Abstract
To facilitate investigations of replication and host cell interactions in the hepadnavirus system, we have developed cell lines permitting the conditional replication of duck hepatitis B virus (DHBV). With the help of this system, we devised conditions for core particle isolation that preserve replicase activity, which was not found in previous preparations. Investigations of the stability of viral DNA intermediates indicated that both encapsidated DNA and covalently closed circular DNA (cccDNA) were turned over independently of cell division. Moreover, we showed that alpha interferon reduced the accumulation of RNA-containing viral particles. The availability of a synchronized replication system will permit the biochemical analysis of individual steps of the viral replication cycle, including the mechanism and regulation of cccDNA formation.
Hepadnaviruses are small DNA viruses that contain a relaxed circular DNA (rcDNA) genome with modified 5′ ends. The 5′ end of minus strand DNA is covalently attached to the viral reverse transcriptase (RT), whereas the 5′ end of the plus strand is linked to an 18-nucleotide-long capped RNA oligomer. After infection, the rcDNA is converted into covalently closed circular DNA (cccDNA), which is the template for the transcription of at least three viral RNA species. The longest transcript, termed pregenomic RNA (pgRNA), is packaged into core particles, which are the site of viral DNA synthesis (20, 22).
Although the general mechanism of replication has been investigated in great detail, major gaps still exist because certain intermediates of the replication cycle are either short lived or difficult to isolate in pure form from persistently infected cells. For example, cccDNA formation occurs primarily during the establishment of infections, precluding the isolation of intermediates that occur during the conversion of rcDNA to cccDNA. Hence, the mechanism responsible for the repair of rcDNA is still enigmatic. Likewise, the mechanism responsible for the assembly of nucleocapsids is not well understood. Biochemical and structural data are consistent with a model in which core monomers form dimers, which are then assembled into icosahedral subviral core particles (27, 30). Specific incorporation of pgRNA and RT into core particles also depends on the interaction of the RT with a packaging signal on pgRNA, which in turn requires the cellular chaperone complex hsp90 (6, 7). However, the assembly of pgRNA- and polymerase-containing cores competent for viral DNA synthesis has so far not been achieved in cell-free systems. Similarly, efforts to isolate pgRNA-containing core particles from infected cells competent for genome replication in vitro have not been reported. Finally, little is known about the stability of DNA replication intermediates, including cccDNA, (i) under normal conditions, (ii) in the presence of cytokines or inhibitors of viral DNA synthesis that are used for antiviral therapy, and (iii) during cell division, which is known to occur during recovery from natural infections (5).
In an effort to address these problems, we have established cell lines that support efficient replication of duck hepatitis B virus (DHBV) in a conditional and synchronized fashion. We demonstrated that these cell lines permit the accumulation and subsequent isolation of core particles at different stages of the replication cycle. Moreover, we developed conditions for the isolation of pgRNA-containing capsids that are competent for the synthesis of complete viral genomes in vitro. Finally, we showed that the accumulation of pgRNA-containing capsids can be regulated by cellular factors induced by the antiviral program of alpha interferon (IFN-α) and investigated how cell division affects the stability of viral DNA intermediates.
MATERIALS AND METHODS
Plasmids.
Plasmid ptetDHBV1S, derived from plasmid pCMVDHBV (3), directs the expression of envelope-deficient 1S DHBV pgRNA under the control of the tet promoter (4). The 1S mutant carries three termination codons in the envelope gene, preventing translation of both envelope proteins p17 and p36 (23). pCDNA6CHI-IFN-α, expressing chicken IFN-α, was derived from pCDNA1-IFN-α/β (a gift from Jesse Summers, University of New Mexico, Albuquerque). The IFN-encoding gene was subcloned into the HindIII and XbaI sites of pCDNA6/VS-hisA (Invitrogen). To obtain cDNA clones of the gene for chicken Mx-1, LMH cells were treated with 1,000 U of chicken IFN-α. For the PCR amplification of Mx-1 cDNA, the primers used were 5′-TACGAAGC TGGAGGAGCCAGC-3′ and 5′-TACCAGGTATTGGTAGGCTTT GTTGAG-3′. The purified 584-bp-long PCR fragment was cloned into the pGEM-T Easy vector (Promega). The identity of the cloned fragment was verified by nucleotide sequence analysis. A similar approach was used to clone a cDNA corresponding to the gene for chicken IRF1. The primers used were 5′-CCAGAAGAGCAGCCTGGACTT-3′ and 5′-TAATACGACTCACTATAGGGATGTGGCAGCTCCTCCACT-3′.
Cell cultures.
The chicken hepatoma cell line LMH (10) and the LMH derivatives dstet8 and dstet5 were maintained in Dulbecco's modified Eagle medium-F12 containing 10% fetal bovine serum, 200 μg of G418 per ml, and 1 μg of tetracycline per ml (dstet5 and dstet8 cells). dstet cells were maintained on plates coated with 0.1% swine skin gelatin type 1 (Sigma). For generation of the dstet cell lines, LMH cells were transfected with plasmids ptetDHBV1S, pUDH15.1 (4), and pRSVneo at a ratio of 5:5:1, respectively, by calcium phosphate coprecipitation. Two days after the transfection, the cells were passaged and cultured in medium containing 400 μg of G418 per ml and 1 μg of tetracycline per ml for 10 days and then in medium containing 200 μg of G418 per ml and 1 μg of tetracycline per ml. Until the isolation of clones, the culture medium was changed daily. To inhibit viral replication, cells were maintained in the presence of 1 mM phosphonoformic acid (PFA) (see Fig. 2 to 4 and 7) and 20 μM lamivudine (3TC) (see Fig. 5 and 6). dstet5 cells were used for all of the experiments described in this report.
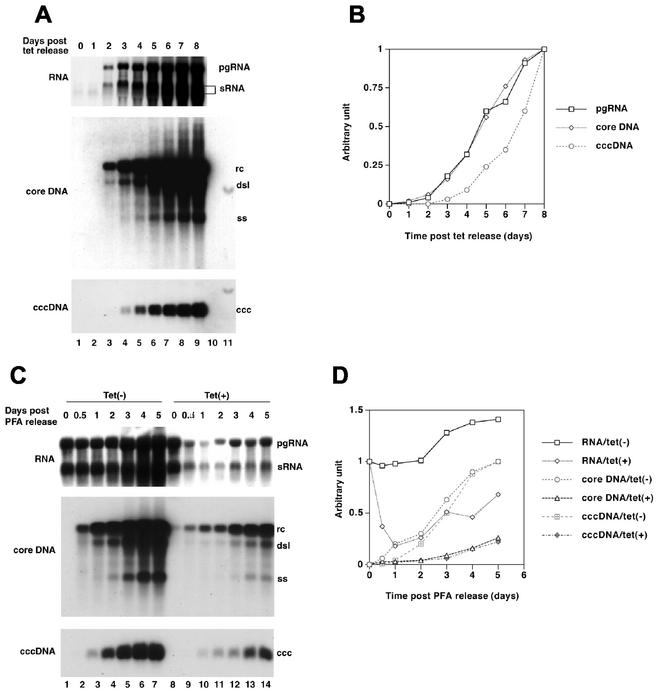
FIG. 2.
Conditional replication of DHBV. (A and B) dstet5 cells were maintained in the presence of tetracycline (Tet; 1 μg/ml) and then without the antibiotic for the indicated number of days. Total RNA (10 μg), cytoplasmic core DNA, and nuclear cccDNA were extracted from dstet5 cells and analyzed by Northern and Southern blot analyses. For core DNA and cccDNA analysis, each lane represents the amount of viral DNA in one-eighth of the cells present in a 60-mm-diameter plate. DHBV RNA and DNA levels were quantitated with a Fuji phosphorimager, and the results were expressed as the fractions of the values obtained on day 8. Unit length DHBV DNA (10 pg, lane 11) served as a standard by which to estimate the copy number of core DNA and cccDNA. (C and D) The cells were incubated for 4 days with PFA in the absence of tetracycline. After removal of the PFA, the cells were incubated for the indicated number of days without (lanes 1 to 7) or with (lanes 8 to 14) the antibiotic. Viral RNA and DNA levels were determined as described above, except that for cccDNA analysis, each lane represents the amount of viral DNA in one-fourth of the cells present in a 60-mm-diameter plate. pgRNA levels were normalized to the value obtained on day 0. Core DNA and cccDNA levels were normalized to the values obtained on day 5. The positions corresponding to the major viral DNA forms are indicated. ss, single-stranded DNA.
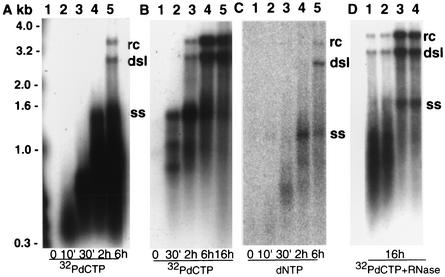
FIG. 4.
In vitro synthesis of DHBV genomes. dstet5 cells were incubated with PFA for 3 days. Core particles were isolated immediately (A and C; D, lanes 1 and 2) or after 6 h of an additional incubation without PFA (B; D, lanes 3 and 4). Endogenous polymerase reactions were performed with dNTPs and [32P]dCTP (A, B, and D) and with dNTPs alone (C) for the indicated time periods. Purified DNA was electrophoresed through 1.2% agarose gels, and the dried gels were exposed to X-ray film (A, B, and D) or the DNA was transferred to a nylon membrane and hybridized with a DHBV-specific probe (C). In panel D, EPRs were performed in the presence of RNase A (10 μg/ml). The nuclease was added together with the dNTPs (lanes 1 and 3) or 10 min before the beginning of the reactions with dNTPs (lanes 2 and 4). ss, single-stranded DNA.
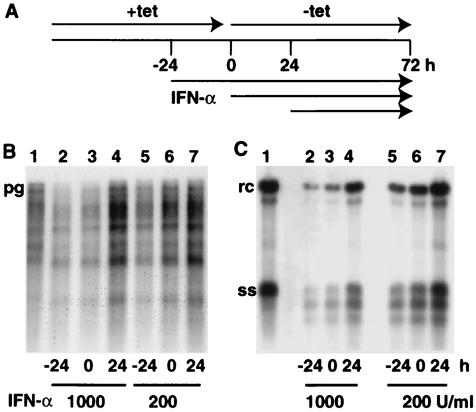
FIG. 7.
Viral DNA synthesis in the presence of IFN-α. (A) Cells were incubated with IFN-α (200 or 1,000 U/ml) for different time periods as indicated and harvested 72 h after the removal of tetracycline (tet). (B and C) Northern and Southern blot analyses of RNA and DNA isolated from core particles obtained from untreated (lane 1) and IFN-treated (lanes 2 to 7) cells. ss, single-stranded DNA.
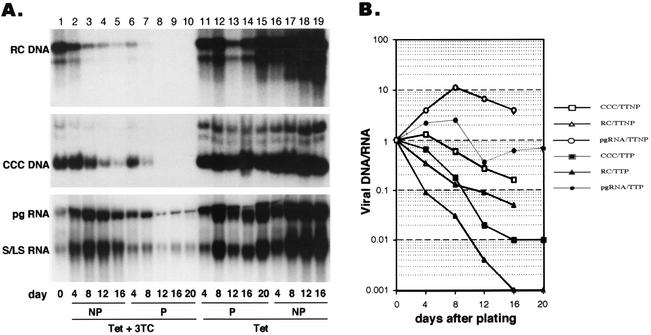
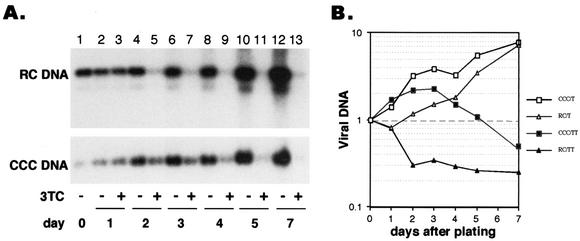
FIG. 5.
Stability of core DNA and cccDNA. (A) Tetracycline (Tet) was removed from dstet5 cells for 7 days before it was added back with (lanes 2 to 10) or without (lanes 11 to 19) 3TC, and DNA and RNA samples were extracted and analyzed. Lane 1 (day 0) shows the DNA and RNA levels present in cells at the end of the 7 days of incubation without tetracycline. Cells were either trypsinized and diluted 1:2 every 4 days (P) or maintained in culture without passage (NP). At each time point when the cells were passaged, a fraction of the cells was used for isolation of viral RNA and DNA (P, lanes 6 to 9 and 11 to 14). The same number of unpassaged cells was harvested at each time point (NP, lanes 2 to 5 and 16 to 19). Lanes 10 and 15 show the samples from the fourth passage as in lanes 9 and 14, respectively, but were incubated for an additional 4 days after they became confluent. The amount of DNA or RNA loaded onto each lane was normalized to the number of cells in the cultures. S/LS, surface and large surface mRNA. (B) The results shown in panel A, lanes 1 to 10, were quantified with a phosphorimager and plotted by using RNA and DNA levels at day 0 (lane 1) as the standard. TT, tetracycline plus 3TC.
FIG. 6.
t1/2s of core DNA and cccDNA. (A) The cells were incubated as described in the legend to Fig. 5, except that the cells were passaged only once and harvested 1 to 5 and 7 days after passage. Following passage, the cells were incubated either with tetracycline alone (lanes 2, 4, 6, 8, 10, and 12) or with tetracycline and 3TC (lanes 3, 5, 7, 9, 11, and 13). Lane 1 shows the input viral DNA at the time of cell passage. The amount of DNA loaded into each well corresponded to one-third of the cells present in a 60-mm-diameter dish. (B) The results were quantitated as described in the legend to Fig. 5. Input viral DNA was used as the standard. T, tetracycline; TT, tetracycline plus 3TC.
Isolation of viral DNA.
Extraction of DNA replicative intermediates and cccDNA was performed essentially as previously described (28).
Isolation of core particles and EPR.
The methods used for immunoprecipitation and endogenous polymerase reaction (EPR) were adaptations of those of Macrae et al. (13). Cells in 60-mm-diameter plates were lysed on ice in 2.0 ml of 50 mM Tris-HCl (pH 7.5)-150 mM NaCl-5 mM MgCl2-0.2% Triton X-100-1 μg of leupeptin per ml-0.7 μg of pepstatin A per ml-1 mM phenylmethylsulfonyl fluoride and collected into two Eppendorf tubes. Cell debris and nuclei were removed by centrifugation at 16,000 ×g for 2 to 4 min at 4°C. The supernatants were incubated with 2 μl of a polyclonal rabbit anti-DHBV core antigen (DHcAg) antibody for 1.5 h and then with 40 μl of a suspension containing Sepharose A (Pharmacia) for 2 h with shaking at 4°C. The beads were washed three times with 1 ml of lysis buffer without the protease inhibitors. Three-quarters of each sample was used for the EPR. In some experiments, cells were lysed in 1.0 ml of lysis buffer and only 1 μl of the polyclonal rabbit anti-DHcAg antibody was used for precipitation of the cores. The antibodies and beads were added at the same time and incubated for 2 h. One-half of the samples were used for the EPR.
For the EPR, the beads were washed three times with 50 mM Tris-HCl (pH 7.5)-75 mM NH4Cl-1 mM EDTA. The beads were resuspended in 30.5 μl of endogenous reaction buffer, which contained 25.8 μl of 50 mM Tris-HCl (pH 7.5), 75 mM NH4Cl, 1 mM EDTA, 20 mM MgCl2, 0.1% β-mercaptoethanol, and 0.5% NP-40; this was followed by the addition of 1.2 μl of 10 mM (each) dATP, dGTP, and dTTP; 3 μl of 10 μM dCTP; and 0.5 μl [32P]dCTP (3,000 Ci/mmol). The reaction mixture was incubated for various times at 37°C. In the experiment whose results are shown in Fig. 4C, 28.8 μl of endogenous reaction buffer and 1.2 μl of 10 mM (each) dATP, dGTP, and dTTP were added. To stop the reaction and remove any unincorporated nucleotides, the samples were washed twice in 1 ml of 50 mM Tris-HCl (pH 7.5)-75 mM NH4Cl-20 mM EDTA-0.1% β-mercaptoethanol-0.5% NP-40. To release the DNA from the core particles, the beads were incubated with 100 μl of 0.5% SDS-0.87 mg of proteinase K-50 mM EDTA for 1 h at 37°C. After the addition of 5 μg of yeast tRNA, the viral DNA was extracted with phenol-chloroform (1:1), precipitated with 2.5 volumes of ethanol, and resuspended in 20 μl of TE buffer.
Analysis of viral RNA.
Total cellular RNA was extracted with Trizol reagent (Invitrogen). Encapsidated viral pgRNA was purified as previously described (17), with some modifications. Briefly, cells were lysed in 600 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl, 1% NP-40) and the nuclei were removed by centrifugation. One-half of the sample was incubated with 6 U of micrococcal nuclease (Pharmacia) and 15 μl of 100 mM CaCl2 and incubated for 15 min at 37°C to digest free nucleic acids. The reaction was stopped with 6 μl of 0.5 M EDTA, and capsids were precipitated with 125 μl of 35% polyethylene glycol 8000 in 1.75 M NaCl. Pellets were resuspended in 50 μl of TNE buffer (10 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA). pgRNA was extracted by the addition of 1 ml of Trizol reagent. Total RNA and encapsidated pgRNA were electrophoresed through a 2.2 M formaldehyde-1% agarose gel, transferred to a nylon membrane, and immobilized by UV cross-linking (Stratagene). Hybridization was performed with a [32P]UTP (800 Ci/mmol)-labeled full-length DHBV minus strand riboprobe in a solution containing 50% deionized formamide; 5× SSC (1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0); 1× Denhardt's solution; 0.02 M sodium phosphate (pH 6.8); 0.2% SDS; 100 μg of sheared, denatured salmon sperm DNA; 100 μg of yeast RNA per ml; and 10 μg of poly(A)n per ml and incubated at 52°C for 24 h. Membranes were exposed to X-ray films and phosphorimager screens for 6 to 24 h.
Production of chicken IFN-α.
pCDNA6CHI-IFN-α was transfected into COS7 cells by calcium phosphate coprecipitation (Clontech Laboratories, Inc.). Seventy-two hours posttransfection, the culture supernatant was harvested and cleared of cellular debris by centrifugation. The antiviral IFN-α titer was determined with temperature-sensitive VSV-tl17 (a gift from David Boettiger, University of Pennsylvania). One unit of IFN-α was defined as the reciprocal of the dilution that resulted in 50% protection of dstet5 cells (16).
RESULTS
Conditional replication of DHBV.
To investigate the biochemical properties of RNA-containing capsids in cells or in vitro, we established a cell line that expressed DHBV under the control of the tetracycline-regulated promoter. To better determine the intracellular accumulation of viral particles, we used 1S mutant DHBV genome that does not express functional envelope proteins and, as a consequence, cannot secrete enveloped viral particles (Fig. 1). Moreover, because of the absence of envelope protein, cccDNA amplification is more efficient with this variant than with the wild type. Although the 1S mutant is cytotoxic in primary hepatocytes, we did not observe any cytopathic effects during the course of the experiments described in this report (23). After the transfection of LMH cells with plasmids ptetDHBV1S and pUDH15.1, encoding the tet repressor with the activating domain of virion protein 16 of herpes simplex virus (4), we obtained several cell lines that expressed DHBV when tetracycline was removed from the culture medium (results not shown). dstet5 and dstet8 cells exhibited the highest levels of viral DNA, and the dstet5 line was used for the studies described in this report.
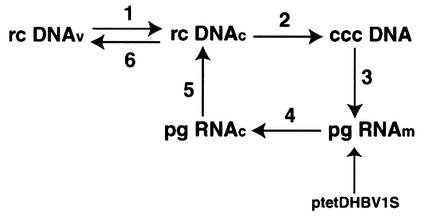
FIG. 1.
Model of hepadnavirus replication. rcDNA in core particles (rc DNAc) derived from virions (rc DNAv) is converted into cccDNA (steps 1 and 2). cccDNA is the template for the transcription of pgRNA (step 3), which functions first as mRNA (pg RNAm) and then as pregenome for DNA synthesis (pg RNAc) in core particles (steps 4 and 5). Core particles assemble into virions and are secreted into the blood (step 6). Alternatively, core particles disassemble, leading to the formation and amplification of cccDNA (step 2). In dstet cells, the replication cycle is limited to steps 2 to 5 and is initiated through the expression of pg RNAm from an integrated copy of ptetDHBV1S.
In a first step, we sought to measure the rate of viral RNA and DNA synthesis and to examine whether RNA-containing particles could accumulate in these cell lines. When tetracycline was removed from the medium of dstet5 cells, pgRNA and the subgenomic surface RNA began to accumulate within 2 days (Fig. 2A and B). Their levels increased at a constant rate during the 8-day observation period, suggesting that cccDNA contributed to the transcription of viral RNA (see below). Expression and accumulation of core DNA correlated well with RNA levels. As expected from the model of reverse transcription, accumulation of cccDNA lagged behind that of rcDNA (Fig. 1 and 2A and B). We estimated that copy numbers of rcDNA and cccDNA increased from approximately 600 and 20 per cell at day 3 to 4,000 and 600 per cell at day 8, respectively.
To determine whether RNA-containing core particles could accumulate and whether cccDNA contributed to viral RNA synthesis, dstet5 cells were incubated in medium containing 1 mM PFA without tetracycline for 4 days. During this time, viral RNA levels reached a steady state (results not shown). While PFA is an inhibitor of the DHBV polymerase, it does not block the protein-priming reaction for reverse transcription (25). Hence, PFA treatment should lead to the accumulation of core particles containing pgRNA and at least four nucleotides of minus strand DNA (24). After removal of the drug, the cells were incubated with and without tetracycline for different times (Fig. 2C and D). Analysis of viral RNA and DNA revealed that, in the absence of tetracycline, viral RNA levels remained steady until day 2 and began to increase thereafter, again suggesting that cccDNA acted as a template for RNA synthesis (lanes 1 to 7). This interpretation was supported by the results obtained when tetracycline was added back to the medium following the removal PFA (lanes 8 to 14). Under these conditions, viral RNA levels dropped rapidly within the first 24 h of incubation and then began to increase simultaneously with cccDNA, confirming that cccDNA functioned as a template for viral RNA synthesis in dstet5 cells. However, in spite of the relatively high copy number of cccDNA, which was estimated to be on the order of 150 per cell at day 5 (lane 14), RNA levels reached only 60% of the levels obtained with the single copy of integrated plasmid DNA (lanes 1 and 8). Hence, either only a small fraction of cccDNA molecules served as templates for RNA synthesis or transcription from cccDNA was inefficient. In the presence of viral RNA synthesis from the tetracycline-inducible promoter (without tetracycline), the amounts of rcDNA-containing cores increased at a constant rate from 12 h to 5 days. In contrast, when tetracycline was added back to the medium (with tetracycline), the rate of rcDNA synthesis was much lower. The levels of the rcDNA present in tetracycline-treated cells harvested 12 h and 1 day after PFA release (lanes 9 to 10) were 30 and 15% of the amounts of DNA present in cells cultured in the absence of the antibiotic (lanes 2 and 3). This result indicated that replication-competent, pgRNA-containing cores accumulated with relatively low efficiency in PFA-treated cells, which was consistent with results obtained previously with DHBV-infected primary hepatocyte cultures (17).
Stability and replication competence of RNA-containing capsids.
To investigate whether PFA caused the formation of core particles that were defective for replication or whether pgRNA-containing cores were unstable in LMH cells, dstet5 cells were incubated without tetracycline in the presence of PFA for 3 days (Fig. 3B and C). The cells were than incubated with tetracycline and PFA to stop viral RNA and DNA synthesis in order to measure the rate of decay of viral RNA. The results of this analysis showed that the half-lives (t1/2s) of total pgRNA and encapsidated RNA were approximately 7.5 and 24 h, respectively.
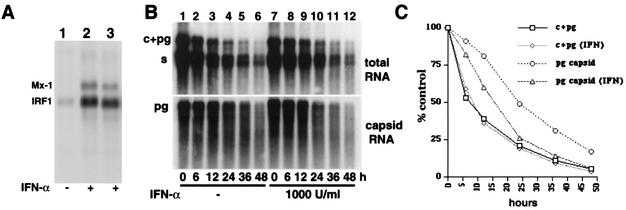
FIG. 3.
Stability of total and packaged pgRNA. (A) Induction of IFN response genes by IFN-α. IFN-α was added to dstet5 cells at a final concentration of 1,000 U/ml for 24 h. Expression of Mx-1 and IRF1 was monitored by Northern blot analysis. (B) dstet5 cells were maintained in the presence of PFA for 3 days and then with (lanes 7 to 12) and without IFN-α (lanes 1 to 6) in the presence of tetracycline and PFA for the indicated time periods. Total and encapsidated RNAs were isolated, electrophoresed through a 1.2% agarose-formaldehyde gel, blotted onto a nylon membrane, and hybridized with a DHBV-specific probe (B). The core (c) and pgRNA signals were quantitated with a Fuji MacBAS phosphorimager and plotted (C).
The model of hepadnavirus reverse transcription predicts that RNA-containing capsids can synthesize complete viral rcDNA in the presence of deoxynucleoside triphosphates (dNTPs) (18, 22). However, our previous attempts to isolate replication-competent pgRNA-containing particles by gel filtration or precipitation with polyethylene glycol, which is commonly used for the concentration of DNA-containing core particles, were not successful (J. Kitson and C. Seeger, unpublished results). As an alternative strategy, we incubated extracts prepared from PFA-treated dstet5 cells with DHcAg antibodies bound to Sepharose beads. The immobilized capsids were able to synthesize rcDNA in an EPR within an incubation time of 2 to 6 h (Fig. 4A). More than 90% of the DNA was synthesized within the first 2 h of incubation. However, only a fraction of the capsids seemed to be competent for the synthesis of full-length viral DNA genomes. Notably, the dominant full-length DNA form was double-stranded linear DNA (dslDNA) and not rcDNA, as observed in cell cultures (Fig. 4A). dslDNA is the result of an in situ priming reaction for plus strand DNA synthesis (21).
The low efficiency of viral DNA synthesis could have resulted from the relatively low concentration of dCTP (1 μM) in the reaction mixture that was used to label the growing DNA strands. However, even when the endogenous reaction was performed in the presence of high concentrations of all four dNTPs, only about 30% of the nascent DNA strands were extended into complete genomes (Fig. 4C). This experiment also demonstrated that DNA synthesis in isolated cores was processive and not merely a minor extension of existing DNA strands. However, cores isolated from dstet5 cells that were incubated for 6 h without PFA exhibited a significantly enhanced efficiency of rcDNA synthesis compared with cores isolated directly from PFA-treated cells (compare Fig. 4A and B). To verify that pgRNA was completely packaged into core particles, we incubated immunoprecipitated core particles with RNase A prior to the addition of dNTPs. The results showed that the presence of the RNase had no effect on DNA synthesis, suggesting that the viral RNA was protected from degradation by the nuclease (Fig. 4D).
In summary, the results demonstrated that pgRNA-containing cores have a t1/2 of approximately 24 h and that they are competent for the replication of complete DHBV genomes in vitro. Moreover, capsids isolated form PFA-treated cells represent a heterogeneous population in which only a relatively small fraction of the capsids can synthesize full-length genomes. Whether RNA-containing capsids were damaged during the extraction procedure or whether cellular factors played a role in the formation of DNA-containing capsids could not be determined.
The stability of core DNA and cccDNA.
Amplification of cccDNA is a hallmark of the hepadnavirus replication cycle and occurs rapidly after the formation of rcDNA (Fig. 2). Little is known about the stability of both rcDNA and cccDNA in infected cells because, under natural conditions, both pools are continuously replenished by de novo reverse transcription and conversion of rcDNA into cccDNA (Fig. 1). To better determine the stability of core DNA and cccDNA, we took advantage of the HBV polymerase inhibitor 3TC to block core DNA synthesis and measured its rate of decay. dstet5 cells were first maintained in the absence of tetracycline for 7 days and then incubated with the antibiotic and 3TC (20 μM) for as long as 20 additional days (Fig. 5). To test the effect of cell division on the decay of core DNA and cccDNA, cells were either passaged every 4 days or maintained as confluent cultures without passage. Under these conditions, the cell number in the unpassaged cultures increased less than 2-fold, compared with a 16-fold increase in the passaged cells during the entire incubation time of 20 days. Independent of the growth condition, both core DNA and cccDNA levels declined, with core DNA being cleared faster than cccDNA (Fig. 5A, lanes 1 to 10, and B). The rate of decline of both DNA forms was increased in the passaged cultures, where the cells were dividing during the incubation with tetracycline and 3TC. However, the approximately 100-fold decline of cccDNA could be attributed to the 16-fold dilution of the DNA to daughter cells, combined with the 6-fold decline in cccDNA observed without cell passage. In contrast to that of cccDNA, cell division appeared to enhance the rate of rcDNA decline about three- to fourfold above the levels expected merely from dilution.
Unexpectedly, viral RNA levels did not decline in proportion to cccDNA levels, which could indicate that only a fraction of the cccDNA molecules served as templates for transcription or that cellular factors limited the rate at which viral RNA was synthesized (Fig. 5A, lanes 2 to 5 and 16 to 19, and B). When the cells were maintained without 3TC (but with tetracycline), viral DNA and RNA continued to accumulate over the entire observation period independently of whether the cells were diluted or kept as confluent cultures (Fig. 5A, lanes 11 to 19). These results confirmed our previous observation indicating that the pool of cccDNA could direct the synthesis of pgRNA (Fig. 2C).
To obtain a better estimate of the t1/2s of core DNA and cccDNA, we measured viral DNA levels in dstet5 cells 1 to 7 days after the cells were passaged in the presence of tetracycline and 3TC (Fig. 6). The decrease in rcDNA levels was biphasic and characterized by an initial rapid decline, reaching 30% of the pretreatment level after 2 days and then declining more slowly during the next 5 days. The apparent t1/2 of core DNA during the first phase was between 24 and 48 h. In contrast to those of core DNA, the levels of cccDNA slowly increased for 3 days before they began to decline, reaching 50% of the pretreatment level at day 7. The apparent t1/2 of cccDNA was estimated to be about 48 h. These results indicated that, during the initial period of drug treatment, the pool of cccDNA was increased through the conversion of rcDNA to cccDNA, suggesting that 3TC did not inhibit cccDNA formation. Moreover, the subsequent decline in cccDNA levels, even when rcDNA was still present and presumably being converted into cccDNA, suggests that a certain threshold level of continued cccDNA synthesis was required to maintain a steady-state cccDNA level. Therefore, cccDNA must have undergone a continuous turnover in these cells.
Regulation of DHBV replication.
Since we had established conditions to analyze the stability of pgRNA- and DNA-containing particles, we sought to determine whether host factors, such as those induced by IFN-α, could influence the stability of viral intermediates and, hence, regulate DHBV replication. To begin our studies, we tested whether IFN-α could induce an antiviral response in dstet5 cells. The cells were incubated with 1,000 U of chicken IFN-α per ml for 16 h or 3 days, and the mRNA levels of two known IFN-induced genes, those that encode Mx-1 and IRF1, were determined by Northern blot analysis. The results showed that IFN-α treatment induced the expression of both genes (Fig. 3A).
The stability of encapsidated pgRNA was only slightly decreased when dstet5 cells were treated with IFN-α, reducing the t1/2 from 24 to 15 h (Fig. 3). In contrast, the stability of total pgRNA was not changed by the cytokine. To examine whether DNA synthesis was sensitive to IFN-α treatment, we incubated dstet5 cells with the cytokine 24 h before and after RNA synthesis was induced (Fig. 7). The results showed that IFN-α led to a reduction of both capsid-associated RNA and DNA levels when the cytokine was present before or at the beginning of DHBV RNA synthesis (−24 and 0 h, respectively). Nevertheless, under these conditions, complete rcDNA synthesis could occur. Almost no inhibition was observed when IFN was added 24 h following the initiation of RNA synthesis (24 h). Surprisingly, IFN treatment also led to the accumulation of two additional DNA bands that migrated faster than single-stranded DNA. Most likely, they represent immature minus strand DNA species, although the possibility cannot be excluded that they are degradation products of viral DNA.
In summary, the results showed that dstet5 cells can respond to IFN-α by inducing the expression of genes known to be activated by the cytokine. As a consequence of IFN-α treatment, the t1/2 of pgRNA-containing core particles was reduced from 24 to 15 h. Although the antiviral program induced by the cytokine in LMH cells was not strong enough to prevent the synthesis of rcDNA, it caused the accumulation of capsids with subgenomic minus strands.
DISCUSSION
Hepadnaviridae is one of the best-studied virus families, in terms both of the mechanism of viral replication and pathogenesis. However, important questions concerning distinct steps of the viral life cycle remain unexplored. The development of a cell line permitting the conditional replication of DHBV should provide an additional tool with which to tackle some of these problems. It will now be possible to isolate distinct intermediates of the replication cycle, such as RNA-containing capsids or capsids containing only minus strands. Moreover, it will be possible to identify compounds that can inhibit specific steps of the viral replication cycle, such as the formation of cccDNA. Because the period for accumulation of intermediates is short, less than 72 h, it will be possible to use certain drugs even when they exhibit some cytotoxicity.
The analysis of DHBV replication revealed that the time for one round of DNA replication is surprisingly long for such a short genome (Fig. 2). Although the processivity of RNA and DNA polymerases can vary, depending on the system and experimental conditions, it is generally on the order of several hundred nucleotides per second (8, 9). However, DHBV synthesis occurred in cells or in vitro at much lower rates, on the order of a few nucleotides per second (Fig. 2 and 4). Although we cannot formally rule out the possibility that residual PFA interfered with the processivity, it seems unlikely because the rates of DNA synthesis were comparable in vivo and in vitro and the rate of plus strand DNA synthesis did not seem to differ from that of minus strand DNA synthesis. Moreover, low rates of DNA synthesis were also observed with a temperature-sensitive mutant of DHBV (19; results not shown).
So far, in vitro replication of hepadnavirus DNA has been unsuccessful, being limited to either the protein-priming reaction carried out by the RT alone or the extension of minus and plus strands in core particles isolated from infected hepatocytes (22, 25). Complete synthesis of double-stranded viral DNA from RNA in purified capsids has not been observed. However, as shown in this report, we have devised conditions for core particle isolation that preserve replicase activity, which was not found in previous preparations. Moreover, the results provide evidence of a maturation step that occurred in LMH cells following the initiation of viral DNA synthesis in the absence of PFA. It resulted in a more efficient DNA synthesis reaction under our selected in vitro conditions (Fig. 4). The results were consistent with the hypothesis predicting that DNA synthesis is required for the assembly of stable, replication-competent capsids. Our results also suggest that viral DNA synthesis can occur in core particles, perhaps without the need for exogenous cellular factors. However, this interpretation has to be considered with caution because we cannot exclude the possibility that our core particle preparations contained cellular proteins that remained bound to cores despite several wash steps that were included in the purification procedure (see Materials and Methods). Nevertheless, it will be interesting to determine whether the apparent dephosphorylation of cores that occurs during the morphogenesis of virus particles in vivo is required for DNA synthesis in vitro (15).
Because PFA cannot block the protein-priming reaction for minus strand DNA synthesis (25), it is likely that we isolated capsids that had already completed the protein-priming reaction and contained short minus strands. On the basis of the DNA levels determined by Southern blot analysis of the products of the EPR (Fig. 4), we estimated that only a small fraction of capsids was competent for the synthesis of full-length genomes. This could be a consequence of the isolation procedure, which may have damaged cores. Alternatively, it could mirror the fidelity of the assembly process that results in the production of defective particles. Interestingly, the ratio of rcDNA to dslDNA was significantly reduced in vitro compared with that under the natural conditions in cells (compare Fig. 2 and 4). Thus, under in vitro conditions, in situ priming of plus strand DNA occurred more efficiently than the transfer of the RNA primer from the 3′ end of minus strand DNA to an internal acceptor site on minus strand DNA (11, 18, 21). Alternatively, in situ priming may be the default pathway for plus strand initiation and primer transfer may require additional factors that are missing from the cell-free system. However, even in vivo, only a fraction of the cores containing complete minus strands (single-stranded DNA) appeared to be capable of rcDNA synthesis (Fig. 2A, lanes 9 to 11). It is conceivable that arrests in DNA synthesis are caused by defects in the process leading to capsid assembly or that they reflect the requirement for one or more cellular factors that were available in limiting amounts in LMH cells. On the basis of these and other observations, it is conceivable that pgRNA and polymerase assemble with the help of core subunits into a metastable procapsid-like structure in which the DNA-priming reaction occurs. DNA elongation could then trigger the assembly of stable capsids.
Previously, Schultz et al. (16) observed that IFN-α treatment of primary hepatocyte cultures infected with DHBV reduced the accumulation of total viral RNA and packaged viral RNA levels. Likewise, Wieland et al. reported that IFN-α treatment reduced the levels of RNA-containing capsids in HBV transgenic mice (26). These studies did not resolve whether IFN-α inhibited assembly or led to rapid degradation of RNA-containing capsids. Our results suggest that the antiviral program induced by IFN-α increased the turnover rate of DHBV RNA in subviral core particles less than twofold and that the stability of total DHBV RNA was not changed in a significant manner under the assay conditions used. In addition, we found that IFN-α can interfere with the accumulation of capsids with complete minus strand DNA (Fig. 7). However, no evidence of an effect of IFN-α on DNA synthesis in vitro was observed (results not shown), suggesting that it might depend on cellular factors that are present in dstet5 cells during viral DNA synthesis. Whether the shorter DNA products represent intermediates of a DNA degradation reaction caused by the disintegration of capsids or whether they are due to a pause or termination of the reverse transcription reaction is not known.
By blocking reverse transcription with a known RT inhibitor, we showed that the amounts of viral cccDNA, like those of core DNA, decreased with time, independently of whether the cells were kept confluent or passaged to induce cell division. Moreover, once rcDNA levels dropped, the rate of cccDNA synthesis became slower than its rate of decay. In dividing cells, the decay rate of both core DNA and cccDNA was higher than in resting cells. However, it was clear that both core DNA and cccDNA decayed even in nondividing cells. The increased decay rate of cccDNA in the passaged cultures could be accounted for by dilution alone, suggesting that cccDNA can survive cell division and, perhaps, partition to daughter cells, similar to episomal DNA of papillomaviruses and herpesviruses (1, 12, 29). In contrast to cccDNA, core DNA declined at a faster rate in dividing cells than would be expected from dilution, suggesting that cell cycle progression or some factor(s) associated with this process influences the stability of DNA-containing nucleocapsids. In any case, our results indicated that both cccDNA and core DNA turn over in the dstet5 system. If this were the case under natural conditions in vivo, it should, at least in theory, be possible to clear infections with antiviral therapies that completely block reverse transcription.
Attempts to determine the exact t1/2s of core DNA and cccDNA were complicated by several factors. For example, we could not determine whether 3TC completely blocked reverse transcription or exclude the possibility that small amounts of core particles were released into the medium even in the absence of envelope proteins. Finally, as described above, rcDNA was converted into cccDNA in the presence of 3TC and hence, cccDNA levels were dependent not only on its rate of decay but also on the available rcDNA. Considering these limitations, the t1/2s of core DNA and cccDNA were estimated to be 24 to 48 and 48 h, respectively. Similar to our results, the t1/2 of cccDNA in DHBV-infected primary duck hepatocyte cultures was estimated to be 3 to 5 days (2). In contrast, cccDNA in woodchuck hepatitis virus-infected hepatocytes did not appear to have a measurable t1/2 (14). Whether these differences reflect specific properties of the DHBV and woodchuck hepatitis virus systems or are the result of differences in experimental designs remains to be resolved.
The development of a cell line permitting conditional replication should facilitate future investigations of specific steps in the hepadnavirus replication cycle. In contrast to previous studies, in which transcription and replication in hepatoma cells occurred from integrated viral DNA, we have created conditions under which cccDNA alone can sustain transcription and replication of a hepadnavirus. Hence, this system mimics more closely the conditions in infected hepatocytes in vivo and provides an opportunity to study the formation and turnover of viral DNA, as well as the regulation of viral transcription from authentic DNA templates. Finally, the system is amenable to the recreation of conditions such as those occurring during antiviral therapy and natural recovery from infections. In this regard, it may permit the testing of novel antiviral strategies and future therapies that can cure HBV infections.
Acknowledgments
We thank Jesse Summers for providing the chicken IFN-α clones and Bill Mason and David Lazinski (Tufts University) for critical reading of the manuscript and helpful suggestions. We acknowledge the help of the Fox Chase Cancer Center tissue culture and DNA sequencing facilities. We thank Tony Whitaker for technical assistance.
This work was supported by grants from the National Institutes of Health and the Commonwealth of Pennsylvania.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Civitico, G. M., and S. A. Locarnini. 1994. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology 203:81-89. [DOI] [PubMed] [Google Scholar]
- 3.Condreay, L. D., C. E. Aldrich, L. Coates, W. S. Mason, and T. T. Wu. 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J. Virol. 64:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo, J.-T., H. Zhou, C. Liu, C. Aldrich, J. Saputelli, T. Whitaker, M. I. Barrasa, W. S. Mason, and C. Seeger. 2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infection. J. Virol. 74:1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubscher, U., G. Maga, and S. Spadari. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71:133-163. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, K. A. 1993. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 62:685-713. [DOI] [PubMed] [Google Scholar]
- 10.Kawaguchi, T., K. Nomura, Y. Hirayama, and T. Kitagawa. 1987. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 47:4460-4464. [PubMed] [Google Scholar]
- 11.Lien, J., D. J. Petcu, C. E. Aldrich, and W. S. Mason. 1987. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J. Virol. 61:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lusky, M., and M. R. Botchan. 1984. Characterization of the bovine papilloma virus plasmid maintenance sequences. Cell 36:391-401. [DOI] [PubMed] [Google Scholar]
- 13.Macrae, D. R., V. Bruss, and D. Ganem. 1991. Myristylation of a duck hepatitis B virus envelope protein is essential for infectivity but not for virus assembly. Virology 181:359-363. [DOI] [PubMed] [Google Scholar]
- 14.Moraleda, G., J. Saputelli, C. E. Aldrich, D. Averett, L. Condreay, and W. S. Mason. 1997. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J. Virol. 71:9392-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pugh, J., A. Zweidler, and J. Summers. 1989. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 63:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz, U., and F. V. Chisari. 1999. Recombinant duck interferon gamma inhibits duck hepatitis B virus replication in primary hepatocytes. J. Virol. 73:3162-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultz, U., J. Summers, P. Staeheli, and F. V. Chisari. 1999. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J. Virol. 73:5459-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeger, C., D. Ganem, and H. E. Varmus. 1986. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science 232:477-484. [DOI] [PubMed] [Google Scholar]
- 19.Seeger, C., E. H. Leber, L. K. Wiens, and J. Hu. 1996. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology 222:430-439. [DOI] [PubMed] [Google Scholar]
- 20.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 23.Summers, J., P. M. Smith, and A. L. Horwich. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, G. H., and C. Seeger. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 26.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 28.Yang, W., W. S. Mason, and J. Summers. 1996. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J. Virol. 70:4567-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, S., and D. N. Standring. 1992. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc. Natl. Acad. Sci. USA 89:10046-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]