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier-Alias, M., K. Patel, H. Dahari, S. Beaucourt, P. Larderie, L. Blatt, C. Hezode, G. Picchio, D. Dhumeaux, A. U. Neumann, J. G. McHutchison, and J.-M. Pawlotsky. 2002. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology 36:211-218. [DOI] [PubMed] [Google Scholar]
- 4.Cerny, A., and F. V. Chisari. 1999. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 30:595-601. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 6.Doughty, A., P. M. Dorothy, and G. W. McCaughan. 2000. Post-transplant quasispecies pattern remains stable over time in patients with recurrent cholestatic hepatitis due to hepatitis C virus. J. Hepatol. 32:126-134. [DOI] [PubMed] [Google Scholar]
- 7.Farrell, P. J. 1995. Epstein-Barr virus immortalizing genes. Trends Microbiol. 3:105-109. [DOI] [PubMed] [Google Scholar]
- 8.Ferri, C., F. Caracciolo, and A. L. Zignego. 1994. Hepatitis C virus infection in patients with non-Hodgkin's lymphoma. Br. J. Haematol. 88:392-394. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh, A. K., R. Steele, K. Meyer, R. Ray, and R. B. Ray. 1999. Hepatitis C virus NS5A protein modulates the cell cycle regulatory genes and promotes cell growth. J. Gen. Virol. 80:1179-1183. [DOI] [PubMed] [Google Scholar]
- 10.Haddad, J., P. Deny, C. Munz-Gotheil, J. C. Ambrosini, J. C. Trinchet, D. Pateron, F. Mal, P. Callard, and M. Beaugrand. 1992. Lymphocytic sialadenitis of Sjögren's syndrome associated with chronic hepatitis C virus liver disease. Lancet 339:321-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn, C. S., Y. G. Cho, B. S. Kang, I. M. Lester, and Y. S. Hahn. 2000. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology 276:127-137. [DOI] [PubMed] [Google Scholar]
- 12.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irivne, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly (A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., K. Yasui, J. Mukaigawa, A. Katsume, M. Kohara, and K. Mitamura. 2001. Acquisition of susceptibility to hepatitis C virus replication in HepG2 cells by fusion with primary human hepatocytes: establishment of a quantitative assay for hepatitis C virus infectivity in a cell culture system. Hepatology 34:566-572. [DOI] [PubMed] [Google Scholar]
- 15.Ivanovski, M., F. Silvestri, G. Pozzato, S. Anand, C. Mazzaro, Q. R. Burrone, and D. G. Efremor. 1998. Somatic hypermutation, clonal diversity, and preferential expression of the VH51p1/VLkv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood 91:2433-2442. [PubMed] [Google Scholar]
- 16.Jin, D.-Y., H. L. Wang, Y. Zhou, A. C. Chun, K. V. Kibler, Y. D. Hou, H. Hung, and K. T. Jeang. 2000. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 19:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1994. Buoyant density of hepatitis C virus recovered from infected hosts: two different features in sucrose equilibrium density-gradient centrifugation related to degree of liver inflammation. Hepatology 19:296-302. [PubMed] [Google Scholar]
- 18.Kato, N., T. Nakazawa, T. Mizutani, and K. Shimotohno. 1995. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem. Biophys. Res. Commun. 206:863-869. [DOI] [PubMed] [Google Scholar]
- 19.Kitay-Cohen, Y., A. Amiel, N. Hilzenrat, D. Buskila, Y. Ashur, M. Fejgin, E. Gaber, R. Safadi, R. Tur-Kaspa, and M. Lishner. 2000. Bcl-2 rearrangement in patients with chronic hepatitis C associated with essential mixed cryoglobulinemia type II. Blood 96:2910-2912. [PubMed] [Google Scholar]
- 20.Kuramoto, I. K., T. Moriya, V. Schoening, and P. V. Holland. 2002. Fluctuation of serum HCV-RNA levels in untreated blood donors with chronic hepatitis C virus infection. J. Viral Hepat. 9:36-42. [DOI] [PubMed] [Google Scholar]
- 21.Lanford, R. E., D. Chavez, F. V. Chisari, and C. Sureau. 1995. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J. Virol. 69:8079-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskus, T., M. Radkowski, L. F. Wang, M. Nowicki, and J. Rakela. 2000. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J. Virol. 74:1014-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerat, H., F. Berby, M. A. Trabaud, O. Vidalin, M. Major, C. Trepo, and G. Inchauspe. 1996. Specific detection of hepatitis C virus minus-strand RNA in hematopoietic cells. J. Clin. Investig. 97:845-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Y.-J., C. L. Liao, and M. M. C. Lai. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 26.Makino, S., C.-K. Shieh, J. G. Keck, and M. M. C. Lai. 1988. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology 163:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariette, X., M. Zerbib, A. Jaccard, C. Schenmetzler, E. Danon, and J. P. Clauvel. 1993. Hepatitis C virus and Sjögren's syndrome. Arthritis Rheum. 36:280-281. [DOI] [PubMed] [Google Scholar]
- 28.Mellor, J., G. Haydon, C. Blair, W. Livingstone, and P. Simmonds. 1998. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J. Gen. Virol. 79:705-714. [DOI] [PubMed] [Google Scholar]
- 29.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 30.Morsica, G., G. Tambussi, G. Sitia, R. Novati, A. Lazzarin, L. Lopalco, and S. Mukenge. 1999. Replication of hepatitis C virus in B lymphocytes (CD19+). Blood 94:1138-1139. [PubMed] [Google Scholar]
- 31.Nakajima, N., M. Hijikata, H. Yoshikura, and Y. K. Shimizu. 1996. Characterization of long-term cultures of hepatitis C virus. J. Virol. 70:3325-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissen, E., M. Hohne, and E. Schreier. 1994. In vitro replication of hepatitis C virus in a human lymphoid cell line (H9). J. Hepatol. 20:437.. [DOI] [PubMed] [Google Scholar]
- 33.Pietschmann, T., V. Lohmann, A. Kaul, N. Krigger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radkowski, M., J. Wilkinson, M. Nowicki, D. Adair, H. Vargas, C. Ingui, J. Rakela, and T. Laskus. 2002. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: evidence of replication. J. Virol. 76:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray, R. B., L. M. Lagging, K. Meyer, and R. Ray. 1996. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J. Virol. 70:4438-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruggieri, A., T. Harada, Y. Matsuura, and T. Miyamura. 1997. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology 229:68-76. [DOI] [PubMed] [Google Scholar]
- 37.Selva-O'Callaghan, A., D. Rodriguez-Pardo, L. Sanchez-Sities, L. Matas-Pericas, R. Solans-Laque, J. A. Bosch-Gil, and M. Vilardell-Tarres. 1999. Hepatitis C virus infection, Sjögren's syndrome, and non-Hodgkin's lymphoma. Arthritis Rheum. 42:2489-2490. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu, Y. K., H. Igarashi, T. Kiyohara, M. Shapiro, D. C. Wong, R. H. Purcell, and H. Yoshikura. 1998. Infection of a chimpanzee with hepatitis C virus grown in cell culture. J. Gen. Virol. 79:1383-1386. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu, Y. K., A. Iwamoto, M. Hijikata, R. H. Purcell, and H. Yoshikura. 1992. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc. Natl. Acad. Sci. USA 89:5477-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, Y. K., A. J. Weiner, J. Rosenblatt, D. C. Wong, M. Shapiro, T. Popkin, M. Houghton, H. J. Alter, and R. H. Purcell. 1990. Early events in hepatitis C virus infection of chimpanzees. Proc. Natl. Acad. Sci. USA 87:6441-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, Z. O., F. Hao, F. Min, O. Y. Ma, and G. D. Liu. 2001. Hepatitis C virus infection of human hepatoma cell line 7721 in vitro. World J. Gastroenterol. 7:685-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung, V. M.-H., and M. M. C. Lai. 2002. Murine retroviral pseudotype virus containing hepatitis B virus large and small surface antigens confers specific tropism for primary human hepatocytes: a potential liver-specific targeting system. J. Virol. 76:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Tosato, G. 2000. Generation of Epstein-Barr virus (EBV)-immortalized B cell lines, p. 7.22.1-7.22.3. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 44.Trestard, A., Y. Bacq, L. Buzelay, F. Dubois, F. Barin, A. Goudeau, and P. Roingeard. 1998. Ultrastructural and physicochemical characterization of the hepatitis C virus recovered from the serum of an agammaglobulinemic patient. Arch. Virol. 143:2241-2245. [DOI] [PubMed] [Google Scholar]
- 45.Weiner, A. J., H. M. Geysen, C. Christophersen, J. E. Hall, T. J. Mason, G. Saracco, F. Bonino, K. Crawford, C. D. Marion, K. A. Crawford, M. Brunetto, P. J. Barr, T. Miyamura, J. McHutchinson, and M. Houghton. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. USA 89:3468-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. C. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zignego, A. L., and C. Brechot. 1999. Extrahepatic manifestations of HCV infection: facts and controversies. J. Hepatol. 31:369-376. [DOI] [PubMed] [Google Scholar]
- 48.Zignego, A. L., F. Giannelli, M. E. Marrocchi, A. Mazzocca, C. Ferri, C. Giannini, M. Monti, P. Caini, G. L. Villa, G. Laffi, and P. Gentilini. 2000. T(14:18) translocation in chronic hepatitis C virus infection. Hepatology 31:474-479. [DOI] [PubMed] [Google Scholar]
- 49.Zuckerman, E., T. Zuckerman, A. M. Levine, D. Douer, K. Gutekunst, M. Mizokami, D. G. Qian, M. Velankar, B. N. Nathwant, and T. L. Fong. 1997. Hepatitis C virus infection in patients with B cell non-Hodgkin's lymphoma. Ann. Intern. Med. 127:423-428. [DOI] [PubMed] [Google Scholar]