Abstract
The large number of protein kinases makes it impractical to determine their specificities and substrates experimentally. Using the available crystal structures, molecular modeling, and sequence analyses of kinases and substrates, we developed a set of rules governing the binding of a heptapeptide substrate motif (surrounding the phosphorylation site) to the kinase and implemented these rules in a web-interfaced program for automated prediction of optimal substrate peptides, taking only the amino acid sequence of a protein kinase as input. We show the utility of the method by analyzing yeast cell cycle control and DNA damage checkpoint pathways. Our method is the only available predictive method generally applicable for identifying possible substrate proteins for protein serine/threonine kinases and helps in silico construction of signaling pathways. The accuracy of prediction is comparable to the accuracy of data from systematic large-scale experimental approaches.
Posttranslational modification of proteins by phosphorylation is the most abundant type of cellular regulation. It affects essentially every cellular process, including metabolism, growth, differentiation, motility, membrane transport, learning, and memory. Defects in protein kinase function result in a variety of diseases, and kinases are a major target for drug design. To ensure signaling fidelity, kinases must be sufficiently specific and act only on a defined subset of cellular targets. Understanding the basis for this substrate specificity is essential for understanding the role of an individual protein kinase in a particular cellular process.
Experimental approaches for determining specificity, such as the use of oriented peptide libraries (1), are expensive and laborious. Identification of in vivo substrates is even more difficult (2). Although identification of novel putative protein kinases via sequence analysis (3, 4) is straightforward, their biological role cannot be generally predicted, and functional genomics approaches are not yet adequate (5, 6). Substrate identification remains one of the rate-limiting steps in understanding the biological roles of novel protein kinases.
The three-dimensional structures of several protein kinases, some with bound substrates and nucleotides, have been determined (7). All protein kinases show a common fold, consisting of two lobes hinged through a short linker region. The active site, where the phosphoryl group is transferred from ATP to the target residue of the substrate, is located in the cleft between the lobes. Active forms of all protein kinase structures have a similar “closed” conformation, and we reasoned that the available structural information could be exploited to develop computational methods that predict substrate specificities of uncharacterized kinases.
On the basis of an analysis of the crystal structures of peptide complexes of protein Ser/Thr kinases (7–11), we identified 20 enzyme residues (“determinants”) that contact the side chains of the residues surrounding the phosphorylation site [only substrate positions (−3), (−2), (−1), (+1), (+2), and (+3) were considered]. Using molecular modeling and sequence analysis of kinases and substrates, we extracted a set of rules that guide the specificity of binding to these positions. We implemented these rules in the web-interfaced program predikin, which performs an automated prediction of optimal substrate peptides by using only the amino acid sequence of the protein kinase as input. To explore the utility of the method, we used predikin to analyze the signaling pathways in two cellular processes in yeast, cell cycle control and DNA damage checkpoints, and predicted new connections in these pathways. Our method should be generally applicable to identifying possible substrate proteins for protein serine/threonine kinases and should aid in unraveling signaling pathways in which these proteins may be involved.
Materials and Methods
Identification of Binding Sites and Sequence Motifs.
The three-dimensional structures of cAMP-dependent protein kinase [protein kinase A (PKA); Protein Data Bank (PDB) ID code 1JBP (9)], phosphorylase kinase [PHK; PDB ID code 2PHK (10)], and cyclin-dependent kinase (CDK) 2 [PDB ID code 1QMZ (11)] with bound substrate peptides were studied to find significant contacts between the catalytic domain and the side chains of the peptide (insightii, Accelrys, San Diego). The locations of key binding residues (determinants) were defined in relation to structural features and conserved sequence motifs (12).
Protein Kinase Models.
As a part of the analysis of substrate specificity determinants, comparative models (insightii) of the kinase catalytic domains with bound substrate peptides were constructed for several kinases [e.g., protein kinase C (PKC)α, PKCζ, PKCμ, calmodulin-dependent kinase (CaMK) 2 (CaMKII), and NIMA]. The choice of template kinase depended on the protein kinase group [AGC (protein kinase A, G, C), CaMK, cyclin-dependent kinase, mitogen-activated protein kinase (MAPK), glycogen synthase kinase 3, CK2 (CMGC), etc. (12)], local similarity, and specific structural features.
The coordinates of ATP or ATP analogues were transferred from the template to the model by superposition. The complexes were energy-minimized using steepest descents and conjugate gradients (discover) to rms deviation of bond lengths of 0.0001 Å, with the coordinates of the backbone atoms of the structurally conserved regions kept constrained. The minimized complexes were examined for likely interactions (<3.5 Å) and steric and electrostatic clashes. The complementarity of an amino acid side chain with a subsite was estimated from the size and charge complementarity and the ability to form favorable interactions (reasonable hydrogen bonds, ionic and van der Waals interactions).
Computer Program.
The program predikin was written with JavaScript. It accepts a protein kinase sequence and outputs predictions of possible heptapeptide substrate sequences. First, it locates (H/Y)RDLKPEN as the characteristic conserved kinase motif and extracts the kinase catalytic domain from the protein sequence provided. Next, it locates other (semi)conserved kinase motifs and, based on the proximity to these motifs, locates the determinant residues. It then applies the specificity rules and predicts an optimal heptapeptide sequence.
To run the program, the user inputs the kinase type and sequence into a form in the browser window. Output consists of the locations of key kinase motifs, the type of kinase, a list of the determinant residues, a list of possible substrate heptapeptide sequences, and commentary text. Substrate data are passed to another window (automatically opened via a link) that contains substrate sequence data formatted for protein database searching.
Searching Protein Databases for Putative Substrates.
Putative substrates were identified using prosite (13), short peptide blast (3), or scansite (14). The searches were usually limited to the same species as the protein kinase. prosite finds all of the sequences containing the permutations of the residues provided. blast will introduce some substitutions and rank the hits according to similarity to the original motif; however, several searches are required to cover all of the sequence choices in a motif. scansite will also introduce substitutions and rank the hits, but only one search is necessary; the simplified user motif method (“Quick Matrix”) was used, where residues making multiple contacts with a subsite pocket were considered “primary” residues and other acceptable residues making fewer contacts were considered “secondary” residues. scansite searches were generally limited to no more than 200 hits, or to scores less than 0.12 (usually, ≈20% of known substrates exactly match the primary residues, and ≈40% exactly match the primary and secondary residues). We found scansite to be the most useful method. The comparison of predictions with the known phosphorylation sites in PhosphoBase was performed using the simplified user motif method in scansite, selecting the substrate protein (according to PhosphoBase) by using the species name and the name of the protein.
Results and Discussion
Development of the Procedure for Specificity Prediction.
The information we used to develop our procedure included the amino acid sequences of protein kinases, the available crystal structures of protein kinases [particularly enzyme-substrate complexes of PKA (9), phosphorylase kinase (10), and CDK2 (11)], and the substrate specificities determined by oriented peptide library experiments (1, 15–17).
The rules were developed in several stages: (i) analysis of residues in contact with the side chains of the substrate in relevant crystal structures; such determinant residues were numbered 1–20 starting at the N terminus (Fig. 1, Table 1, and Table 3, which is published as supporting information on the PNAS web site, www.pnas.org); (ii) locations of determinants were defined in protein kinase sequences relative to conserved sequence motifs (12); (iii) correlations between substrate specificity information (oriented peptide library data), and the determinant residues were sought, taking into account complementarity with the substrate in terms of size, polarity, charge, and hydrogen-bonding potential.
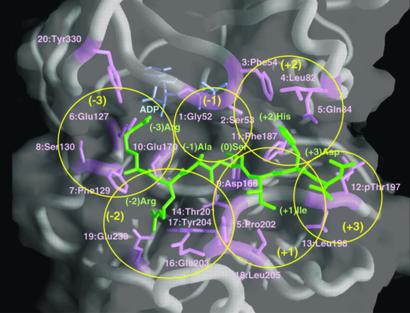
Figure 1.
Substrate-binding site in the crystal structure of PKA (9). The main chain of the protein is shown in worm representation, and the determinant residues (magenta), substrate peptide [green, individual substrate residues are labeled (−3) to (+3)], and ADP (blue) are shown in stick representation. The protein kinase surface is shown in transparent representation, determinant residues are marked 1–20, and individual protein kinase subsites are marked with yellow circles. The figure was produced with GRASP (43).
Table 1.
Comparison of predicted and known substrate specificities for selected protein kinases
| Protein kinase | Predicted optimal heptapeptide substrate sequence* | Optimal sequence based on oriented peptide library experiments | Consensus of known substrates |
|---|---|---|---|
| Protein kinases used to develop the rules | |||
| PKA | (RK)(RKS)X(ST)(FLMI)(AVI)(ILMARK) | (RH)(RKH)(RV)S(IFM)(IVF)(FIM) (1) | (RK)(RK)(SLRGAP)(ST)(LSVR)(STPV)(SAVGE) (20) |
| PKCα | (RKQH)(RKM)X(ST)(FLY)(RKHQ)(AVFLRK) | (RK)(KQR)GS(FMIV)(KRF)(KFR) (16) | (RKS)(RKAG)(LSAR)(ST)(FLVRK)(RKAS)(RKS) (20) |
| PKCζ | (RF)(RKM)X(ST)(FLV)(RKHQ)(AVFLRK) | R(QKY)(GKM)S(FM)(FMA)(YK) (16) | RQGSFFA (16) |
| CaMKII | (RK)(QMRFST)X(ST)(FLY)(EDS)(LIM) | (RQ)(QM)(QMK)S(FIML)(DEI)(LIMK) (15) | (RKG)(QARKLS)(AQLG)(ST)(VLIF)(SADG)(SEMD) (20) |
| PKCμ | (RK)(QMTAS)X(ST)(VILMF)(AVI)(ALIMPFRK) | R(QKEM)(MLK)S(VML)(AV)(FY) (16) | RTASVAF (16) |
| PHK | (RK)(QRMS)X(ST)(VILMF)(QNRKH)(ALIMPFRKHQ) | (RK)(MRQF)(MFL)S(FIM)(LI)(FLI) (15) | R(AQSTL)(ILR)(ST)(VITA)(RHY)(RKFS) (20) |
| NIMA | (RF)(RV)X(ST)R(EDS)(AVFLRK) | (FLM)(RK)(RK)S(RIVM)(RIMV)(MIFV) (15) | FRSSIRR (15) |
| CK2 | (RKQHDSE)(EDS)X(ST)E(DE)(EDS) | (DE)(ED)(ED)S(EDA)(EA)(EA) (15) | (DSE)(ESD)(EGS)(ST)(DE)(ED)E (20) |
| CDK2 | (NGSL)(PVALS)X(ST)P(KRMI)(RKQSL) | H(PH)RSPRK (1) | (TL)(PVLSH)(LATS)(ST)P(PR)(KRL) (20) |
| Protein kinases with peptide library information, used to test predictions | |||
| PKCγ | (RKQH)(RKM)X(ST)(FLY)(RKHQ)(AVFLRK) | R(KRQ)(GK)S(FKM)(KR)(RK) (16) | — |
| PKCδ | (RF)(RKM)X(ST)(FLV)(RKHQ)(AVFLRK) | R(KRQ)(GA)S(FMV)(FKM)Y (16) | MRQSVAV (16) |
| AKT | (RK)(RKS)X(ST)(FLV)(STNGAP)(AVFLRK) | RT(YFGM)S(FM)(GTS) (17) | R(RTSL)(SPRT)(ST)(YSF)(PGAST)(EAND) (19) |
| Slk1 (Bck1) | (QNRKH)(VSAR)X(ST)(AVILMY)(SQN)(AVLIFRK) | (RK)(FR)(GR)S(LIFM)(RIMF)(RFM) (1) | (LA)(AV)X(ST)(FL)(TA)(TG) (1) |
| Chk1 | (RK)(SALF)X(ST)(VA)(AVI)(MFLIRK) | R(YMP)(RF)S(FINM)(SA)(IFL) (19) | RSPSMPE (19) |
| Chk2 | (RK)(ILMA)X(ST)(FLMID)(QNRKH)(ILMARK) | R(YKA)(YE)S(FI)(FIR)(YRF) (19) | R(STI)(FHKP)(ST)(DFM)(LVPS)(WLKE) (19) |
| CK1γ | (QNRKH)(SED)X(ST)(VAILMY)(SQN)(ILMARK) | E(TAG)GSI(IYF)(FY) (15) | (SDET)(SEAV)(SLDE)(ST)(ELSVI)(ESTD)(ESGD) (20) |
| CK1δ | (QNRKH)(SED)X(ST)(VAILMY)(QNRKH)(AVLIMPFTSQN) | E(TAG)GSI(IFY)(IGY) (15) | (SDET)(SEAV)(SLDE)(ST)(ELSVI)(ESTD)(ESGD) (20) |
| ERK1 | (RKF)(PVALS)X(ST)P(PFLI)(RKQSL) | (GPE)(PLI)(LMP)SP(GPF)(PFG) (15) | (PAVTL)(PV)(LT)(ST)P(PSR)(PARKFG) (15) |
| CDK5 | (NGSL)(PVALS)X(ST)P(KRMI)(RKQSL) | H(HP(KG)SP(KR)(HRK) (15) | (EG)(TVH)(KA)(ST)P(VPE)K (15) |
| Protein kinases without peptide library information, used to test predictions | |||
| PKG | (RK)(RKS)X(ST)(AVIP)(AVI)(AVFLRK) | — | R(RK)(RLI)(ST)(RASIK)(SALK)(EPT) (20) |
| βARK | (RKQHE)(QMRFSTY)X(ST)(EQT)(DE)(QNRKHVLIMF) | — | (NTK)(NVT)(DS)(ST)(ENDQ)(EDNG)(RQEDN) (18) |
| S6K(1) | (RK)(RKS)X(ST)(VAIL)(VLIMFC)(AVFLRK) | — | R(RAS)(LS)(ST)(SVL)(SRL)(SRAG) (20) |
| S6K(2) | (RKQH)(RKS)X(ST)(AVILMY)(EDS)(AVFLRK) | — | R(LTA)(SL)(ST)(LVH)(SR)(SA) (18) |
| CaMKI | (RK)(QMRFST)X(ST)(VA)(STNDH)(RK) | — | RRLSDSN (18) |
| AMPK | (RK)(RQMS)X(ST)(VA)(AGVLIMPFTC)(QNRKH) | — | R(SNT)(MEQN)(ST)(FIGK)(LAI)(HFLA) (18) |
“−”, not reported.
X, any amino acid.
The prediction methodology relies on several postulates supported by our analyses and available data: (i) all protein Ser/Thr kinases adopt a similar fold with little scope for conformational differences in the binding cleft, and bind substrates in a similar extended conformation with few bridging water molecules involved (supported by the available crystal structures and the proximity of the substrate binding residues to conserved sequence motifs and structural elements); (ii) residues at the analogous position in a subsite pocket bind the same substrate residue(s) (supported by the available crystal structures); (iii) determinant residues can be identified in any kinase sequence by locating the conserved structural motifs (supported by sequence analyses).
Not all determinants affect specificity, or are involved in substrate binding in all kinases (9–11). The correlations between the determinant residues and substrate specificity were extrapolated to cover other possible sequence variations (Table 4, which is published as supporting information on the PNAS web site). The use of structural data (experimental and comparative models of kinases) implies that that the developed rules are only partially empirical.
General Descriptions of Individual Subsites.
(−3) subsite (determinants 6–8, 10, and 20).
The specificity at (−3) primarily depends on determinant 6. For kinases in the AGC and CaMK groups, Glu or Asp in this position results in a preference for Arg or Lys at (−3). Determinant 7, when hydrophobic, generates the specificity for Pro or Met at (−3). CMGC kinases have residues other than Glu or Asp at 6 and are less specific; determinant 10 consequently has a bigger influence on specificity [e.g., 10 His indicates a (−3) preference for Glu in CK2 (11)].
(−2) subsite (determinants 10, 14–17, and 19).
Binding in the (−2) pocket of AGC and CaMK kinases primarily involves determinant 17 [Phe, effecting hydrophobic specificity; or Tyr, with the phenolic and phenyl groups both possible ligands (for Gln/Arg/Lys and Met/Phe, respectively)]. This specificity is modified in a predictable manner by the nature of 16 [ranging from Glu in PKA (Arg specificity) to Gly in CaMKII (broad specificity)]. Determinant 17 is Trp in CMGC kinases and occludes much of the (−2) pocket, so that binding is restricted to Val, Pro, or Gly; the exception is CK2, where 15 Arg causes a preference for Glu at (−2).
(−1) subsite (determinants 1 and 2).
The (−1) residue makes few contacts with the enzyme. A small residue such as Ala, Gly, or Pro is often found at (−1), although its side chain makes no contacts with the kinase.
(+1) subsite (determinants 11, 13, 15, and 18).
AGC and CaMK kinases frequently prefer large hydrophobic residues, e.g., Phe, when 11, 13, and 18 are large hydrophobic residues, and 15 is Pro (they all contact the phenyl ring). Variations altering specificity include hydrophilic residues at 11, smaller residues at 13 and 18, and Leu or Ala at 15. Tyr at 18 (titin) or at 13 (NIMA) indicate a preference for (+1) Arg. In CDKs and mitogen-activated protein kinases (MAPKs), 15 Leu is diagnostic of the obligatory Pro specificity at (+1).
(+2) subsite (determinants 2, 3, 4, 5, and 11).
In some cases, the binding involves charge–charge interactions, and the specificity is well defined [e.g., PKCα (for Arg/Lys), CK2 (for Glu/Asp)]. Other combinations can result in broad specificity (e.g., PKA, phosphorylase kinase). Determinants 4 and 5 do not bind substrates in CDKs, because this region is involved in cyclin binding (11).
(+3) subsite (determinants 12 and 13).
The subsite is rarely very specific; exceptions include PKCα and CK2 (18). Determinant 12 is often phosphorylated and can result in a preference for Arg/Lys. Determinant 13 (if large enough) is shared with the (+1) site.
No reliable structural information is currently available for subsites N-terminal to (−3) and C-terminal to (+3), although specificity occasionally extends outside this region. It is common that more than one amino acid can successfully bind in a subsite; for example, a typical (+1) subsite can accommodate residues such as Phe, Leu, Met, Ile, and Val to a similar extent. Sometimes even side chains with different properties (e.g., hydrophobic and hydrophilic groups) can bind with similar probabilities. Because the specificity at one subsite can depend on other subsites [links are observed between the (−3) and (−1), (−1), and (+2), (−2) and (+1), and (+1) and (+3) subsites], conditional rules are necessary. Determinants shared by two subsites can contribute to either subsite, and in some cases, both.
Our methodology does not currently cover Tyr kinases; examination of Tyr kinase structures shows that the (−1) residue in the bound peptide bypasses the (−1) and (−2) pockets and binds to what normally is the (−3) pocket, therefore different rules apply to these enzymes. Our rules can be used for protein kinases other than AGC, CaMK, and CMGC groups (e.g., dual specificity kinases, CK1, prokaryote protein kinases); however, the results are less reliable, because little structural information is available.
Computer Program.
We incorporated the rules described above into the web-based computer program predikin, which uses only sequences of protein kinases as input (available on www.biosci.uq.edu.au/kinsub/home.htm; functional within INTERNET EXPLORER 5). The user inputs the amino acid sequence of a protein, and predikin extracts the sequence of the protein kinase catalytic domain (if present), locates the sequence motifs, identifies the determinant residues, and writes a prediction for subsites (−3) to (+3) with relevant comments. A search for possible target proteins by using the predicted optimal heptapeptide sequence can be carried out with prosite (13), blast (3), or scansite (14).
Accuracy of Predictions.
The substrate sequences predicted by predikin are optimal and analogous to those generated by an oriented peptide library experiment. The predictions agree well with the peptide library results (Table 1).
Peptide library data previously suggested that a complete complement of possible interactions is unnecessary for productive substrate binding; this observation has ramifications for using an optimal motif for substrate searches in protein databases (1, 14–17, 19). In the example of PKA [hydrophobic residues are predicted for (+1), (+2), and (+3)], the optimal motif is likely to be unsuitable as a protein kinase substrate, because it would be too hydrophobic to be found on the surface of the protein. Small residues, such as Gly, Ala, Ser, and Pro, are frequently found in PKA substrates at positions (−1), (+1), (+2), and (+3), even though in most cases they would make few contacts with the enzyme; they appear beneficial by reducing hydrophobicity and increasing the likelihood of location in a flexible loop. PKA consequently displays a broad specificity, and the substrates are difficult to predict based on searches with the optimal motif.
Substrate searches based on peptide library data (14) showed up to 70% of published sites [PhosphoBase (20)] predicted at the top 5% level. We performed an analogous test based on predikin predictions (see Materials and Methods) and found comparable statistics on sensitivity and specificity for protein kinases as diverse as PKCα, CaMKII, CK1, and CK2. The substrates not found by either method appear suboptimal, poorly matching the motifs, and many may not be functional in vivo. The accuracy is comparable to secondary structure predictions, as well as systematic large-scale experimental methods (21–24). In individual cases, we observe that the predikin predictions can resemble real substrates better than the peptide library results do (e.g., Slk1, Chk2, and CK1; Table 1), and scansite searches using our predicted motif can produce better results than the equivalent searches using experimental peptide library matrices (14) [e.g., in the first 100 hits, our predicted motif finds 3 and 10 known substrates, and peptide library-based motif only 2 and 1 known substrates for PKCα (16) and CK2 (18), respectively]. Analyses show different predictive powers for individual subsites of the AGC and CaMK class kinases; 70% for (−3) and (−2), 55% for (+1), and 40% for (+2) and (+3) [these numbers are based on direct comparisons of predicted residues to the ones found in real substrates of Ser/Thr kinases (18); no predictions were generally made for the (−1) site]. The predictive power also depends on the specificity; (−3) and (−2) predictions of Arg specificity appear particularly reliable, likely due to the larger number of interactions the Arg residue makes with the protein kinase at these subsites.
predikin can locate phosphorylation sites in a single protein sequence with high reliability. However, the key utility of predikin is to be able to predict novel putative substrates through searching protein databases. The number of hits can range from 1 to 600 for different kinases (using prosite and a single organism; it is most commonly between 100 and 200). The user has the flexibility to restrict the degeneracy of the profile if too many hits are obtained, but the probability of finding known positives increases with the number of hits considered. Only ≈20% of hits appear unlikely as substrates due to incorrect cellular localization or other considerations. These results indicate that predikin can be used even for genome-wide analyses, especially if some additional filtering tools are applied (14). However, any searches have to be performed prudently, and the following caveats need to be considered: (i) some protein kinases are inherently not very specific for reasons outlined earlier; in the cell, apparently weak kinase specificity may be boosted by anchoring, adaptor and scaffold proteins, e.g., the A kinase anchor proteins for PKA (25); (ii) the specificity of some kinases depends on subsites outside the (−3) to (+3) range; (iii) determinant residues can be phosphorylated, affecting specificity (e.g., determinant 12 is a phospho-threonine in many protein kinases); (iv) substrates can be prephosphorylated in adjacent sites [e.g., many phosphorylation sites in CK1 substrates are surrounded by phospho-serines (18)]; (v) small residues such as Gly, Ala, Ser, and Pro are often present in substrate sequences even though they make few, if any, contacts with the enzyme; residues binding in other sites appear to be able to provide adequate interactions for the substrate to bind successfully; (vi) occasionally, residues inconsistent with our predictions (or peptide library results) are found at (+3), together with small bend-forming residues at (+2) [e.g., protein kinase G (PKG) and CaMKII]; because the amino acids present at (+3) appear to follow the predictions for (+2) instead, the (+3) residues may in these cases in fact bind to the (+2) pocket; and (vii) the substrate sequence must be accessible to the kinase and capable of adopting an extended conformation.
To test the utility of predikin in finding novel putative substrates and constructing signal transduction pathways, we analyzed the protein kinases involved in the cell cycle control and DNA damage checkpoint pathways in yeast. We identified phosphorylation sites for substrates with unmapped sites and many plausible phosphorylation events within the pathways and between proteins known to interact (Table 2; Figs. 2 and 3; Tables 5 and 6, which are published as supporting information on the PNAS web site).
Table 2.
Examples of substrates predicted for selected protein kinases in the cell cycle control and DNA damage checkpoint pathways in S. cerevisiae
| Protein kinase | Predicted substrate | Predicted phosphorylation site | scansite score (14) | Substrate function | Evidence for kinase–substrate association |
|---|---|---|---|---|---|
| Hsl1 | Kss1 | RPV83SIDK | 0.084 | Mitogen-activated protein kinase, filamentous growth | Pathway downstream of CDC28 (26, 27) |
| Swe1 | QFS457TVYQ | 0.184 | CDC28 inhibitor | Known substrate (28) | |
| Kel1 | QEL1001TISK | — | Cell morphology | Physical association (with Hsl1, Hsl7, Swe1) (44) | |
| Hsl7 | HLD771SINK | — | Swe1 inhibition | Physical association (with Hsl1 and Swe1) (28) | |
| Dun1 | Mre11 | KMK308SISL | 0.083 | Maintaining telomere length, DNA repair | Physical association (with Rad50, Xrs2) (23) |
| Rad9 | RML1203TIDL | 0.063 | Cell cycle arrest | Linked pathway (23) | |
| Rad24 | QME165SFSE | 0.063 | Cell cycle arrest | Linked pathway (23) | |
| Rad50 | RQS336SLQS | 0.083 | DNA repair | Physical association (with Mre11, Xrs2) (23) | |
| Xrs2 | KLT78SLNK | 0.190 | Maintaining telomere length, DNA repair | Known substrate, physical association (with Rad50, Mre11) (23) | |
| Rfx1 | KSK242TIEE | 0.147 | Repressor of DNA-inducible genes | Known substrate (40) | |
| Rfx1 | KVL259SMDS | 0.210 | Repressor of DNA-inducible genes | Known substrate (40) | |
| Adr1 | KSQ731TIEL | 0.083 | Regulatory protein | Known substrate (40) | |
| Adr1 | RRA230SFSA | 0.083 | Regulatory protein | Known site (40) | |
| Rir1 (Sml1) | KQT91TKQF | 0.064 | DNA-inducible ribonucleoside reductase | Known substrate (35) | |
| Dun1 | QQS488SVSL | 0.123 | Checkpoint kinase | Autophosphorylation |
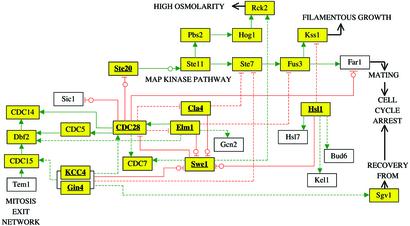
Figure 2.
Schematic diagram of signaling connections linked to cell cycle control in S. cerevisiae. Yellow boxes, protein kinases; solid and dashed green arrowed connections, known and predicted activatory phosphorylations, respectively; solid and dashed red blocked connections, known and predicted inhibitory phosphorylations, respectively; circles, predicted sites in known substrates; open black arrows, general connections between processes. The joined boxes represent complexes. For the protein kinases analyzed (bold and underlined), all known interactions shown were also successfully predicted with PREDIKIN.
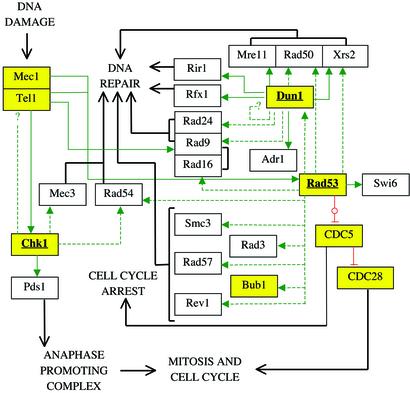
Figure 3.
Schematic diagram of signaling connections linked to DNA damage checkpoints in S. cerevisiae, drawn as in Fig. 2. For the protein kinases analyzed (bold and underlined), all known interactions shown were also successfully predicted with PREDIKIN.
Yeast Cell Cycle Control Pathways.
Under normal circumstances, yeast cells passage through G1, S, G2, and M (mitosis) phases of the cell cycle. Many of these processes are controlled by a single “master switch” protein kinase CDC28, a CDK that uses different cyclins at different stages of the cell cycle (26). For example, the transition from G2 to M and the simultaneous septin ring formation requires the complex of CDC28 with cyclin B (27). A group of septin-localized protein kinases controls CDC28 activity; the dual specificity kinase Swe1 is a negative regulator (antimitotic; phosphorylates and inhibits CDC28), whereas KCC4, Gin4, Hsl1, Cla4, and Elm1 are positive regulators (by inhibiting Swe1 activity). The cell cycle is shut down in situations such as pheromone-induced cell cycle arrest (leading to mating), high osmolarity, and starvation (leading to filamentous growth); signaling in each of these situations involves a central MAPK cascade with a shared MAPKKKK (Ste20) and MAPKKK (Ste11), before branching to a specific MAPK (e.g., Kss1 in filamentous growth, Fus3 in mating) (26). The known pathway connections are depicted with solid lines in Fig. 2. The results of predictions for selected cell cycle control kinases are listed below.
CDC28.
CDC28 is predicted to have a broad specificity, consistent with its many known substrates and diverse roles (not all known substrates have functions related to cell budding). Our analysis predicts phosphorylation sites in several proteins known to be substrates (with hitherto unknown sites), and several new substrates are predicted, suggesting regulatory connections to various related pathways such as pheromone-induced cell cycle arrest and mitotic exit network (e.g., CDC7, Elm1, and Cla4; Fig. 2).
Swe1.
Our analysis confirms CDC28 as Swe1 substrate (27) and predicts new substrates, including CDC7 and Rck2 in related pathways, and CDKs Ctk1 and Kin28. Phosphorylation of Rck2, a Hog1 substrate in the antimitotic high osmolarity pathway (26), would, for example, be predicted to activate the kinase.
KCC4, Gin4, and Hsl1.
These closely related kinases are predicted to have a similar specificity, except that Hsl1 differs at (+2). We predict that KCC4, Gin4, and Hsl1 phosphorylate Swe1 at a site that would interfere with ATP and substrate binding. The predicted site in CDC28, on the other hand, is close to the cyclin-binding site. An analogous site is present in Sgv1, involved in recovery from pheromone-induced cell arrest (28). We identified two other potential KCC4/Gin4 substrates that would link the various cell cycle control pathways, CDC15 (29) and Ste7 (18). Hsl1 is known to bind and phosphorylate Hsl7, the complex then phosphorylating Swe1; we can predict the likely sites in these proteins. Other predicted Hsl1 substrates could contribute to promoting mitosis and suppressing the MAPK pathways.
Cla4 and Ste20.
These two enzymes are predicted to have the same specificity; the opposing functions (Cla4 inhibits Swe1 and Ste20 activates Ste11) must therefore result from different cellular localization and regulation. The major predicted site in Swe1 is shared with KCC4/Gin4/Hsl1. Predictions confirm Ste11 as Ste20 substrate, leading into a MAPK cascade.
Elm1.
The best site in the known substrate Swe1 (30) is shared with KCC4/Gin4/Hsl1 and Cla4/Ste20. New predicted substrates include Fus3, Gcn2 (18), and Dbf2 (26), which could regulate the connected pathways.
Yeast DNA Damage Checkpoint Pathways.
Damage to double-stranded DNA triggers a number of cellular events including cessation of cell cycle and mitosis, apoptosis, and DNA repair. Protein kinases play a central role, both as sensors of damage (lipid kinase-like kinases Mec1 and Tel1 in yeast, ATM, and ATR in humans) and downstream signal transducers (CaMK group kinases Chk1, Rad53, and Dun1 in yeast; Chk1 and Chk2 in humans) (31). Although many components have been identified in checkpoint and DNA repair pathways, only a few kinase substrates have been established. The known pathway connections are depicted with solid lines in Fig. 3. The results of predictions for selected checkpoint kinases are listed below.
Chk1.
Yeast Chk1 is predicted to have similar specificity as its human namesake (19). Several phosphorylation sites are predicted in the known substrate Pds1 [phosphorylation of Pds1 leads to its ubiquitination and destruction by the anaphase promoting complex (APC) (32)]. Other predicted substrates with related functions (33) include the sensor protein kinase Tel1, the checkpoint protein Mec3 (32), and DNA repair protein Rad54 (34).
Rad53.
The predicted specificity of Rad53 is similar but not identical to Chk1. No phosphorylation sites are predicted in its known binding partner Rad9. New predicted substrates include Xrs2 (35) and Rad50 [involved in double-strand DNA break repair and telomeric function through association with Mre11 (36)]; several proteins associated with radiation damage (34); DNA polymerase Rev1 (37); checkpoint kinase Bub1 (38); Rad53-associated protein Smc3 [involved in chromatin adhesion and DNA recombination (39)]; and Dun1 [previously proposed to act downstream of Rad53 (40)].
Dun1.
Several proteins known to associate with Dun1 (23) are predicted to be its substrates; Mre11 and Rad50 [members of the Mre11-Rad50-Xrs2 complex (36)], and Rad9 and Rad24 (involved in nucleotide excision repair and S-phase regulation). Sites are predicted in the known substrates Rfx1 (Crt1) (41) and Adr1 (40). The site in the known substrate Rir1 is successfully predicted, although the (+1) residue is suboptimal (35).
Conclusion
Using the available three-dimensional structural information, we developed a set of rules that govern substrate specificity of classical protein Ser/Thr kinases, CMGC, and dual specificity kinases, and incorporated these rules in a web-interfaced computer program predikin for specificity prediction. Our analyses show that families of kinases with overall sequence similarities or similar regulatory mechanisms do not necessarily have similar substrate specificities, therefore one should be careful inferring functions based on sequence comparisons. Our results suggest that it is possible to make rational predictions of the optimal substrates based on protein kinase sequence alone. We show in an example involving yeast signal transduction pathways that such methodology aids in identifying the substrates of known and novel protein kinases deduced from genome sequences, the components of signaling networks, and therefore has the potential of identifying new therapeutic targets.
The important distinctive feature of our method is that it can make predictions of specificity. Methods exist to suggest sites in proteins that may be phosphorylated by characterized kinases, based on analyses of known substrates (42) or peptide library results (14). Our tests show that our method shows similar accuracy of identifying such sites, but additionally it can predict sites phosphorylated by uncharacterized protein kinases for which no information other than amino acid sequence is available.
Flawless substrate prediction using our methodology is not achievable. Substrate recognition in the cell depends not only on the internal molecular specificity of a protein kinase for a certain peptide sequence, but on other cellular mechanisms, particularly specific localization. To increase the probability of correct identification of substrates, specificity information should be integrated with other available information such as cellular localization, functional information and structural information for substrate proteins, and used with filtering tools such as dual motif searches (14). The significance of the method is its utility. The method works extremely well predicting a phosphorylation site in a protein known to be phosphorylated. The more general utility is the identification of new substrates; even by partially narrowing the list of candidates to be tested experimentally, substantial savings can be made on cost and duration of experimental research.
Our results also show the potential that similar methodology is extended to other proteins which recognize short amino acid motifs, such as modular signal transduction domains (SH2, FHA).
Supplementary Material
Acknowledgments
Early work on this project was supported by the Australian Research Council (B.K.). B.K. is a Wellcome Senior Research Fellow in Medical Science in Australia.
Abbreviations
- PKA
protein kinase A
- CDK
cyclin-dependent kinase
- MAPK
mitogen-activated protein kinase
- CMGC
CDK, MAPK, glycogen synthase kinase 3, CK2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 2.Hardie D G. Protein Phosphorylation. Oxford: Oxford Univ. Press; 1999. [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting C P, Schultz J, Milpetz F, Bork P. Nucleic Acids Res. 1999;27:229–232. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop A C, Ubersax J A, Petsch D T, Matheos D P, Gray N S, Blethrow J, Shimizu E, Tsien J Z, Schultz P G, Rose M D, et al. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Klemic J F, Chang S, Bertone P, Casamayor A, Klemic K G, Smith D, Gerstein M, Reed M A, Snyder M. Nat Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 7.Knighton D R, Zheng J, Ten Eyck L F, Ashford V A, Xuong N-H, Taylor S S, Sowadski J M. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 8.Bossemeyer D, Engh R A, Kinzel V, Postingl H, Huber R. EMBO J. 1993;12:849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madhusudan, Trafny E A, Xuong N-H, Adams J A, Ten Eyck L F, Taylor S S, Sowadski J M. Protein Sci. 1994;3:176–187. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe E D, Noble M E, Skamnaki V T, Oikonomakos N G, Owen D J, Johnson L N. EMBO J. 1997;16:6646–6658. doi: 10.1093/emboj/16.22.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown N R, Noble M E, Endicott J A, Johnson L N. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 12.Hanks S K, Quinn A M. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann K, Bucher P, Falquet L, Bairoch A. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaffe M B, Leparc G G, Lai J, Obata T, Volinia S, Cantley L C. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 15.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, et al. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa K, Toker A, Johannes F J, Songyang Z, Cantley L C. J Biol Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 17.Obata T, Yaffe M B, Leparc G G, Piro E T, Maegawa H, Kashiwagi A, Kikkawa R, Cantley L C. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 18.Pearson R B, Kemp B E. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill T, Giarratani L, Chen P, Iyer L, Lee C H, Bobiak M, Kanai F, Zhou B B, Chung J H, Rathbun G A. J Biol Chem. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- 20.Kreegipuu A, Blom N, Brunak S. Nucleic Acids Res. 1999;27:237–239. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 22.Gavin A C, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick J M, Michon A M, Cruciat C M, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 23.Ho Y, Gruhler A, Heilbut A, Bader G D, Moore L, Adams S L, Millar A, Taylor P, Bennett K, Boutilier K, et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 26.Hunter T, Plowman G D. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 27.Mendenhall M D, Hodge A E. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cid V J, Shulewitz M J, McDonald K L, Thorner J. Mol Biol Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sreenivasan A, Kellogg D. Mol Cell Biol. 1999;19:7983–7994. doi: 10.1128/mcb.19.12.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgington N P, Blacketer M J, Bierwagen T A, Myers A M. Mol Cell Biol. 1999;19:1369–1380. doi: 10.1128/mcb.19.2.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durocher D, Jackson S P. Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Liu D, Wang Y, Qin J, Elledge S J. Genes Dev. 2001;15:1361–1372. doi: 10.1101/gad.893201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myung K, Datta A, Kolodner R D. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 34.Moore C W. Mutat Res. 1978;51:165–180. doi: 10.1016/s0027-5107(78)80016-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhao X, Rothstein R. Proc Natl Acad Sci USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamoto Y, Taggart A K, Zakian V A. Curr Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 37.Haracska L, Prakash S, Prakash L. J Biol Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 38.Roberts B T, Farr K A, Hoyt M A. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S T, Xu B, Kastan M B. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge S J. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 41.Huang M, Zhou Z, Elledge S J. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 42.Blom N, Gammeltoft S, Brunak S. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 43.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 44.Philips J, Herskowitz I. J Cell Biol. 1998;143:375–389. doi: 10.1083/jcb.143.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.