Abstract
Spliced leader RNA transcription is essential for cell viability in trypanosomes. The SL RNA genes are expressed from the only defined RNA polymerase II-dependent promoter identified to date in the trypanosome genome. The SL RNA gene promoter has been shown by in vitro and in vivo analyses to have a tripartite architecture. The upstream most cis-acting element, called PBP-1E, is located between 70 and 60 bp upstream from the transcription start site. This essential element functions along with two downstream elements to direct efficient and proper initiation of transcription. Electrophoretic mobility-shift studies detected a 122-kDa protein, called PBP-1, which interacts with PBP-1E. This protein is the first sequence-specific, double-stranded DNA-binding protein isolated in trypanosomes. Three polypeptides copurify with PBP-1 activity, suggesting that PBP-1 is composed of 57-, 46-, and 36-kDa subunits. We have cloned the genes that encode the 57- and 46-kDa subunits. The 46-kDa protein is a previously uncharacterized protein and may be unique to trypanosomes. Its predicted tertiary structure suggests it binds DNA as part of a complex. The 57-kDa subunit is orthologous to the human small nuclear RNA-activating protein (SNAP)50, which is an essential subunit of the SNAP complex (SNAPc). In human cells, SNAPc binds to the proximal sequence element in both RNA polymerase II- and III-dependent small nuclear RNA gene promoters. These findings identify a surprising link in the transcriptional machinery across a large evolutionary distance in the regulation of small nuclear RNA genes in eukaryotes.
Keywords: spliced leader RNA genes‖snRNA transcription
The trypanosomatids are a family of parasitic protozoa that cause a wide range of diseases in humans and domestic cattle in many areas of the world. Their evolutionary divergence from other unicellular eukaryotes and from metazoans is illustrated by their surprising array of unusual molecular phenomena. Most strikingly, mRNA maturation of nuclear-encoded protein-coding transcripts requires a transsplicing reaction that places a 39-nt capped RNA sequence (the spliced leader or SL) upstream from the start site of an ORF (1). Transsplicing of a short RNA onto mRNAs is not limited to trypanosomes; parasitic worms, nonparasitic worms, euglenoids, and primitive chordates assemble many of their mRNAs by adding an independently synthesized short RNA to the body of the message (2–6). A unique feature of the trypanosomatids is that SL RNA addition is essential for formation of every nuclear-encoded mature mRNA (7). Many mRNA-coding genes in trypanosomes are tandemly arrayed and appear to be transcribed together as long polycistronic primary RNAs (8–10). Transsplicing of the SL to a region upstream of a translational start site, along with polyadenylation, converts the polycistronic premRNAs into mature, translatable mRNA (11–13).
The transspliced SL sequence is derived from the 5′-end of a small nuclear RNA (snRNA), the SL RNA, which is a short (≈120 nt) nonpolyadenylated RNA (11). Approximately 200 copies of SL RNA gene are present in the parasite genome, and most of them exist as repetitive units. Individual SL RNA repeats have their own promoter. In Leptomonas seymouri, in vivo and in vitro studies have shown that a 90-bp upstream region, followed by a 70-bp internal region, is sufficient for proper initiation and termination of SL RNA (14–16).
To date, the only RNA polymerase II (RNAP II)-dependent promoter identified in trypanosomes is the SL RNA gene promoter (28). Accordingly, many groups have compared and contrasted the promoter elements in various trypanosome species (15, 17–21). In most cases, SL RNA promoters consist of three closely spaced short elements that are located upstream of and proximal to the transcription start site. Mutational analysis shows that nucleotides near the start site (+1), at position −10 to −1 bp in L. seymouri (22), −11 to −2 bp in T. brucei (19), and −10 to +10 bp in Leishmania amazonensis (23) serve to direct correct transcription initiation of the SL RNA. An upstream −80- to −60-bp element in the promoter is essential for efficient SL RNA expression in vivo in all trypanosomes analyzed. A trinucleotide GAC sequence is conserved within this upstream promoter element. By using a combination of in vitro transcription assays, electrophoretic mobility-shift assays (EMSAs), and DNA footprinting analysis, we have begun to define the proteins that regulate SL RNA transcription. Because the SL RNA gene promoter is the only defined trypanosome RNAP II-dependent promoter, transcriptional analysis of the SL RNA gene will help uncover the transcriptional mechanisms used by these protozoan parasites.
Central to our understanding of the SL RNA transcription machinery is the identification of PBP-1 (24). This transcription factor is a sequence-specific double-stranded DNA-binding protein that interacts with the essential −80- to −60-bp region of the SL RNA gene promoter. In this report, PBP-1 has been purified to apparent homogeneity. It is composed of three polypeptides, with molecular masses of 57, 46, and 36 kDa. Density sedimentation and gel filtration analysis previously demonstrated that the PBP-1 activity cofractionated with a 122-kDa protein. Therefore, PBP-1 is likely a heterotrimer, containing one copy of each polypeptide. p57 and p46 have been cloned and sequenced. p46 is a unique protein with multiple dileucine residues. Threading analysis suggests that the leucines localize to a concave face of the protein consistent with protein–protein interactions. The p57 subunit of PBP-1 is orthologous to the human small nuclear RNA-activating protein (SNAP)50, which is an essential component of the SNAP/proximal sequence element-binding transcription factor (PTF) complex. This complex participates in the transcription initiation of all human snRNA genes (25). Therefore, these findings demonstrate a conserved role for SNAP50 in RNA transcription across great evolutionary distances.
Materials and Methods
Protein Purification.
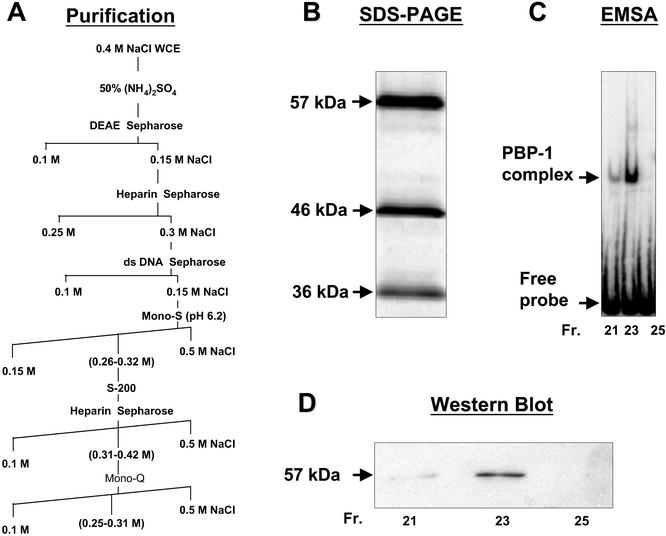
L. seymouri was grown and harvested, and the PBP-1-enriched lysate was prepared as described (24). A 50% ammonium sulfate precipitation step was used to concentrate protein before DEAE chromatography. A detailed purification scheme is shown in Fig. 1. EMSAs that used a 66-bp 32P-labeled probe corresponding to the −83- to −17-bp region of the L. seymouri SL RNA gene promoter were performed as described in ref. 24 and used to monitor PBP-1 activity during purification. The final Mono-Q chromatography (Amersham Pharmacia) step yielded a homogeneous preparation of PBP-1 protein, as determined by silver staining of SDS/polyacrylamide gels after electrophoresis.
Figure 1.
Purification of PBP-1. (A) The biochemical steps used in the PBP-1 purification. The final Mono-Q Sepharose column yielded the three polypeptides shown in a silver-stained 10% SDS/PAGE gel (B). (C) Fractions 21, 23, and 25 from the Mono-Q column contain the peak PBP-1 activity. (D) An immunoblot of the proteins eluted in these fractions and probed with anti-p57 antiserum.
Peptide sequence was obtained from each of the three polypeptides following in-gel trypsin digestion of a Coomassie blue-stained preparative SDS/PAGE. Trypsin fragments were separated by HPLC, and selected peptides were sequenced by Edman degradation at the Harvard Microchemistry Facility (Cambridge, MA).
Isolation of the 46- and 57-kDa Protein-Coding Genes.
To generate a DNA probe for the 57-kDa protein gene, we designed degenerate oligonucleotides containing sense and antisense sequences corresponding to the termini of the largest peptide (21 aa) obtained from the Edman analysis. Reverse transcription of L. seymouri mRNA followed by PCR amplification with degenerate oligonucleotides produced a 54-bp DNA that precisely encoded the four internal amino acids present in the peptide sequence. This DNA was used as a 32P-labeled probe to screen a L. seymouri genomic DNA library constructed in λ-EMBL3 by partial Sau3A digestion. The 9-kb insert identified in a positive plaque was subcloned into pUC19. DNA sequencing identified an ORF of 476 aa that contained the three peptides obtained from Edman degradation. RT-PCR analysis showed that the p57 transcript possesses a 30-nt 5′-untranslated region (data not shown).
The p46 protein-coding gene was isolated in a similar manner. Northern analysis demonstrated that p46 is translated from a 1.6-kb mRNA (data not shown).
Purification of Histidine-Tagged p57 and Ab Production.
The entire ORF of p57 was subcloned into pET15b (Novagen) and expressed as an amino-terminal His-fusion protein. Polyclonal Ab was raised against insoluble (His)p57 in rabbits (Cocalico, Reamstown, PA). To improve antisera specificity, Abs were purified by using a maltose-binding protein-p57 fusion polypeptide as antigen (26). Western blot analyses confirmed that the affinity-purified antisera specifically reacted with both recombinant and parasite p57.
Immobilized Template Preparation and Preinitiation Complex (PIC) Formation.
The WT-SL RNA gene promoter was uniquely labeled at the upstream, 5′-end by using a 5′-biotin labeled oligonucleotide (the −150- to −130-bp region of the SL RNA gene) and an unlabeled oligonucleotide, (the +100- to +120-bp region) in PCRs. The promoter substitution mutation (S-Mut SL) was made by recombinant PCR. This template contained a biotin group at its upstream 5′-end and a substitution within the −72- to −62-bp region of the promoter.
The promoter deletion mutation (Δ-Mut SL) was synthesized by replacement of the −60- to −10-bp-promoter region with a random 30-bp sequence that contained a BglII restriction site. (The 30-bp sequence was 5′-biotinGTTAAGGCCAGATCTATACGCTATAACAGA.) The −10- to −1-bp region contains an initiator element that is important for correct transcription initiation. Templates were gel-purified before use.
Biotinylated DNAs were immobilized on streptavidin-coated resin (Dynabeads M-280, Dynal, Great Neck, NY) (27). In brief, beads were washed twice with buffer S (10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/1 M NaCl/0.01% Nonidet P-40), concentrated with a magnetic particle concentrator, resuspended to a concentration of 4 mg/ml, incubated with ≈50 fmol/μg biotinylated template DNA at room temperature (15 min), washed twice with buffer S, and blocked (15 min at room temperature) with blocking buffer (50 mg/ml BSA/5 mg/ml polyvinylpyrrolidone in transcription buffer). DNA bound resin was washed three times at room temperature with buffer B (150 mM K-glutamate/10 mM Hepes-KOH, pH 7.9/2.5 mM MgCl2/2.5 mM DTT/3% polyethylene glycol 3350/0.5% Nonidet P-40/1 μM each of pepstatin, leupeptin, and PMSF) and resuspended to a final concentration of 10 mg/ml.
PICs were formed on immobilized templates by mixing 100 μg of nuclear extract (20 μl) with 2 μg of pUC19 DNA (90-μl total volume) and incubating at room temperature for 10 min. After centrifugation (9,000 × g, 1 min), the supernatant was mixed with 10 μl of immobilized template and incubated (28°C, 30 min) to allow assembly of PICs.
Transcription reactions were performed by using purified PICs that were washed (four times with 500 μl of buffer B) and collected by using the magnetic particle concentrator to remove all loosely associated proteins by adding ribonucleotides, creatine phosphate, and creatine phosphokinase in transcription buffer (28°C, 30 min).
Proteins associated with the PICs were detected by Western blot analyses. PICs were released from the resin by addition of 40 μl of restriction buffer and cleaved with a restriction enzyme between the biotinylated base and the upstream promoter region. Kpn l digestion released the WT and S-Mut SL RNA promoters; Bgl ll digestion released the Δ-Mut promoter. Resin was removed from the PICs by using a magnetic particle concentrator, and supernatants were boiled in SDS/sample buffer and separated on a 7% SDS/PAGE gel. Resolved proteins were transferred to poly(vinylidene difluoride) membranes, immunoblotted by using anti-carboxyl-terminal domain (28) and anti-p57 Abs, and detected by using ECL reagents (Amersham Pharmacia).
Results
Purification and Subunit Characterization of PBP-1.
PBP-1 is the first double-stranded DNA-binding protein identified in trypanosomes. It binds to the −74 to −60 region of the SL RNA gene promoter, as shown by EMSA analysis. By using the model trypanosomatid L. seymouri, in vitro and in vivo studies demonstrated that mutations in this promoter element blocked SL RNA expression. Biophysical analysis confirmed that PBP-1 was a spherical molecule with a molecular mass of 122 ± 16 kDa. By characterizing PBP-1, we hoped to gain insight into RNAP II-dependent transcription in this group of organisms that are evolutionarily very distant from other well studied eukaryotes. Accordingly, we undertook a large-scale purification of PBP-1. Fig. 1A shows the purification scheme, and Fig. 1B shows the protein profile of the fractions from the Mono-Q column that correlates with the peak of EMSA activity (Fig. 1C). PBP-1 is composed of three subunits, with molecular masses of 57, 46, and 36 kDa. Because PBP-1 is a 122-kDa protein (24), it likely contains one molecule of each polypeptide. To understand the function of these polypeptides, the genes corresponding to the p57 and p46 proteins were cloned and analyzed. Fig. 1D shows the p57 in the active fractions on a Western blot.
A Unique Polypeptide Is a Component of PBP-1.
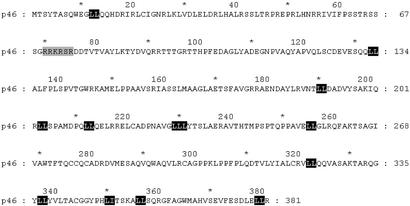
The p46 sequence is shown in Fig. 2. It is a 381-aa protein (calculated molecular mass of 42.3 kDa) with a net positive charge at neutral pH (pI = 8.9). Database searches revealed that p46 was a unique protein. We produced His-tagged and maltose-binding protein-tagged recombinant p46 in Escherichia coli and His-tagged recombinant p46 in baculovirus for functional assays. In each case, only insoluble protein was obtained. In addition, Abs produced against either insoluble protein or synthetic peptides were not of sufficient specificity for further studies. Molecular modeling, by using a 3D fold-based homology search algorithm, ProFit by Proceryon Biosciences (New York) (37) revealed that p46 has a 3D surface replete with hydrophobic leucine residues on one face of the protein. These data correlate with the finding that p46 is associated with other proteins in a multisubunit complex. Furthermore, a molecular threading profile analysis [ProSup by Proceryon Biosciences (38)], which calculates the most thermodynamically stable conformation of a polypeptide, suggests p46 is a member of the 1DML family of proteins. The founding member of this family is the herpes simplex UL42 protein (29). Because UL42 functions as a DNA-binding protein during DNA polymerase movement along the replication fork (30), we infer that p46 also functions as a component of the DNA-binding activity of PBP-1.
Figure 2.
The p46 subunit of PBP-1 is a previously uncharacterized protein. The 381-aa sequence of the L. seymouri p46 protein is shown. All dileucines are in black. The gray-shaded sequence is a nuclear localization signal.
The 57-kDa Subunit of PBP-1 Is Orthologous to an Essential Subunit of SNAPc.
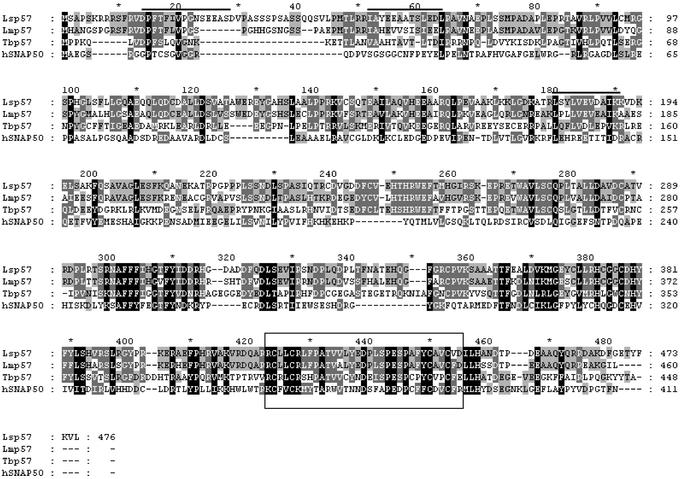
The 476-aa sequence of L. seymouri p57 is shown in line one of Fig. 3. Lsp57 is acidic at neutral pH (pI = 5.4). Bioinformatic analysis demonstrated that this protein is conserved among trypanosomatids. The L. major and Trypanosoma brucei p57 sequences are included in Fig. 3. Among these three sequences, the p57 proteins are 55–75% similar (36–70% identical). Surprisingly, the trypanosomal 57-kDa protein is strikingly similar to the SNAP50 subunit of the human SNAPc/PTF complex (hSNAP50) (31, 32) (see Fig. 3). There is an overall 43% similarity (23% identity) between Lsp57 and hSNAP50 (the expected “E” value is <e−15); the homology is significantly higher in the carboxyl-terminal third of the proteins. SNAPc is a multisubunit transcription factor required for expression of most, if not all, snRNA genes in human cells (25). A C4-type zinc finger is located near the carboxyl end of both the trypanosome and human proteins. The function of this region is unknown.
Figure 3.
Multiple sequence alignment of the p57 orthologs from L. seymouri (Lsp57), L. major (Lmp57, accession no. AC058781), T. brucei (Tbp57, TRYP9.0.000918; Sanger Center, Cambridge, U.K.), and HeLa SNAP50 (hSNAP50, accession no. HSU71300). The alignment was obtained by using the clustalw program. The peptides overlined above the L. seymouri sequence were obtained from peptide sequencing. The C4-type zinc finger conserved among the proteins is boxed. SNAP50 has an additional zinc finger located upstream from the boxed region. The black shade means that the residues either are identical or conserved substitution in all four sequences. The medium shade means identical or conserved substitution in three of four sequences. The light shade means the same in two sequences.
p57 Is Required for SL RNA Transcription.
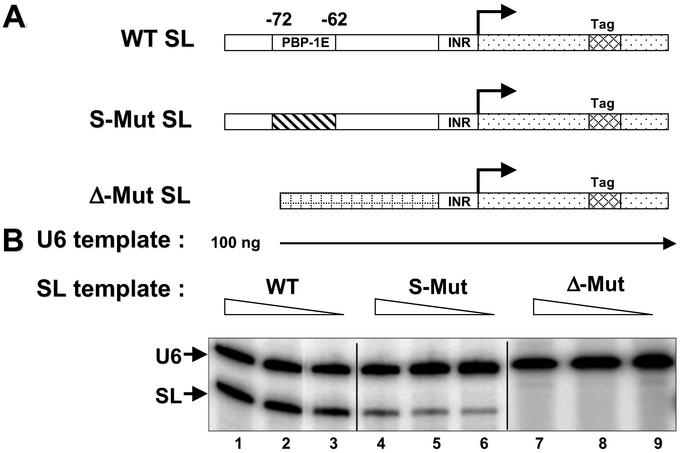
The finding that trypanosomal p57 is orthologous to hSNAP50 indicated that Lsp57, within the PBP-1 complex, may be an essential component of the SL RNA transcription machinery. To test this hypothesis, we used an in vitro transcription system (15) that can be programmed with exogenous SL RNA gene templates. Fig. 4A is a diagram of the DNA templates used in assays. Fig. 4B shows a titration of SL RNA transcription by using varying amounts of WT and two mutant templates. Transcription from the WT promoter was robust, whereas transcription from a template with a base-substituted PBP-1E was diminished significantly. Removal of the entire −72- to −10-bp region (Δ-Mut SL) abolished transcription. These data recapitulate what was observed in vivo (14).
Figure 4.
Efficient transcription requires an intact PBP-1 element. (A) Schematic of the templates used for the transcription assays. Shown are a 20-bp tag in the coding region (cross-hatched), the transcribed region (dotted), the −10- to −1-bp initiator region (INR), the 10-bp substitution of the PBP-1 element (diagonal lines), and the sequences that replace the large Δ-Mut (checkered). (B) Transcription activity assays. In addition to the tagged SL gene, all reactions contain a similarly tagged U6 snRNA gene. Equal amounts of U6 snRNA transcription in each reaction ensured consistent extract activity. WT SL templates amounts were 50 (lane 1), 20 (lane 2), and 10 ng (lane 3). Identical titrations of S-Mut template or Δ-Mut are shown in lanes 4–6 and 7–9, respectively.
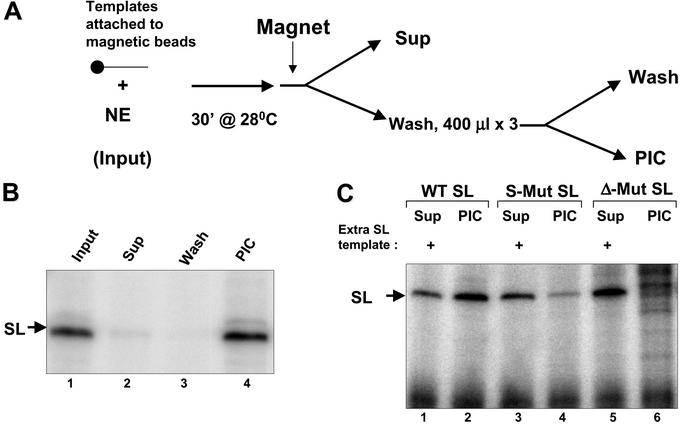
To investigate whether the p57 is involved in SL RNA transcription, we developed a method to isolate transcriptionally competent PICs on an immobilized template. The immobilized DNA templates were incubated with nuclear extract, and complexes were washed with 150 mM K-glutamate and 0.5% Nonidet P-40 to remove all loosely associated proteins (Fig. 5A). The isolated PICs were transcriptionally competent when incubated with rNTPs (Fig. 5B). A template lacking the entire upstream region failed to form any PICs whereas a template with a mutation only in the PBP-1 element formed unstable PICs (Fig. 5C). Our ability to establish PICs on the SL gene promoter is a powerful tool for defining the transcription complex that assembles at this promoter. It also suggests a structural similarity between trypanosomal gene promoters and those found in other eukaryotic cells.
Figure 5.
Transcription on immobilized templates. (A) Experimental design for the purification of a functional RNAP II-containing PIC. Promoter-containing templates, immobilized on magnetic beads, were incubated with nuclear extract, and PICs were captured by using a magnetic particle concentrator. (B) Lane 1 shows transcription from WT SL RNA template attached to the magnetic beads. Lanes 2–4 show transcription from the supernatant (Sup), wash, and purified PICs on the WT SL RNA template. (C) Lanes 2, 4, and 6 show the transcripts produced from purified PICs formed on WT, S-Mut, or Δ-Mut SL templates, respectively. Lanes 1, 3, and 5 show residual transcriptional activity in the supernatants. WT SL RNA template (100 ng) was added along with rNTPs and transcription buffer for supernatant assays.
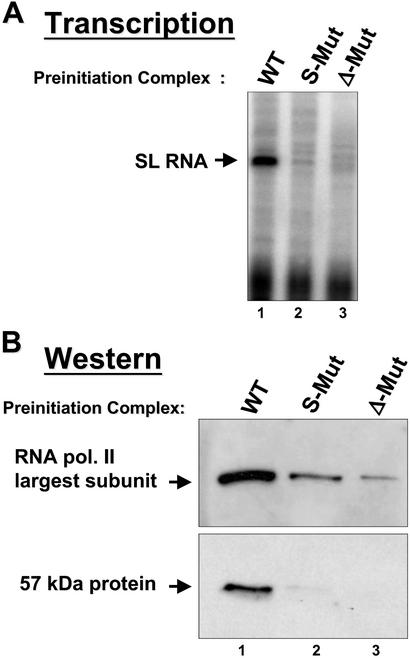
To examine whether p57 is a component of the PIC, we performed transcription assays and Western blot analysis. Fig. 6A shows a direct comparison of transcription from PICs assembled on the WT and mutated promoters. Only in the presence of the WT promoter is the p57 protein retained in a PIC complex, as detected by Western blot analysis (Fig. 6B). Therefore, robust and correctly initiated transcription from the SL RNA gene promoter requires p57, likely as a component of a functional PBP-1 complex. As expected, the association of RNAP II with the SL RNA gene promoter is correlated with the association of p57 in the PIC (Fig. 6B).
Figure 6.
The 57-kDa subunit of PBP-1 is a component of the PIC. (A) Transcription from purified PICs on WT (lane 1), S-Mut (lane 2), or Δ-Mut SL (lane 3) templates. (B) Protein immunoblots from each PIC, probed with a polyclonal Ab specific for the carboxyl-terminal domain of the L. seymouri RNAP II largest subunit (Upper) or a polyclonal Ab specific for the p57 subunit of PBP-1 (Lower).
Discussion
We have purified the first transcription factor for RNAP II-dependent gene expression in trypanosomes. Cloning of two of the three subunits of this protein, PBP-1, revealed that the trypanosome transcription machinery is related to, but distinct from, that found in other eukaryotes. We also developed an immobilized SL RNA gene promoter template assay that allows analysis of the only known RNAP II promoter in trypanosomes. A relatively small (270 bp) region of the SL RNA gene promoter was sufficient to nucleate functional PICs by using nuclear extract. The inability of the mutated promoter templates to support efficient PIC formation confirmed the authenticity of the PICs. Finally, we demonstrated that the p57 subunit of PBP-1, along with RNAP II, is present in the purified PICs. Considering the dearth of identified RNAP II-dependent promoters in trypanosomes, we expect that studies by using immobilized PICs at the SL RNA gene promoter will enable us to understand important aspects of the trypanosome RNAP II transcription machinery.
One of our important findings regarding SL RNA gene transcription is its apparent similarity to snRNA transcription in higher eukaryotes. snRNA gene promoters contain a few basal elements and a single distal sequence element. Depending on subtle changes in the configuration of these elements, either RNAP II or III is used for transcription (25). In the case of plants, the spacing of two basal elements dictates which RNA polymerase is assembled into the initiation complex (33). In Drosophila, RNA polymerase specificity is affected by minimal (5 bp) changes in the major proximal sequence element (PSE), named the PSEA (34). In vertebrates, the presence of a single PSE element (along with the distal sequence element), recruits RNAP II to the promoter, whereas the addition of a TATA box downstream from the PSE forces the assembly of an RNAP III-containing complex (25). The snRNA gene promoter in trypanosomes seems to be yet another variation on the theme of differential RNA polymerase recruitment. We have shown that the SL RNA is transcribed by RNAP II and that there is an important basal element (the PBP-1 element) that contributes to PIC assembly. Moreover, we have directly shown that the PIC within the SL RNA gene promoter contains RNAP II (28). Extensive evidence from several laboratories has shown that the U2, U4, and U6 snRNA genes in trypanosomes are transcribed by RNAP III. Preliminary EMSA data suggests that the L. seymouri PBP-1 protein binds specifically within the upstream promoter region of the homologous U snRNA genes (our unpublished observations). Thus, a structural link likely exists among snRNA promoters in trypanosomes as is the case in distantly related metazoans. Clearly, a more extensive comparison among the snRNA promoters in trypanosomes will illuminate this potentially important observation.
The protein factor that binds to the PSE sequence in human snRNA genes is SNAPc/PTF. SNAPc/PTF has been extensively analyzed in the human system (32, 35). It contains five subunits, SNAP190 (PTFα), SNAP50 (PTFβ), SNAP45 (PTFδ), SNAP43 (PTFγ), and SNAP19. No single subunit, on its own, binds stably to the PSE region of the promoter. Similarly, recombinant L. seymouri p57 protein, produced in either bacteria or baculovirus, fails to bind the SL RNA or the U2 snRNA promoter when used on its own in an EMSA (our unpublished data).
In human cells, a miniSNAPc functions in snRNA gene promoter binding, as assayed by EMSA. MiniSNAPc contains full-length SNAP50 and SNAP43 proteins and approximately one-fourth of the amino-terminal region of SNAP190 (36). Moreover, deletion of the carboxyl-terminal 100 aa in SNAP43 did not disrupt miniSNAPc function or the direct association of SNAP50 with SNAP43. Therefore, a minimal, unembellished version of SNAPc exists as a ≈130-kDa trimeric protein. The trypanosome PBP-1 protein contains an ortholog of the SNAP50 protein as part of a trimeric, ≈122-kDa complex. It is likely that the evolutionary progenitor of SNAPc is a PBP-1 type of complex. The conservation of a zinc finger between the human SNAP50 and the p57 subunit of PBP-1 suggests a key functional role for this motif. This motif likely functions in a protein–protein or protein–DNA interaction that is necessary for PIC assembly.
It is intriguing that the p46 component of the L. seymouri PBP-1 complex seems to be a unique protein. Although the human SNAPc/PTF exists as a 200-kDa native complex (32), in L. seymouri the PBP-1 complex is much smaller (≈122 kDa), as observed by gel filtration chromatography and glycerol gradient sedimentation (24). In human miniSNAPc, two unusual Myb domains in the truncated SNAP190 are required for PSE binding. However, our analysis of PBP-1 reveals that the largest two of the three subunits do not contain any Myb domains. This observation raises an exciting possibility of involvement of a different protein, not an SNAP190 ortholog, for interaction with DNA. Molecular modeling of p46 suggests a structural similarity to the herpes simplex UL42 protein. Because UL42 binds to DNA in a nonspecific fashion and interacts with DNA polymerase, we speculate that p46, as part of the PBP-1 complex, has a similar function. Indeed a 46-kDa polypeptide was specifically crosslinked to a 37-bp-promoter probe (from −83 to −46 bp), when partially purified PBP-1 was irradiated with UV (24). It will be interesting to see whether p46 serves to help stabilize the DNA–PBP-1 interaction at the SL RNA gene promoter and possibly assists in recruiting RNA polymerases.
Acknowledgments
We thank Gwen Gilinger, Y. Ramanathan, and members of the V.B. laboratory for critical reading of the manuscript. We thank Elizabeth Coates and Nicholas Murgolo for molecular modeling analysis. V.B. is supported by National Institutes of Health Grant AI29478 and is a Burroughs Wellcome New Investigator in Molecular Parasitology.
Abbreviations
- snRNA
small nuclear RNA
- SL RNA
spliced leader RNA
- RNAP II
RNA polymerase II
- PIC
preinitiation complex
- PSE
proximal sequence element
- S-Mut
substitution mutation
- Δ-Mut
deletion mutation
- EMSA
electrophoretic mobility-shift assay
- PTF
proximal sequence element-binding transcription factor
- SNAP
snRNA-activating protein
- SNAPc
SNAP complex
Footnotes
References
- 1.Pays E, Vanhamme L, Berberof M. Ann Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- 2.Brehm K, Jensen K, Frosch M. J Biol Chem. 2000;275:38311–38318. doi: 10.1074/jbc.M006091200. [DOI] [PubMed] [Google Scholar]
- 3.Bruzik J P, Van Doren K, Hirsh D, Steitz J. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 4.Frantz C, Ebel C, Paulus F, Imbault P. Curr Genet. 2000;37:349–355. doi: 10.1007/s002940000116. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen T W. Exp Parasitol. 1989;69:413–416. doi: 10.1016/0014-4894(89)90191-4. [DOI] [PubMed] [Google Scholar]
- 6.Rajkovic A, Davis R E, Simonsen J M, Rottman F M. Proc Natl Acad Sci USA. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonen L. FASEB. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 8.Kooter J M, van der Spek H J, Wagter R, d'Oliveira C E, van der Hoeven F, Johnson P J, Borst P. Cell. 1987;51:261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- 9.Rudenko G, Le Blanco S, Smith J, Lee M G S, Rattray A, Van der Ploeg L H T. Mol Cell Biol. 1990;10:3492–3504. doi: 10.1128/mcb.10.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tschudi C, Ullu E. EMBO J. 1988;7:455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 12.LeBowitz J H, Smith H, Rushce L, Beverley S M. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 13.Matthews K R, Tschudi C, Ullu E. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 14.Hartree D, Bellofatto V. Mol Biochem Parasitol. 1995;71:27–39. doi: 10.1016/0166-6851(95)00034-x. [DOI] [PubMed] [Google Scholar]
- 15.Huie J, He P, Bellofatto V. Mol Biochem Parasitol. 1997;90:183–192. doi: 10.1016/s0166-6851(97)00146-1. [DOI] [PubMed] [Google Scholar]
- 16.Lücke S, Xu G, Palfi Z, Cross M, Bellofatto V, Bindereif A. EMBO J. 1996;15:4380–4391. [PMC free article] [PubMed] [Google Scholar]
- 17.Agami R, Aly R, Halman S, Shapira M. Nucleic Acids Res. 1994;22:1959–1965. doi: 10.1093/nar/22.11.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldring A, Karchi M, Michaeli S. Exp Parasitol. 1995;80:333–338. doi: 10.1006/expr.1995.1041. [DOI] [PubMed] [Google Scholar]
- 19.Günzl A, Ullu E, Dörner M, Fragoso S, Hoffmann K, Milner J, Morita Y, Nguu E, Vanacova S, Wünsch S, et al. Mol Biochem Parasitol. 1997;85:67–76. doi: 10.1016/s0166-6851(96)02816-2. [DOI] [PubMed] [Google Scholar]
- 20.Saito R M, Elgort M G, Campbell D A. EMBO J. 1994;13:5460–5469. doi: 10.1002/j.1460-2075.1994.tb06881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M C, Sturm N, Saito R, Roberts T, Campbell D. Mol Biochem Parasitol. 1998;94:265–281. doi: 10.1016/s0166-6851(98)00083-8. [DOI] [PubMed] [Google Scholar]
- 22.Luo H, Gilinger G, Mukherjee D, Bellofatto V. J Biol Chem. 1999;274:31947–31954. doi: 10.1074/jbc.274.45.31947. [DOI] [PubMed] [Google Scholar]
- 23.Agami R, Shapira M. Nucleic Acids Res. 1992;20:1804. doi: 10.1093/nar/20.7.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo H, Bellofatto V. J Biol Chem. 1997;272:33344–33352. doi: 10.1074/jbc.272.52.33344. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez N. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 27.Ranish J A, Yudkovsky N, Hahn S. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilinger G, Bellofatto V. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuccola H J, Filman D J, Coen D M, Hogle J M. Mol Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
- 30.Gottlieb J, Marcy A I, Coen D M, Challberg M D. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry R W, Ma B, Sadowski C L, Kobayashi R, Hernandez N. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon J B, Murphy S, Bai L, Wang Z, Roeder R G. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waibel F, Filipowicz W. Nature. 1990;346:199–202. doi: 10.1038/346199a0. [DOI] [PubMed] [Google Scholar]
- 34.Jensen R C, Wang Y, Hardin S B, Stumph W E. Nucleic Acids Res. 1998;26:616–622. doi: 10.1093/nar/26.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry R W, Sadowski C, Kobayashi R, Hernandez N. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 36.Ma B, Hernandez N. J Biol Chem. 2001;276:5027–5035. doi: 10.1074/jbc.M009301200. [DOI] [PubMed] [Google Scholar]
- 37.Hendlich M, Lackner P, Weitckus S, Floeckner H, Froschauer R, Gottsbacher K, Casari G, Sippl M J. J Mol Biol. 1990;216:167–180. doi: 10.1016/S0022-2836(05)80068-3. [DOI] [PubMed] [Google Scholar]
- 38.Domingues F S, Koppensteiner W A, Jaritz M, Prlic A, Weichenberger C, Wiederstein M, Floeckner H, Lackner P, Sippl M J. Proteins. 1999;37(S3):112–120. [PubMed] [Google Scholar]