Abstract
The late RNA synthesis in alphavirus-infected cells, generating plus-strand RNAs, takes place on cytoplasmic vacuoles (CPVs), which are modified endosomes and lysosomes. The cytosolic surface of CPVs consists of regular membrane invaginations or spherules, which are the sites of RNA synthesis (P. Kujala, A. Ikäheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kääriäinen J. Virol. 75:3873-3884, 2001). To understand how CPVs arise, we have expressed the individual Semliki Forest virus (SFV) nonstructural proteins nsP1 to nsP4 in different combinations, as well as their precursor polyprotein P1234 and its cleavage intermediates. A complex of nsPs was obtained from P123 or P1234, indicating that the precursor stage is essential for the assembly of the polymerase complex. To prevent the processing of the polyprotein and its cleavage intermediates, constructs with the mutation C478A (designated with a superscript CA) in the active site of the protease domain of nsP2 were used. Uncleaved polyproteins containing nsP1 were membrane bound and palmitoylated, and those containing nsP3 were phosphorylated, reflecting properties of authentic nsP1 and nsP3, respectively. Similarly, polyproteins containing nsP1 or nsP2 had enzymatic activities specific for the individual proteins, indicating that they were correctly folded in the precursor state. Uncleaved P12CA was localized almost exclusively to the plasma membrane and filopodia, like nsP1 alone, whereas P12CA3 and P12CA34 were found on cytoplasmic vesicles, some of which contained late endosomal markers. In immunoelectron microscopy these vesicles resembled CPVs in SFV-infected cells. Our results indicate that the nsP1 domain alone is responsible for the membrane association of the nonstructural polyprotein, whereas the nsP1 domain together with the nsP3 domain targets it to the intracellular vesicles.
The alphaviruses replicate in the cytoplasm of both invertebrate and vertebrate cells. The virus enters the cell by adsorptive endocytosis, followed by fusion of the virus envelope with endosomal membranes (34). The virus nucleocapsid is disassembled by ribosomes, which have affinity for the capsid protein (52, 63). The capped positive-strand RNA genome of about 11.5 kb is then translated to yield a polyprotein, P1234, of about 2,500 amino acids (aa), the precursor of nonstructural proteins nsP1 to nsP4. The parental 42S RNA genome is copied to complementary minus-strand RNA by a short-lived RNA polymerase consisting of the catalytic subunit nsP4 and polyprotein P123, the initial cleavage products of P1234 (31, 50). By inhibition of cleavage between nsP2 and nsP3, minus-strand RNA synthesis was demonstrated also to occur by the nsP1-P23-nsP4 combination (30).
Early work with Semliki Forest virus (SFV) and Sindbis virus showed that the parental RNA was converted into an RNase-resistant, membrane-associated form soon after infection, suggesting that the synthesis of minus strands takes place in association with membranes (8, 12, 17). The cellular structures associated with the minus-strand RNA synthesis have been difficult to recognize, since the minus-strand polymerase is short lived, with a half-life of less than 10 min, and the cleavage of P123 to nsP1-nsP2-nsP3 converts it into a stable plus-strand polymerase. Even early in infection, maximally 20% of synthesized RNAs are minus strands (47). More is known about the late plus-strand RNA polymerase, which is associated with virus-induced cytoplasmic vacuoles (CPVs). CPVs are modified endosomes and lysosomes with a diameter of 200 to 1,000 nm. The surface of CPVs consists of small invaginations with a diameter of about 50 nm, which have been designated spherules (13, 14, 16). Cryo-immunoelectron microscopy (cryo-immuno-EM) at 5 to 6 h postinfection (p.i.) has shown that all four nsPs are associated with spherules together with nascent RNA, indicating that they are the sites of synthesis of plus-strand RNAs (23).
Previously we have characterized the properties of individually expressed SFV nsPs. The N-terminal nsP1 of 537 aa is a methyltransferase and guanylyltransferase involved in the capping of the viral positive-strand RNAs (1, 2, 26). It is also tightly membrane bound and palmitoylated (3, 24, 40). The N-terminal half of nsP2 (799 aa) is an RNA helicase (15), nucleoside triphosphatase (NTPase) (45), and RNA triphosphatase (58), while the C-terminal half is a papain-like protease responsible for the autocatalytic processing of the nonstructural polyprotein P1234 (18, 35, 59). Work with temperature-sensitive mutants suggests that the C-terminal part also regulates the synthesis of the subgenomic 26S RNA (reference 56 and references therein). The functions of the nsP3 phosphoprotein (32, 42, 60, 61) in RNA replication are poorly understood (20). According mostly to genetic evidence, nsP4 (614 aa in SFV) is the catalytic subunit of the alphavirus RNA polymerase (20, 54).
In order to understand the role of the P1234 polyprotein and its intermediate cleavage products in the membrane association and targeting of the SFV replication complex, we have expressed cleavable and uncleaved nonstructural polyproteins in insect cells as well as in mammalian cells. We show that uncleaved polyproteins had enzymatic activities typical for nsP1 and nsP2 and that those carrying nsP3 were phosphorylated, suggesting that they were properly folded. Uncleaved P123 and P1234 were associated with large vesicles, some of which were late endosomes or lysosomes resembling structures seen late in SFV-infected cells.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown in Dulbecco's modified minimal essential medium supplemented with 10% inactivated fetal calf serum (PAA Laboratories GmbH) and 100 U of penicillin and streptomycin per ml. High Five BTL-Tn-5B1-4 insect cells (Tn5 cells) (Invitrogen) were used for baculovirus infections. Construction of baculoviruses Bac1234, Bac123, Bac23, Bac12CA34, Bac12CA3, Bac2CA3, and Bac34 has been described by Merits et al. (35), and that of baculovirus BacnsP1 has been described by Laakkonen et al. (26). The viruses were propagated and assayed as described previously (35). The identities of expressed proteins were verified by Western blotting with antibodies against SFV nsPs (26). The modified recombinant vaccinia virus Ankara (MVA) encoding T7 RNA polymerase, kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.), was propagated and used as described previously (61).
Plasmid constructions.
For transient expression of SFV nsPs, nonstructural polyproteins, and their cleavage-deficient mutants in HeLa cells, the inserts from constructs encoding P123, P1234, P12CA3, P12CA34, P2CA3, P34 (35), P12, P12CA (59), and pTSF1 (39) were ligated with BamHI- and EcoRV-digested expression plasmid pcDNA4/TO (Invitrogen). Positive clones were selected and designated pcP123, pcP1234, pcP12CA3, pcP12CA34, pcP2CA3, pcP34, pcP12, pcP12CA, and pcnsP1 (Fig. 1). To obtain recombinant adenoviruses expressing SFV polyprotein precursors P12 and P123 and cleavage-deficient polyproteins P12CA, P12CA3, and P2CA3, the inserts from expression plasmids pcP12, pcP123, pcP12CA, pcP12CA3, and pcP2CA3 were cloned into BglII- and XbaI-digested vector pShuttle-CMV (19). To construct an adenovirus expressing SFV nsP4 with the authentic N terminus, the nsP4-coding sequence was first fused with ubiquitin-coding sequence. Recombinant adenoviruses were created and assayed essentially as described previously (19). The virus stocks were checked for protein expression and amplified in HEK 293 cells. For expression in animal cells, cytomegalovirus (CMV) promoter-containing plasmids often gave satisfactory yields of polyproteins, but more efficient expression was achieved with adenovirus vector.
FIG. 1.
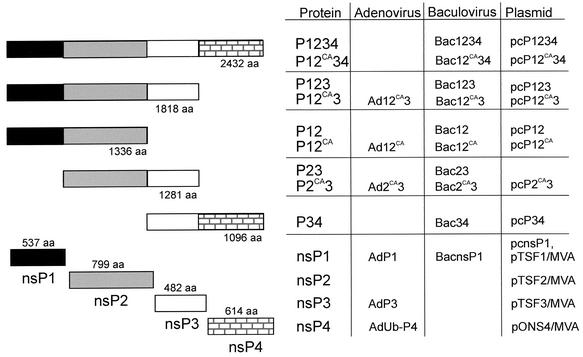
Constructs expressing SFV nsPs and polyproteins. The proteins are shown schematically on the left. Different shadings represent cleavage products (nsPs) within the nonstructural coding region. Constructs containing inactivated nsP2 protease C478ACA produce nonprocessed polyproteins, whereas wild-type polyproteins are processed to individual nsPs (see also Fig. 2C). Recombinant adeno- and baculovirus systems as well as transfection of plasmids were utilized for expression of the proteins under study.
Radioactive labeling and cell fractionation.
The four individual nsPs were expressed pairwise or all together by using MVA encoding T7 RNA polymerase. HeLa cells were infected with MVA, followed by transfection with derivatives of pGEM vector (Promega) pTSF1, pTSF2, pTSF3, and pONS4 (39), in which the nsP-coding genes are under control of the T7 promoter. MVA infection, transfection, and [35S]methionine labeling of cells were carried out as described previously (61). Postnuclear supernatant was used for immunoprecipitations.
Tn5 cells (1.8 × 107 in a 10-cm-diameter petri dish) were infected with recombinant baculovirus stock at 10 PFU/cell, followed by incubation at 28°C in High Five medium (Invitrogen) for 44 h. For labeling with [35S]methionine, the cells were incubated in methionine-free Grace medium for 30 min at 28°C and pulse-labeled with 500 μCi of [35S]methionine (1,000 Ci/mmol; Amersham) in methionine-free medium for 2 h at 28°C. The cells were chased with a 20-fold excess of unlabeled methionine for 1 h at 28°C. Cells were harvested, washed and swollen in RS buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl), disrupted in a Dounce homogenizer, and fractionated into 15,000 × g membrane (P15) and the supernatant (S15) fractions as described previously (33). Membrane flotation was carried out as described previously (24), using postnuclear supernatant as the starting material. The floated fractions were collected and subjected to immunoprecipitation with anti-nsP1, anti-nsP2, anti-nsP3, or anti-nsP4 antiserum. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 7.5% gel and visualized by phosphorimaging analysis. To assay the mode of membrane association, P15 fractions were exposed to 1 M NaCl, 50 mM EDTA, and Na2CO3 at pH 11.5 or 12, followed by resedimentation at 15,000 × g and SDS-PAGE analysis as described previously (24).
For palmitate labeling, Tn5 cells were infected with Bac1234, Bac12CA34, Bac123, Bac12CA3, and BacnsP1 at 10 PFU/cell. Cells were labeled at 44 h p.i. with 300 to 400 μCi of [9,10(n)-3H]palmitic acid (52 Ci/mmol; Amersham) per 60-mm-diameter plate in Sf-900 II serum-free-medium for 8 h at 28°C. Cells were collected, and part of the P15 fraction was immunoprecipitated with anti-nsP1 or anti-nsP3 antiserum as described previously (26). Protein samples before and after immunoprecipitation were analyzed in SDS-7.5% polyacrylamide gels, transferred to nitrocellulose, and visualized by phosphorimaging analysis.
For labeling with [32P]orthophosphate, 4 × 107 Tn5 cells on 15-cm-diameter petri dishes were infected with recombinant baculoviruses as described above. At 40 h p.i. the cells were washed, and phosphate-free Grace medium was added for 1 h, followed by labeling with carrier-free [32P]orthophosphate (2.5 mCi/dish) for 3 h. Cells were harvested in 1 ml of cold phosphate-buffered saline, washed twice with phosphate-buffered saline, and lysed by addition of 1 ml of hot 2% SDS. The lysate was boiled for 3 min and diluted 1:10 with ice-cold NET buffer (1% NP-40, 50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.02% Na3N, and 100 U of Trasylol per ml). Labeled nsP3 and polyproteins were immunoprecipitated with anti-nsP3 antibody as described previously (42), subjected to SDS-PAGE, and visualized by autoradiography.
Immunoprecipitation.
For native immunoprecipitation, the postnuclear supernatant or P15 fractions were diluted in NET buffer supplemented with protease inhibitor cocktail (Boehringer, Mannheim, Germany). After preclearing with preimmune serum and protein A-Sepharose, the samples were immunoprecipitated overnight with monospecific anti-nsP antiserum (23), followed by protein A Sepharose precipitation of the immunocomplexes. The precipitates were washed four times with NET buffer containing 400 mM NaCl. For SDS-PAGE, the precipitates were dissolved in Laemmli buffer as described previously (42). For second immunoprecipitations, the proteins were released from the beads by treatment with 1% SDS and 10 mM dithiothreitol, diluted with NET buffer, and then reimmunoprecipitated with antisera against nsP1 to nsP4. A PhosphorImager (Molecular Dynamics) was used for visualization of proteins.
Enzyme assays.
Tn5 cells (4 × 107) infected with the recombinant baculoviruses Bac1234, Bac12CA34, Bac123, Bac12CA3, Bac12, Bac12CA, and Bac2CA3 were used as the source of enzyme for NTPase and RNA triphosphatase assays. The same number of cells infected with Bac1 and Bac34 were used as a control. The infected cells were collected at 44 h p.i. and lysed in 1 ml of NET buffer. PNS was used for immunoprecipitation with anti-nsP1, anti-nsP2, or anti-nsP3, followed by addition of protein A-Sepharose (Pharmacia). After washing, the precipitates were incubated in 100 μl of 10-fold NTPase assay buffer (200 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 50 mM KCl, 1.5 M NaCl, 20 mM dithiothreitol) for 10 min at 30°C. The Sepharose particles were removed by sedimentation. The supernatant fraction was diluted 1:10 with distilled water, and NTPase and RNA triphosphatase assays were carried out as described previously (58).
Postnuclear supernatants from Tn5 cells infected with Bac1234, Bac123, Bac12CA34, Bac12CA3, Bac12, and Bac12CA were immunoprecipitated as described above and assayed for methyltransferase activity as described previously (26), using 10 μl per reaction mixture and corresponding extracts from wild-type Autographa californica nuclear polyhedrosis virus-infected cells as controls. The formation of the covalent guanaylate complex was assayed in the presence or absence of 100 mM S-adenosylmethionine (AdoMet) as described previously (1). The reactions were stopped by boiling in the presence of 1% SDS, the reaction products were immunoprecipitated with anti-nsP1 antibody essentially as described previously (42), and the products were analyzed by SDS-PAGE in 7.5% gels followed by autoradiography.
Immunofluorescence and confocal microscopy.
For indirect immunofluorescence microscopy, HeLa cells were grown on coverslips in 35-mm-diameter dishes. Cells were transfected with plasmids pcP123, pcP1234, pcP12CA3, pcP12CA34, pcP2CA3, pcP34, pcP12, pcP12CA, and pcnsP1, encoding the respective proteins under control of the CMV promoter. FuGENE 6 transfection reagent (Roche) was used according to the manufacturer's instructions, followed by incubation for 24 to 28 h at 37°C. Individual nsPs were expressed in combination by MVA-aided transfection (61) for 6 h. The cells were fixed with 4% paraformaldehyde in cytosolic buffer (10 mM MES [morpholineethanesulfonic acid] [pH 6.1], 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, and 0.32 M sucrose) and permeabilized with 0.1% Triton X-100 for 1 min. The aldehyde groups were quenched with 50 mM NH4Cl. Cells were treated with specific anti-nsP antibodies (23) or antibody against human lysosome-associated membrane glycoprotein 2 (lamp2) (21). This was followed by treatment with secondary, species-specific antibodies conjugated with fluorescein isothiocyanate or rhodamine (tetramethyl rhodamine isocyanate; Jackson ImmunoResearch Laboratories). Rhodamine-conjugated concanavalin A (Sigma) was used for the plasma membrane staining. Labeled cells were analyzed with a Bio-Rad MRC-1024 confocal microscope with an American Laser Corporation laser as the source for the argon-krypton ion laser beam.
EM.
HeLa cells grown on coverslips were infected with recombinant adenoviruses coding for nsP3, P12CA, or P12CA3 or with SFV. The cells were fixed 28 h after adenovirus infection or after 5 h of infection with SFV. For ultrastructural analysis, the cells were fixed with 2.5% glutaraldehyde for 20 min at room temperature, postfixed with 1% reduced osmium tetroxide for 1 h and 1% uranyl acetate in 0.3 M sucrose for 1 h at 4°C, and processed for Epon embedding as described previously (49). For immunolabeling, cells were fixed with PLP fixative (2% formaldehyde, 0.01 M periodate, and 0.075 M lysine-HCl in 0.075 M phosphate buffer, pH 7.4) for 2 h at room temperature. Cells were permeabilized with 0.01% saponin (Sigma) and immunolabeled by using anti-nsP3 or anti-nsP1 rabbit antiserum (23) and 1.4-nm-diameter gold particle-conjugated Fab′ fragments against rabbit immunoglobulin G (Nanoprobes). Nanogold was silver enhanced with an HQ Silver kit (Nanoprobes) for 0.5 to 4 min and gold toned with 0.05% gold chloride (5). After washing, the cells were processed for Epon embedding as described above. Sections were cut parallel to the coverslip, picked up on single-slot copper grids, poststained with uranyl acetate and lead citrate, and examined with a Jeol JEM-1200EX II transmission electron microscope at 80 kV.
RESULTS
Expression of SFV nsPs.
We have shown previously that in SFV-infected cells the majority of the nonstructural proteins (nsP1 to nsP4) are associated with a membrane fraction sedimenting at 15 000 × g (P15) (42), which contains all virus-specific RNA polymerase activity of the cell (6, 7, 43). Immunoprecipitation with a monospecific anti-nsP antiserum coprecipitated all other nsPs from the P15 fraction, suggesting that they were associated with the RNA replication complex (23). Of the four nsPs, only nsP2 is found in significant amounts in the 15,000 × g supernatant (S15) and nuclear fraction (41).
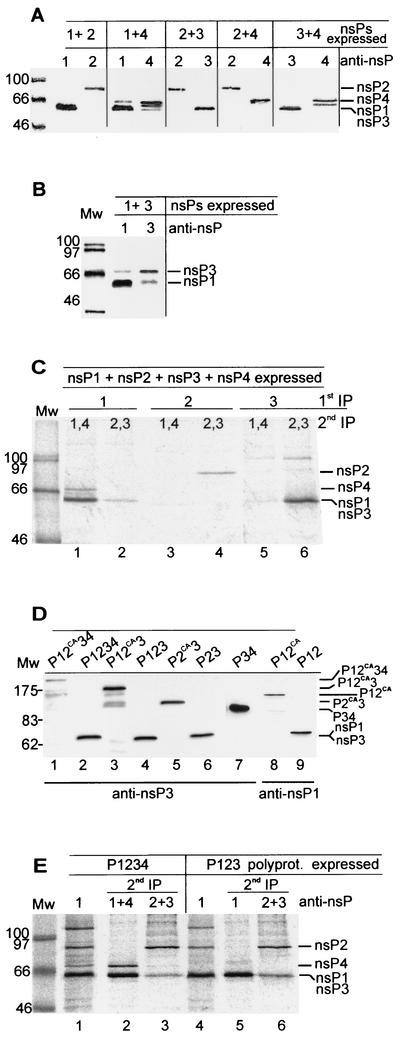
To study how the nsPs form the replication complexes, we expressed the four nsPs pairwise or all together by using recombinant vaccinia virus MVA encoding T7 polymerase. The cells were labeled with [35S]methionine 3 to 5 h after transfection. After a chase period, immunoprecipitations were carried out under nondenaturing conditions to detect interactions between the expressed nsPs. As shown in Fig. 2A and B, some coimmunoprecipitation was observed only in coexpression of the pairs nsP1-nsP3 and nsP1-nsP4. These results suggest that mature nsP2 does not interact with any of the other nsPs. There was also no interaction between nsP3 and nsP4. Yeast two-hybrid analysis of the same combinations failed to suggest interactions between the nsPs (unpublished data).
FIG. 2.
Expression of SFV nsPs and polyproteins. (A and B) Immunoprecipitation of SFV nsPs from transfected HeLa cells. Recombinant vaccinia virus (MVA)-infected cells were transfected with two plasmids encoding nsPs (combinations are indicated at the top) and labeled with [35S]methionine from 3 to 5 h posttransfection, followed by a 30-min chase. The postnuclear supernatant was subjected to immunoprecipitation with appropriate anti-nsP antibodies as indicated, and proteins were analyzed by SDS-PAGE in 10% gels. Expression of nsP4 often results in a double band, where the upper one represents full-length nsP4. In panel B, 0.1% bisacrylamide was used for the nsP1-nsP3 combination to enable their separation in electrophoresis. (C) Coexpression of nsP1, nsP2, nsP3, and nsP4 in transfected HeLa cells as in panels A and B. SDS-PAGE after two successive immunoprecipitations is shown. First precipitations were done with anti-nsP1 (lanes 1 and 2), anti-nsP2 (lanes 3 and 4), and anti-nsP3 (lanes 5 and 6), followed by second precipitations with a combination of anti-nsP1 and anti-nsP4 (lanes 1, 3, and 5) and anti-nsP2 and anti-nsP3 (lanes 2, 4, and 6). (D) Expression of SFV nonstructural polyproteins by the baculovirus system. Tn5 cells were infected with the indicated baculovirus-SFV polyprotein recombinants at 10 PFU/cell. Cells were harvested at 44 h after infection and subsequently lysed by boiling in Laemmli sample buffer. Samples were analyzed by SDS-PAGE in 7.5% gels and detected by immunoblotting with anti-nsP3 (lanes 1 to 7) or anti-nsP1 (lanes 8 and 9) antibodies by using the ECL procedure (Amersham Pharmacia Biotech). (E) Immunoprecipitation-recapture experiment with cleavable P1234 and P123 polyproteins. Tn5 cells were infected with baculoviruses encoding SFV P1234 and P123 at 5 PFU/cell. Cells were labeled with [35S]methionine at 42 to 44 h after infection, chased for 1 h, and collected. The P15 fraction was subjected to two successive immunoprecipitations. Samples were first immunoprecipitated with anti-nsP1 under nondenaturing conditions (lanes 1 and 4). The precipitates were denatured and subjected to a second immunoprecipitation with anti-nsP1 plus anti-nsP4 (lane 2), anti-nsP2 plus anti-nsP3 (lanes 3 and 6), or anti-nsP1 (lane 5). Analysis was done by SDS-PAGE in 10% gels. Molecular weight markers (Mw), in thousands, are shown on the left and the positions of nsPs or polyproteins are shown on the right in all panels.
Finally, all four nsPs were expressed simultaneously, followed by labeling and two successive immunoprecipitations. After the first precipitation under native conditions, the precipitates were denatured and exposed to reimmunoprecipitations with all four anti-nsP antisera (Fig. 2C). When the first precipitation was carried out with anti-nsP1 antiserum, small amounts of nsP4 and nsP3 coprecipitated (Fig. 2C, lanes 1 and 2), while anti-nsP2 antiserum in the first precipitation did not precipitate other nsPs detectable in the second precipitate (lanes 3 and 4). Somewhat unexpectedly, anti-nsP3 failed to precipitate nsP1, which was detectable as a faint band in the pairwise expression of nsP1 and nsP3 (Fig. 2C, lanes 5 and 6; see also Fig. 2B).
Expression of SFV nonstructural polyproteins.
To express the polyprotein precursors of nsPs, we used the baculovirus system, which allows the production of large proteins, such as P1234, which consists of 2,432 aa. Recombinant baculoviruses expressing P1234, P123, P12, P23, and P34 (Fig. 1) were used to infect Trichoplusia ni (Tn5) cells. The expression of SFV-specific nsPs was detected in cells from 20 h p.i. on, until the cells died at 72 to 96 h p.i. Recombinant protein synthesis reached its maximal level at 40 to 48 h p.i. Cells were harvested at 44 h p.i., and the postnuclear supernatant was subjected to SDS-PAGE followed by immunoblotting with anti-nsP antisera. The cleavable precursor proteins carrying the nsP2 protease (35) were processed to yield their constituent nsPs, as shown for nsP3 (Fig. 2D, lanes 2, 4, and 6) and nsP1 (lane 9), while P34 remained uncleaved (Fig. 2D, lane 7). Crude fractionation of postnuclear supernatant showed that most of the cleavage products of P123 and P1234 were in the 15, 000 × g pellet (P15) fraction. To study the possible interactions of nsPs, we carried out double immunoprecipitations like those described above. The first precipitation was done for the undenatured P15 fraction of infected Tn5 cells with anti-nsP1 antiserum (Fig. 2E, lanes 1 and 4), followed by second immunoprecipitations after denaturation of the first precipitate. In the case of P1234 expression, nsP1 and nsP4 (Fig. 2E, lane 2) and nsP2 and nsP3 (lane 3) were detected, indicating that they had formed a complex. Similarly, anti-nsP1 antiserum precipitated nsP1, nsP2, and nsP3 from cells expressing P123 (Fig. 2E, lanes 5 and 6). Similar results were obtained for floated membrane fractions made from postnuclear supernatant of P1234- and P123-expressing cells (not shown). All these results indicate that the polyprotein intermediate is necessary for the successful assembly of the replication complex, especially to ensure that the soluble nsP2 remains associated with the polymerase after proteolytic processing.
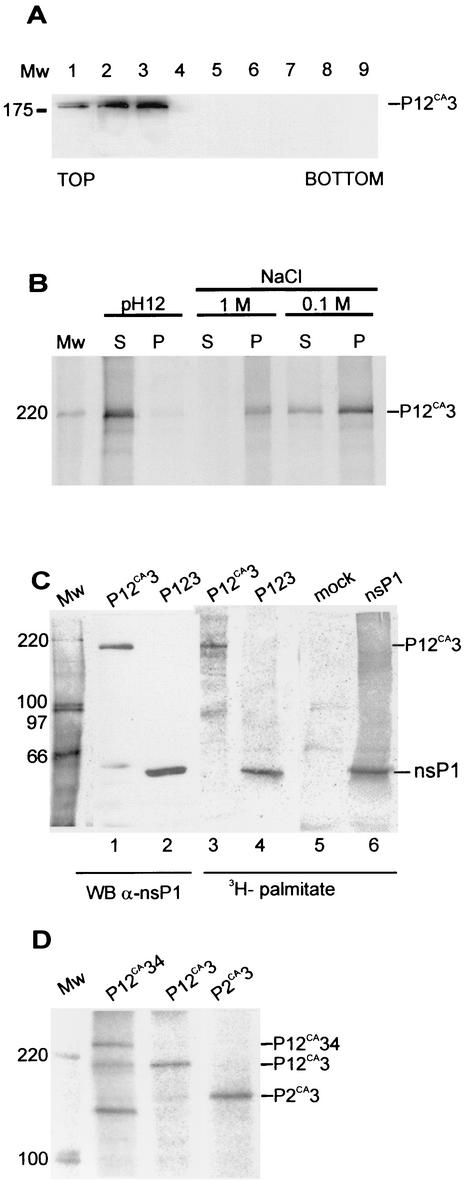
To study whether the polyproteins have properties identified for their constituents we made constructions for expression of uncleaved nonstructural polyproteins. These were created by mutating the active-site cysteine 478 of nsP2 to alanine (P2CA) to yield polyproteins P12CA34, P12CA3, P12CA, and P2CA3 (Fig. 1). The uncleaved polyproteins migrated in SDS-PAGE at the expected positions (Fig. 2D), but some degradation products could be seen with the two largest proteins (Fig. 2D, lanes 1 and 3). Flotation assays were used to find out whether the polyproteins were associated with cytoplasmic membranes. The postnuclear supernatant was subjected to flotation in discontinuous sucrose gradients (Fig. 3A). During centrifugation, P12CA3 migrated from 60% sucrose at the bottom of the tube to the top fractions at the 50%-10% interface, indicating that it was associated with membranes. When the membranes were collected and exposed to treatment with a high salt concentration, which detaches peripherally associated membrane proteins, most of the P12CA3 remained in the sedimenting fraction (Fig. 3B). The protein was detached from the membranes only after treatment with alkaline solution, at pH 12. In this respect, P12CA3 behaved similarly to nsP1 expressed alone in HeLa cells (24, 26). Similar results were obtained with uncleaved P12CA34 and P12CA, whereas P2CA3 and P34 did not float (data not shown), indicating that the presence of nsP1 is needed for stable membrane association of the polyprotein.
FIG. 3.
Membrane association, palmitoylation, and phosphorylation of polyproteins. (A) Bac12CA3-infected Tn5 cells were labeled as for Fig. 2E. Postnuclear supernatant was subjected to flotation in a discontinuous sucrose (weight/weight) gradient consisting of 0.675 ml of 10%, 3.5 ml of 50%, and 0.625 ml of 60% sucrose on top of a 0.5-ml 67% sucrose cushion. Fractions of 0.5 ml were collected from the top and analyzed by immunoblotting with antiserum against nsP1. The sample was initially included in the 60% sucrose layer (fractions 8 and 9), and upon centrifugation, membrane-bound proteins floated to the 10 to 50% sucrose interface (fractions 2 and 3). (B) Fractions 2 and 3 were pooled, and aliquots were subjected to treatments with 50 mM Tris-HCl (pH 7.5)-100 mM NaCl (TN buffer), 1 M NaCl in TN buffer, and 50 mM Na2CO3 at pH 12. After 30 min of incubation on ice, the samples were centrifuged at 70,000 × g for 30 min. The pellet (P) and supernatant (S) fractions were immunoprecipitated with anti-nsP3 antiserum, followed by SDS-PAGE in a 7.5% gel and autoradiography. (C) Palmitoylation of SFV P12CA3. Tn5 cells were infected with Bac12CA3 and labeled at 44 h p.i. with 300 to 400 μCi of [9,10(n)-3H]palmitic acid for 8 h. Bac123- and Bac1-infected and mock-infected cells served as controls. Cells were collected, the P15 fraction was prepared, and protein expression was verified by immunoblotting (WB) (lanes 1 and 2) with anti-nsP1 antibody. Part of the P15 fraction was immunoprecipitated by anti-nsP1 antiserum. Samples were analyzed with SDS-PAGE in a 7.5% gel, and incorporated [3H]palmitate was visualized with a PhosphorImager. (D) In vivo phosphorylation of SFV P12CA34, P12CA3, and P2CA3 proteins. Tn5 cells were infected with Bac12CA34, Bac12CA3, or Bac2CA3, labeled with [32P]orthophosphate at 41 to 44 h p.i., harvested, and lysed by boiling in 1% SDS. Proteins were immunoprecipitated with anti-nsP3 antibody and analyzed with SDS-PAGE and a PhosphorImager. In all panels, positions of proteins are indicated on the right and those of molecular weight (Mw) markers, in thousands, are indicated on the left.
Characterization of the nonstructural polyproteins.
To get information on the proper folding of the polyproteins, we studied the posttranslational modifications and enzymatic activities previously identified for the individual nsPs. The tight membrane association of P12CA34, P12CA3, and P12CA suggested that these proteins may be palmitoylated like nsP1 in SFV-infected cells as well as after expression alone (4, 24, 26, 40). Results of in vivo labeling with tritiated palmitic acid showed that P12CA3 and P12CA were indeed palmitoylated (Fig. 3C). Next we wanted to know whether the polyproteins would be also phosphorylated like nsP3 (32, 42, 60, 61). To this end, P12CA34, P12CA3, and P2CA3 expressed in Tn5 cells were labeled with [32P]orthophosphate, followed by immunoprecipitation with anti-nsP3 antiserum and SDS-PAGE analysis. Labeled proteins migrating at the positions of P1234, P123, and P23 were detected (Fig. 3D). Thus, we conclude that acylation and phosphorylation can take place at the polyprotein stage.
Next, we tested the polyproteins for the enzymatic activities of nsP1 and nsP2. We have shown previously that nsP1 has a unique methyltransferase activity involved in the capping of viral mRNAs. In this unique reaction, GTP is methylated to position 7 of the guanine, yielding m7GTP (2, 26). The same protein is also a guanylyltransferase, which forms a covalent complex with [α-32P]GMP when incubated in the presence of AdoMet and [α-32P]GTP (1). P12CA34, P12CA3, and P12CA had methyltransferase activity (not shown), as well as guanylyltransferase activity, in the presence of AdoMet (Fig. 4A).
FIG.4.
.(A) Guanylyltransferase activity of SFV polyproteins. P15 fractions from cells infected with Bac12CA34, Bac1234, Bac12CA3, Bac123, or Bac12CA were subjected to guanylyltransferase assay in the presence (+) or absence (−) of 100 μM AdoMet. Reactions were stopped by boiling with 1% SDS and followed by immunoprecipitation with anti-nsP1 antibody. Samples were analyzed by SDS-PAGE in a 7.5% gel, followed by autoradiography. Molecular weight (Mw) standards in thousands are shown on the left. (B) NTPase activity of SFV nsP2 and nonstructural polyproteins expressed by baculovirus vector, as in panel A. nsP2 expressed and purified from Escherichia coli was used as a positive control. Proteins were immunoprecipitated (IP) from the postnuclear supernatant with the antibody indicated at the bottom and incubated with [α-32P]GTP. The NTPase reaction products were analyzed with thin-layer chromatography. The positions of the reaction products are indicated with arrows. Baculo wt, baculovirus vector.
The amino-terminal half of nsP2 has NTPase and RNA triphosphatase activities (45, 58), which were also found in the immunoprecipitates of P12CA34, P12CA3, P12CA, and P2CA3, as shown for NTPase (Fig. 4B). The carboxy terminus of nsP2 of Sindbis virus (11, 18), as well as SFV, has protease activity (35, 59). We have recently shown that uncleavable SFV polyproteins with active protease domains but with mutations in the cleavage sites have protease activity in trans (unpublished). Thus, the enzymatic activities of authentic nsP2 and nsP1 were already expressed in their precursor proteins, suggesting that the different domains of the polyprotein were properly folded.
Subcellular localization of nsPs and their precursors.
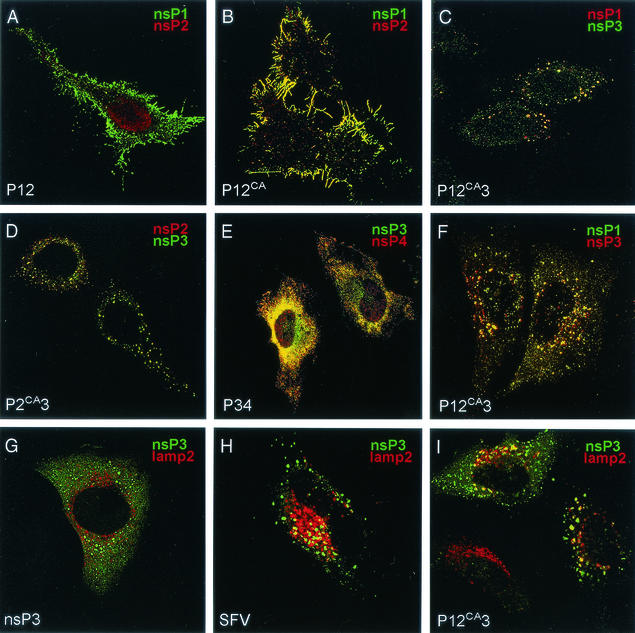
When individual nsPs are expressed in vertebrate cells, each of them has a typical subcellular distribution. nsP1 is found mostly at the cytoplasmic side of the plasma membrane and in filopodium-like extensions (24, 25, 40), whereas nsP2 is transported almost quantitatively to the nucleus (39, 41, 44). nsP3 is distributed in the cytoplasm to vesicle-like structures of variable size (38, 61), while nsP4 is dispersed diffusely in the cytoplasm (39). Here, the localization of nsP pairs was studied by using double-immunofluorescence staining and confocal microscopy. In conformity with the immunoprecipitation results, we found no colocalization of nsP1 plus nsP2, nsP2 plus nsP3, nsP2 plus nsP4, or nsP3 plus nsP4 (not shown). Only some colocalization was seen in cells expressing nsP1 plus nsP3. We also found no colocalization of nsP1 and nsP4, which we expected on the basis of the coimmunoprecipitation studies. This reflects the fact that only a fraction of expressed proteins actually interact. Neither was significant colocalization of nsPs detected when all four nsPs were expressed simultaneously. Expression of polyproteins was carried out in HeLa cells by using either plasmids or adenovirus vectors in which the coding gene was under control of the CMV promoter. Expression of wild-type P12 resulted in separation of nsP1 and nsP2. The former was found at the plasma membrane and filopodia, whereas nsP2 was transported almost quantitatively to the nucleus (Fig. 5A). This is in agreement with our previous results, which showed that P12 is rapidly cleaved into nsP1 and nsP2 (35). In contrast, expression of P12CA resulted in staining of the plasma membrane and filopodium-like extensions with both anti-nsP1 and anti-nsP2 antibodies, with negligible nuclear staining (Fig. 5B). A similar result was obtained when P12CA was expressed more efficiently by adenovirus vector (not shown). Therefore, the effect of nsP1 dominates in the determination of the localization of the uncleaved P12CA.
FIG. 5.
Immunolocalization analysis of SFV nonstructural polyproteins. Transfected (A, B, C, and E) or recombinant adenovirus-infected (D, F, G, and I) HeLa cells were fixed with 4% paraformaldehyde 28 h after transfection or infection. SFV-infected cells (H) (50 PFU/cell) were fixed at 5 h p.i. Cells were permeabilized and double labeled with rabbit and guinea pig antibodies against nsP1 and nsP2 (A and B), nsP1 and nsP3 (C and F), nsP2 and nsP3 (D), nsP3 and nsP4 (E), or nsP3 and lysosomal marker lamp2 (G, H, and I). Rhodamine Red X-conjugated anti-rabbit (red) and fluorescein-conjugated anti-guinea pig (green) antisera were used as secondary antibodies. Merged confocal microscopy images are shown.
Expression of P2CA3 resulted in diffusely distributed cytoplasmic vesicle-like structures (Fig. 5D) resembling those seen in cells expressing nsP3 alone (Fig. 5G). These structures (38, 61) did not costain with the endolysosomal marker lamp2 (Fig. 5G). Interestingly, no nuclear staining was detected with anti-nsP2 antibodies, showing that nsP3 prevented the nuclear transport of the nsP2 domain. P34 had a disperse cytoplasmic granular distribution (Fig. 5E), in accordance with flotation experiments, which showed that this protein is not membrane bound. Moreover, there was a complete overlap of anti-nsP3 and anti-nsP4 staining, in agreement with results showing that P34 cannot be cleaved without nsP2 protease (35). Expression of wild-type P123 resulted, to a large extent, in dissociation of the nsPs from each other. Anti-nsP2 antibodies stained the nuclei, anti-nsP1 stained the plasma membrane, and anti-nsP3 stained cytoplasmic spots (not shown). Expression of P12CA3 gave a different staining pattern. Large vesicular structures mainly under the plasma membrane were stained with antibodies against nsP1 and -3 (Fig. 5C). There was no staining of filopodia similar to those seen abundantly in cells expressing P12CA. A higher expression level with adenovirus vector resulted in widespread cytoplasmic vesicular staining (Fig. 5F). Double staining with anti-nsP3 and anti-lamp2 showed that a fraction of these vesicles were late endosomes or lysosomes (Fig. 5I) similar to those detected in SFV-infected HeLa cells at 5 h p.i. (Fig. 5H). Thus, the presence of nsP3 as part of the uncleavable P12CA3 polyprotein directed it to a new intracellular localization compared with P12CA. P12CA34 polyprotein could be expressed only from a plasmid, as it was too large for our adenovirus expression system. The localization was similar to that shown for P12CA3 in Fig. 5C (not shown).
Immuno-EM.
To have a better understanding of the morphology of the structures associated with the expressed polyproteins, we examined P12CA-, P12CA3-, or nsP3-producing cells by EM. Recombinant adenovirus-infected cells were fixed and permeabilized with saponin, followed by treatment with anti-nsP1 or anti-nsP3 antibodies and nanogold with Fab′ fragment against immunoglobulin G. The nanogold was visualized by silver enhancement followed by Epon embedding. P12CA was localized mostly at the plasma membrane and in tubular surface structures, apparently broken filopodia (data not shown). P12CA3 was found in large patches at the cytoplasmic side of the plasma membrane (Fig. 6A), as well as on the cytoplasmic side of large vesicles or vacuoles, which were often filled with membranes. The diameter of the vacuoles ranged from 200 to 500 nm (Fig. 6B). When the same staining procedure was applied to SFV-infected cells at 5 h p.i., similar but more numerous vacuoles were stained (Fig. 6C). Their diameter ranged between 400 and 900 nm (Fig. 6D), and some of them contained small vesicles (Fig. 6D) resembling spherules seen abundantly in SFV-infected cells which had not been permeabilized with saponin treatment.
FIG. 6.
Immunogold localization of P12CA3 expressed by adenovirus vector (A and B) and nsP3 or nsP1 in SFV infection (C and D). HeLa cells were infected with adenovirus 12CA3 or SFV and fixed with PLP fixative for 2 h at 28 or 5 h p.i., respectively. Proteins were visualized by treating saponin-permeabilized cells with anti-nsP3 (A, B, and C) or anti-nsP1 (D) rabbit antiserum and nanogold-conjugated Fab fragments. P12CA3 was seen in patches at the plasma membrane (A) and on the outer surface of electron-lucent cytoplasmic vesicles (marked with an asterisk in panel B). In SFV-infected cells nsP1 and nsP3 were on the cytoplasmic side of the limiting membrane of CPVs (C and D) and in patches at the plasma membrane (C). Note that some spherules remained intact even after saponin treatment (arrows in D). Arrows indicate areas positive for immunostaining. Bars, 500 nm (A and D), 200 nm (B), and 2 μm (C). PM, plasma membrane; N, nucleus.
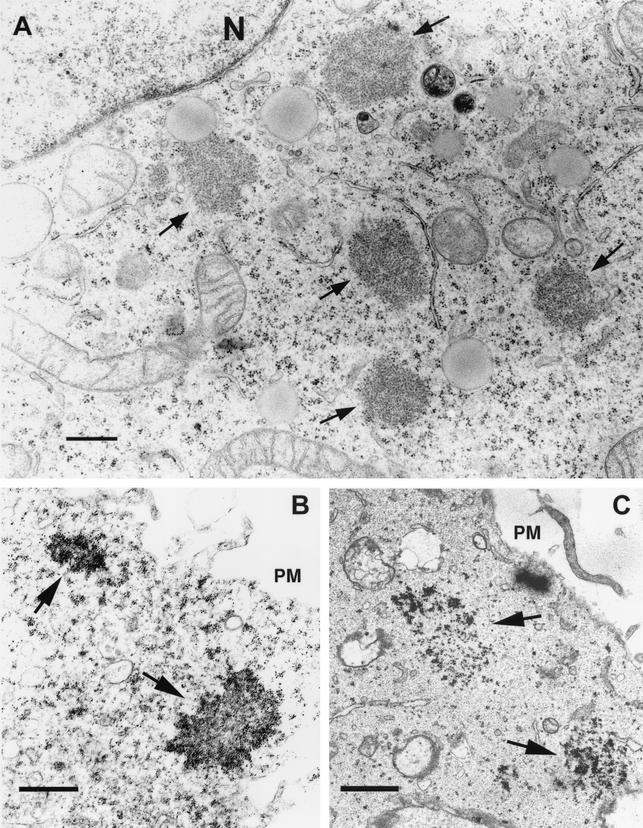
Since nsP3 as part of the P12CA3 polyprotein caused a dramatic change in the targeting of the polyprotein, we investigated cells expressing nsP3 alone. Numerous large electron-dense patches were seen in cells expressing nsP3 by the adenovirus vector (Fig. 7A). When the nsP3-expressing cells were immunostained with anti-nsP3 antiserum, large patches with irregular boundaries were seen (Fig. 7B). Similar irregularly lined patches of nsP3 were seen also in SFV-infected cells, suggesting that a fraction of this protein also aggregates during virus infection (Fig. 7C). We conclude that nsP3, which alone is not a membrane-bound protein, as part of P123 directs this polyprotein to intracellular vesicles which resemble CPV structures seen in SFV-infected cells.
FIG. 7.
Aggregate-like structures of nsP3. HeLa cells were infected with adenovirus AdP3 (A and B) or with SFV (C) and fixed at 28 or 5 h p.i., respectively, using 2.5% glutaraldehyde (A) or treatment supporting the localization by the nanogold technique (B and C) as for Fig. 6. In SFV-infected cells nsP3 was also found abundantly on the cytoplasmic side of the limiting membrane of CPVs and to a lesser extent on the plasma membrane (see Fig. 6C). Putative aggresomes of nsP3 are indicated by arrows. Bars, 500 nm. PM, plasma membrane; N, nucleus.
DISCUSSION
The association of RNA synthesis with intracellular membranes is a typical feature of positive-strand RNA viruses replicating in animal, plant, or insect cells. Information on the targeting of the RNA replication complexes has accumulated steadily during the last 10 years. Poliovirus replication complexes are derived from the endoplasmic reticulum (ER) membranes by the cellular COP II coat machinery (46). Expression of poliovirus nonstructural proteins in the absence of RNA synthesis results in membrane alterations similar to those seen in infected cells (references 55 and 57 and references therein). The RNA synthesis of arteriviruses takes place on ER-derived double-membrane vesicles, to which the replication complex is anchored by the joint action of nonstructural proteins nsp2 and nsp3. Coexpression of these two proteins is necessary and sufficient for the development of the double-membrane vesicles (53). The nonstructural protein 1a of brome mosaic virus, a member of the alphavirus superfamily, expressed in the yeast Saccharomyces cerevisiae is targeted to the cytoplasmic side of the ER (10), where it induces the formation of small perinuclear vesicles or spherules, 50 to 70 nm in diameter, similar to those detected in brome mosaic virus-infected plant cells (48). The RNA synthesis of cowpea mosaic virus, a plant virus member of the picornavirus-like superfamily, takes place on ER-derived small membrane vesicles induced by nonstructural proteins 32K and 60K (9). Spherule-like replication complexes are also formed in flock house virus-infected Drosophila cells on the outer mitochondrial membrane. The single replicase protein A harbors both the targeting address and membrane-binding signal of the RNA replication complex (37).
The alphavirus replication complex is localized to the cytosolic surface of late endosomes and lysosomes, creating CPVs (20). Here we have approached their biogenesis in a stepwise manner. First, we studied whether the individual nsPs interact with each other when expressed pairwise. Interactions were detected by native immunoprecipitation between nsP1 and nsP4 and, to a lesser extent, between nsP1 and nsP3. Second, only upon expression of SFV P123 or P1234 did the nsPs form a membrane-bound complex (Fig. 2). These results are in accordance with those demonstrating that the synthesis of Sindbis virus minus-strand RNA takes place only in cells expressing the nsPs through the polyprotein intermediate (28, 29, 30). Thus, we suggest that interactions of nsP1 with nsP3 and nsP4 at the polyprotein stage enable the assembly of the membrane-bound replication complex. After proper assembly, the components of the complex remain associated even after proteolytic cleavages of the polyprotein. This view is supported by studies of temperature-sensitive Sindbis virus mutants and their revertants, which have suggested interactions between nsP1 and nsP4 (51, 62). However, only a fraction of the synthesized polyproteins form replication complexes in SFV-infected cells (23) or in cells expressing cleavable P123 or P1234, as shown in this study.
Third, since the synthesis of minus-strand RNA is an early event catalyzed only by the P123-nsP4 polymerase, we studied the properties of P1234 and its cleavage intermediates by using constructs in which the nsP2 protease activity was inhibited by a mutation, C478A, in the active site of the enzyme. The enzymatic activities of uncleaved polyproteins, produced in insect cells, indicated that their components had folded correctly. P12CA, P12CA3, and P12CA34 had methyltransferase and guanylyltransferase activities typical for nsP1 (1, 26, 36). Polyproteins P12CA, P2CA3, P12CA3, and P12CA34 had NTPase and RNA triphosphatase activities typical for nsP2 (58). Since all wild-type polyproteins are cleaved both in vitro and in vivo (35), they also possess the protease activity associated with nsP2 (59). At least P12CA and P12CA3 were also palmitoylated like nsP1, suggesting that this posttranslational modification can take place in SFV-infected cells already at the polyprotein stage. Similarly, P12CA3, P12CA34, P2CA3, and P34 were phosphorylated, suggesting that this modification may precede cleavage of nsP3 from the precursor.
Next, we examined the requirements for the intracellular targeting of nonstructural polyprotein by using its truncated uncleaved forms. Flotation experiments showed that P12CA34, P12CA3, and P12CA were associated with the isolated membrane fraction, whereas P2CA3 and P34 were not. Thus, the nsP1 domain of the polyprotein is alone responsible for the membrane association of the polymerase complex. This is in conformity with our previous results where we expressed all four nsPs and found that only nsP1 was associated with membranes by two different mechanisms. Palmitoylation of cysteine residues 418 to 420 of nsP1 made the association very tight, resembling that of integral membrane proteins (24, 40). However, the mutation of these residues to alanines did not abolish membrane binding of nsP1 but made it sensitive to treatment with high salt concentrations (24). Most importantly, palmitoylation of nsP1 was not essential for virus replication and induction of CPVs similar to those in wild-type SFV-infected cells (4). Subsequently, we identified a 20-amino-acid region in the middle of nsP1 which was responsible for the peripheral binding of nsP1 to membranes and liposomes rich in anionic phospholipids. Interaction with anionic phospholipids turned out to be essential for the methyltransferase and guanylyltransferase activities of nsP1 (3). Determination of the nuclear magnetic resonance solution structure of the synthetic lipid-binding peptide revealed an amphipathic helix with hydrophobic amino acid residues on one side and polar residues on the other side. Studies on interaction of the binding peptide with liposomes suggested that nsP1 binds monotopically to the outer leaflet of the lipid bilayer and extends to a depth of 9 to 10 carbon atoms of the acyl chains (27).
To find out the role of the nonstructural polyprotein in targeting of the cleavage intermediates we expressed the uncleaved derivatives separately. Expression of P12CA resulted in the accumulation of the protein at the plasma membrane and filopodium-like structures (Fig. 5B). The staining pattern was strikingly similar to that described for expression of nsP1 alone (24, 25). However, when a cleavable P12 was expressed, nsP1 was localized on the cytoplasmic side of the plasma membrane and in filopodium-like structures, while nsP2 was almost exclusively in the nucleus (Fig. 5A). The result was similar to that obtained by pairwise expression of nsP1 and nsP2. Thus, the targeting of nsP1 to the plasma membrane and filopodia overrides the nuclear attraction of the nsP2 domain in P12CA. A different pattern was seen for P12CA3. As determined by confocal microscopy, the polyprotein was localized to the plasma membrane and to vesicles of variable size, some of which costained with lamp2, a marker of late endosomes. No staining of filopodia similar to those in P12CA-expressing cells was seen. As determined by immuno-EM, P12CA3 was localized in large patches on the cytoplasmic side of the plasma membrane, as well as on the surface of vesicles (200 to 500 nm in diameter), many of which were filled with membranes (Fig. 6). Thus, the presence of the nsP3 domain in the nonstructural polyprotein dramatically affected its intracellular targeting, through an unknown mechanism. Our previous light microscopic studies have suggested that when expressed alone, nsP3 is associated with vesicles whose identity remained unknown (38, 61; A. Salonen, unpublished data). However, when cells expressing nsP3 were examined by immuno-EM, the protein was not associated with membrane-bound compartments. The label was rather found in spots of different sizes, which had irregular boundaries (Fig. 7). These observations, together with the finding that nsP3 did not float with membranes, indicate that it was not associated with vesicles. The large patches of nsP3 resembled EM images of aggresomes, which consist of overexpressed or otherwise misfolded protein aggregates (22).
Thus, nsP3 alone appears to precipitate in the cytosol, whereas as part of P123, it targets the polyprotein to the endosomal apparatus. Our finding that P23 was not membrane associated either indicates that the targeting of P12CA3 (and P12CA34) must be dependent on the properties of both nsP1 and nsP3 domains in the polyproteins. Thus, we propose that during SFV infection the P123 intermediate is necessary for the endolysosomal targeting of the replication complex. This would mean that premature proteolytic cleavages at sites between nsP1 and nsP2, or between nsP2 and nsP3, prevent proper targeting of the polymerase complex.
When the immuno-EM technique was applied to SFV-infected cells, staining of genuine CPVs was quite similar to that seen in P12CA3-expressing cells, suggesting that the polyproteins target the RNA polymerase complex to the surface of the endolysosomal vesicles. We assume that the specific attraction of the amphipathic peptide of nsP1 to phosphatidylserine-rich membranes, such as the cytoplasmic leaflet of the plasma membrane, plays an important role in this process (23). However, only the contribution of the nsP3 domain within the polyprotein enables the targeting to endosomes and lysosomes. One possible hypothesis is that the presence of the nsP3 domain in P123 overrides the attraction of the joined P12 domain to the plasma membrane and filopodia, facilitating its release for endosomal targeting.
Acknowledgments
We thank Tero Ahola and Marja Makarow for critical reading of the manuscript and Airi Sinkko for excellent technical assistance.
These studies were supported by the Academy of Finland (grant 8397) and the Sigrid Juselius Foundation.
REFERENCES
- 1.Ahola, T., and L. Kääriäinen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., P. Laakkonen, H. Vihinen, and L. Kääriäinen. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 71:392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola, T., A. Lampio, P. Auvinen, and L. Kääriäinen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 11:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahola, T., P. Kujala, M. Tuittila, T. Blom, P. Laakkonen, A. Hinkkanen, and P. Auvinen. 2000. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J. Virol. 74:6725-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai, R., M. Geffard, and A. Calas. 1992. Intensification of labelings of the immunogold silver staining method by gold toning. Brain Res. Bull. 28:343-345. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1988. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J. Virol. 62:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 65:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burge, B. W., and E. R. Pfefferkorn. 1967. Temperature-sensitive mutants of Sindbis virus: biochemical correlates of complementation. J. Virol. 1:956-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carette, J. E., K. J. Van Lent, S. A. MacFaralne, J. Wellink, and A. Van Kammen. 2002. Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J. Virol. 76:6293-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, M. X., and M. J. Schlesinger. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280-284. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, R. M., and T. Sreevalsan. 1970. Membrane binding of input arbovirus ribonucleic acid: effect of interferon or cycloheximide. J. Virol. 6:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, R. M., J. G. Levin, P. M. Grimley, and I. K. Berezesky. 1972. Membrane-associated replication complex in arbovirus infection. J. Virol. 10:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez de Cedron, M., N. Ehsani, M. L. Mikkola, J. A. García, and L. Kääriäinen. 1999. RNA helicase activity of Semliki Forest virus replicase protein nsP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 16.Grimley, P. M., I. K. Berezesky, and R. M. Friedman. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 2:1326-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimley, P. M., J. G. Levin, I. K. Berezesky, and R. M. Friedman. 1972. Specific membranous structures associated with the replication of group A arboviruses. J. Virol. 10:492-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, T. C., S. Zhou, L.T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kääriäinen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71:187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson, K., and S. R. Carlsson. 1998. Sorting of lysosomal membrane glycoproteins lamp-1 and lamp-2 into vesicles distinct from mannose 6-phosphate receptor/g-adaptin vesicles at the trans-Golgi network. J. Biol. Chem. 273:18966-18973. [DOI] [PubMed] [Google Scholar]
- 22.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 23.Kujala, P., A. Ikäheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kääriäinen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laakkonen, P., T. Ahola, and L. Kääriäinen. 1996. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 271:28567-28571. [DOI] [PubMed] [Google Scholar]
- 25.Laakkonen, P., P. Auvinen, P. Kujala, and L. Kääriäinen. 1998. Alphavirus replicase protein nsP1 induces filopodia and rearrangement of actin filaments. J. Virol. 72:10265-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laakkonen, P., M. Hyvönen, J. Peränen, and L. Kääriäinen. 1994. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J. Virol. 68:7418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lampio, A., I. Kilpeläinen, S. Pesonen, K. Karhi, P. Auvinen, P. Somerharju, and L. Kääriäinen. 2000. Membrane-binding mechanism of an RNA virus capping enzyme. J. Biol. Chem. 275:37853-37859. [DOI] [PubMed] [Google Scholar]
- 28.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, G. P., M. W. La Starza, W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416-427. [DOI] [PubMed] [Google Scholar]
- 33.Magden, J., N. Takeda, T. Li, P. Auvinen, T. Ahola, T. Miyamura, A. Merits, and L. Kääriäinen. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75:6249-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merits, A., L. Vasiljieva, T. Ahola, L. Kääriäinen, and P. Auvinen. 2001. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 82: 765-773. [DOI] [PubMed] [Google Scholar]
- 36.Mi, S., and V. Stollar. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184:423-427. [DOI] [PubMed] [Google Scholar]
- 37.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peränen, J., and L. Kääriäinen. 1991. Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J. Virol. 65:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peränen, J., M. Rikkonen, and L. Kääriäinen. 1993. A method for exposing hidden antigenic sites in paraformaldehyde-fixed cultured cells, applied to initially unreactive antibodies. J. Histochem. Cytochem. 41:447-454. [DOI] [PubMed] [Google Scholar]
- 40.Peränen, J., P. Laakkonen, M. Hyvönen, and L. Kääriäinen. 1995. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology 208:610-620. [DOI] [PubMed] [Google Scholar]
- 41.Peränen, J., M. Rikkonen, P. Liljeström, and L. Kääriäinen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 64:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peränen, J., K. Takkinen, N. Kalkkinen, and L. Kääriäinen. 1988. Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein. J. Gen. Virol. 69:2165-2178. [DOI] [PubMed] [Google Scholar]
- 43.Ranki, M., and L. Kääriäinen. 1979. Solubilized RNA replication complex from Semliki Forest virus-infected cells. Virology 98:298-307. [DOI] [PubMed] [Google Scholar]
- 44.Rikkonen, M., J. Peränen, and L. Kääriäinen. 1992. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 189:462-473. [DOI] [PubMed] [Google Scholar]
- 45.Rikkonen, M., J. Peränen, and L. Kääriäinen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rust, R. C., L. Landmann, R. Cosert, B. L. Tang, W. Hong, H-P. Hauri, Egger, D., and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawicki, D. L., and S. G. Sawicki. 1980. Short-lived minus-strand polymerase for Semliki Forest virus. J. Virol. 34:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 49.Seemann, J., E. J. Jokitalo, and G. Warren. 2000. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell 11:635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology. 276:148-160. [DOI] [PubMed] [Google Scholar]
- 52.Singh, I., and A. Helenius, A. 1992. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol. 66:7049-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 54.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suhy, D. A., T. H. Giddings, and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kääriäinen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79:309-319. [DOI] [PubMed] [Google Scholar]
- 57.Teterina, N. L., D., K. Bienz, D. Egger, D. M. Brown, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kääriäinen. 2000. Identification of a novel function of the alphavirus capping apparatus—RNA 5′ triphosphatase activity of nsP2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 59.Vasiljeva, L., L. Valmu, L. Kääriäinen, and A. Merits. 2001. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem. 276:30786-30793. [DOI] [PubMed] [Google Scholar]
- 60.Vihinen, H., and J. Saarinen. 2000. Phosphorylation site analysis of Semliki Forest virus nonstructural protein 3. J. Biol. Chem. 275:27775-27783. [DOI] [PubMed] [Google Scholar]
- 61.Vihinen, H., T. Ahola, M. Tuittila, A. Merits, and L. Kääriäinen. 2001. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 276:5745-5752. [DOI] [PubMed] [Google Scholar]
- 62.Wang, Y.-F., S. G. Sawicki, and D. L. Sawicki. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 65:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wengler, G., D. Wurkner, and G. Wengler. 1992. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology 191:880-888. [DOI] [PubMed] [Google Scholar]