Abstract
Protein folding in the cell involves the action of different molecular chaperones and folding-facilitating enzymes. In the endoplasmic reticulum (ER), the folding status of glycoproteins is stringently controlled by a glucosyltranferase enzyme (GT) that creates monoglucosylated structures recognized by ER resident lectins (calnexin/calreticulin, CNX/CRT). GT serves as a folding sensor because it only glucosylates misfolded or partly folded glycoproteins. Nevertheless, the molecular mechanism behind this recognition process remains largely unknown. In this paper we explore the structural determinants for GT recognition by using a single domain model protein. For this purpose we used a family of chemically glycosylated proteins derived from chymotrypsin inhibitor-2 as GT substrates. Structural characterization of species showing higher glucose acceptor capacity suggests that GT recognizes solvent accessible hydrophobic patches in molten globule-like conformers mimicking intermediate folding stages of nascent glycoproteins. It was further confirmed that BiP (binding protein, a chaperone of the heat shock protein 70 family) preferentially recognized neoglycoproteins displaying extended conformations, thus providing a molecular rationale for the sequential BiP-CNX/CRT interaction with folding glycoproteins observed in vivo.
The folding status of glycoproteins is strictly censored in the endoplasmic reticulum (ER), before they attain their final destination to membranes, organelles, or the extracellular environment (1). Proteins that fail to fold properly are initially retained in the ER and eventually degraded in the proteasomes. The quality control system involves the action of UDP-Glc:glycoprotein glucosyltranferase (GT) and glucosidase II (GII) enzymes and the ER resident lectins calnexin (CNX) and calreticulin (CRT). Proteins that are incompletely folded are glucosylated by GT, thus generating Glc1Man9GlcNAc2 structures. On binding those structures, CNX and/or CNR retain the monoglucosylated glycoproteins in the ER. Eventually, the Glc units added by GT are removed by GII, causing the release of glycoproteins from the lectin anchors. Glucosylation–deglucosylation cycles catalyzed by the opposing activities of GT and GII continue until native conformations are attained. Glycoproteins then become substrates of GII but not of GT and are thus free to pursue their travel through the secretory pathway.
The CNX/CRT-monoglucosylated glycan interaction is one of the mechanism by which cells retain incompletely folded glycoproteins in the ER and, in addition, it enhances folding efficiency by preventing protein aggregation and allowing intervention of additional ER chaperones and folding accessory proteins. The key element in this mechanism is GT as it is the only component exclusively acting on incompletely folded glycoproteins. This feature is based on the recognition of two structural determinants not exposed in native conformations. One of them is the innermost GlcNAc unit, which is buried within the protein scaffold in native but solvent exposed in nonnative conformations (2). In addition, there is an ill-defined determinant in incompletely folded polypeptides that is also recognized by GT. This protein determinant has eluded identification so far, mainly because of the difficulties intrinsic to the study of partly folded conformations of possible intermediates along the folding pathway of nascent polypeptides. It has been suggested that hydrophobic residues exposed in incompletely folded conformers could be the protein elements recognized by GT as exposure of those residues is the only structural feature common to all nonnative conformers. Although GT was described to be retained by hydrophobic surfaces as Sepharose 4B-linked hydrophobic nonapeptides or by phenyl-Superose columns (2, 3), no suitable experimental systems have been used so far to confirm that binding to hydrophobic surfaces in folding intermediates was the event that actually triggered glycoprotein glucosylation. Along with this hypothesis of hydrophobic surface binding, an excessive dynamic mobility of folding intermediates has been invoked to explain GT selectivity (4).
The analysis of a nascent polypeptide of a model protein, chymotrypsin inhibitor 2 (CI2), revealed that secondary and tertiary structures in molten globule-like intermediate fragments are formed in parallel, but side chain packing and cooperativity only develop in the near full-length polypeptide (5, 6). The shortest fragments were disordered because of the absence of long-range interactions [from CI2(1–25) to CI2(1–40); ref. 7]. As the proteins grew from its N terminus, elements of secondary and tertiary structure are formed in a highly concerted manner, and fragment CI2(1–53) containing 83% of the sequence information already displayed substantial native-like structure (6). The formation of the final tertiary structure in the full-length protein, CI2(1–64), relied on the presence of the last three residues. An NMR detailed analysis of fragment CI2(1–40) revealed the presence of local nonnative hydrophobic clustering in parts of its sequence, arising as a result of the burial of local hydrophobic residues with no classical elements of secondary structure being formed (8).
To address the structural determinants of GT substrates we used the CI2 nascent polypeptides fragments but converted into neoglycoproteins (GCI2s). The structural analysis of GCI2s of increasing length provides an ideal model system for the analysis of the successive conformations adopted by nascent glycoproteins as they emerge into the ER lumen. As the neoglycoproteins used span a wide range of hydrophobicity and conformational states, they allowed the study of GT selectivity in a highly controlled system. Here we show that GT recognition of its substrates is mediated by exposed hydrophobic surfaces, preferentially in molten globule-like conformers displaying native-like structure but lacking side chain packing and folding cooperativity, characteristic of compactly folded proteins
Materials and Methods
Materials.
All chemicals were purchased from Sigma, with the exception of sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate that was purchased from Pierce and NAP-5 desalting columns from Amersham Pharmacia. Rat liver GT and BiP were purified as described (3, 9).
Expression and Purification of CI2.
Truncated forms and full length CI2 were purified according to ref. 6. Briefly, two short forms of CI2 having 40 and 53 amino acid residues, termed CI2(1–40) and CI2(1–53), were produced by cleavage of the wild type or of a mutant protein with a Met residue at position 53 with CNBr. The shortest protein, with 25 amino acids, was prepared by solid phase peptide synthesis. We also introduced the substitution E7C in the four polypeptides to chemically glycosylate the proteins.
Synthesis of Neoglycoproteins.
Because substrates of GT are high mannose-type glycoproteins, thyroglobulin/pronase-derived high mannose type glycopeptides were covalently linked to the new engineered Cys-7 by using sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Sulfo-SMCC). Shortly, 1–2 mg of glycopeptide were mixed with 1 mg Sulfo-SMCC in PBS buffer, pH 7.2. The glycopeptides were desalted after 1 h at room temperature by gel filtration in a Sephadex G-10 column (80 × 1 cm) eluted with 7% 2-propanol in water. Fractions containing oligosaccharides were pooled and dried. In parallel, 2–3 mg of each CI-2 were dissolved in 300 μl of PBS buffer, pH 7.2/6 M urea/10 mM DTT/5 mM EDTA and incubated for 90 min at 37°C. They were then desalted with a NAP-5 column and added to the glycopeptides. After 4 h at room temperature, the initial volume (800 μl) was reduced to 300 μl with a Speedvac, and the incubations were left overnight at the same temperature. The coupling efficiency was usually above 50% and was highly dependent on using reagents at high concentrations. Finally, mixtures were dissolved in 5% acetonitrile and glycosylated forms were purified from nonglycosylated ones by reverse-phase HPLC with a Vydac C8 column using a 25–50% acetonitrile gradient supplemented with 0.05% trifluoroacetic acid. The purified glycosylated forms were dissolved in 20 mM Hepes buffer (pH 7.4) [for GCI2(1–25) and GCI2(1–40)] or the same buffer with 6 M urea [for GCI2(1–53) and GCI2(1–64)] and then dialyzed against assay buffer containing decreasing urea concentrations.
Spectroscopic Characterization of GCI2s.
CD spectra were measured with a Jasco J810 spectropolarimeter. We used 15 μM of GCI2s in far-UV and 50 μM in near-UV CD. Intrinsic fluorescence and 8-anilinonaphthalene-1-sulfonic acid (ANS) binding were measured with an Aminco Bowman Series 2 spectrofluorimeter with 15 μM GCI2s. We used 50 μM ANS with the excitation wavelength set at 360 nm and emission at 460 nm. All spectra were recorded at 25°C in 20 mM Hepes buffer, pH 7.4.
Activity Assays.
GT assays were carried out at 37°C in 50 μl of 40 mM Hepes buffer, pH 7.4/20 mM KCl/5 mM CaCl2/15 μM UDP-Glc/100,000 cpm UDP-[14C]Glc. Reactions were started by the addition of 3 μg of rat liver GT and stopped after 10 min by the addition of 400 μl of concanavalin A-binding buffer at 100°C. After further incubation at 100°C for 5 min, samples were ice-cooled and supplemented with 50 mg of concanavalin A-Sepharose. After 25-min incubations at room temperature, samples were centrifuged for 2 min at 14,000 rpm in an Eppendorf microcentrifuge. Radioactivity bound to the resin was quantified after 12 washes with 400 μl of concanavalin A-binding buffer. Under these conditions, blanks were <100 cpm per tube and radioactivity recovery was >95%. BiP ATPase activity was assayed as described in ref. 9.
Results
Strategy for the Production of Nascent Neoglycoproteins.
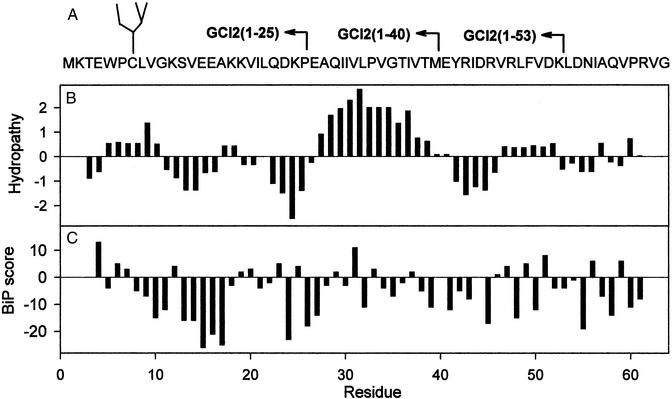
To generate a family of nascent neoglycoprotein fragments we used an approach similar to that used before for the nonglycosylated species (6). In addition, we introduced a point mutation (E7C) and a glycopeptide (Man9GlcNAc2-Asn) was covalently linked to the resulting sulfhydryl group by means of a bifunctional reagent (see Materials and Methods). The position of the resulting single Cys residue was chosen to minimize any disturbing effect on structures, because the original glutamic acid residue was highly exposed to the solvent in a type III turn, with no effects on the conformation or stability of the full-length protein [CI2(1–64)] (10). The primary sequence of GCI2(1–64), the location of glycopeptide chemical coupling (Cys-7), and the C-terminal truncations that yielded shorter GCI2s as well as a hydropathy plot of the nonglycosylated species are depicted in Fig. 1. Predicted BiP binding scores are shown in Fig. 1C (11). Scores >5 are assumed to allow BiP binding. It is worth remarking the high hydrophobicity introduced by residues 27–40. However, a synthetic peptide spanning this hydrophobic stretch was completely insoluble aqueous solutions, indicating that those residues must be buried in the otherwise highly soluble fragment CI2(1–40) (5).
Figure 1.
Primary structure of GCI2s. (A) Amino acid sequence of GCI2(1–64). The position of the covalently linked glycopeptide and lengths of truncated neoglycoproteins are indicated. (B) Hydropathy plot of GCI64 by using a window of seven residues. (C) Predicted BiP binding scores of GCI2(1–64). Scores higher than 5 are assumed to allow BiP binding.
The Neoglycoproteins Follow a Conformational Pathway That Parallels That of the Nonglycosylated Chain.
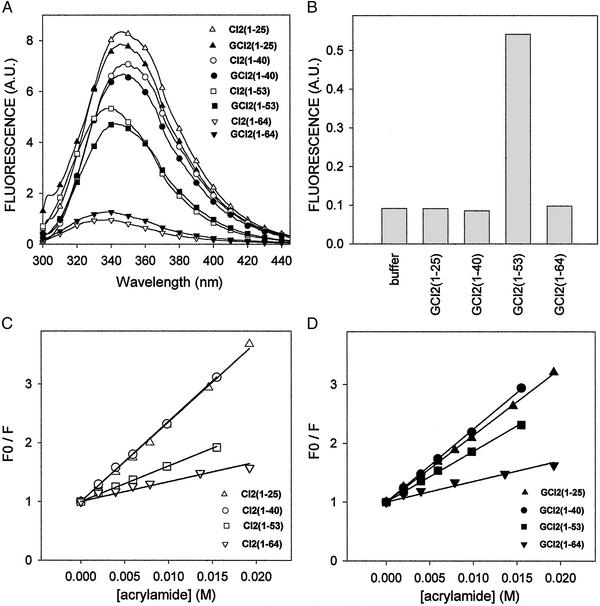
Trp-5, the unique tryptophan residue present in CI2s, is buried in the major hydrophobic core of CI2(1–64), and can be used as a probe to follow the folding and conformational states of CI2s and GCI2s (6). Comparison of the Trp fluorescence of glycosylated and nonglycosylated derivatives is particularly relevant when studying possible conformational modifications introduced by covalently linking a glycopeptide to CI2s as the Trp and Cys are only two residues apart. The full-length folded form GCI2(1–64) had a fluorescence emission maximum at around 339 nm (Fig. 2A, Table 1). This maximum was shifted to 347 nm for the shortest GCI2 fragments [GCI2(1–25) and GCI2(1–40)] with a concomitant increase of intensity, revealing the exposure of Trp-5 to the solvent in those structures. GCI2(1–53), the minimum fragment where secondary and tertiary structures are formed concertedly (6), displayed an intermediate fluorescence intensity with a maximum at ≈342 nm. No major differences between glycosylated and nonglycosylated forms were observed except for CI2(1–53) and GCI2(1–53), where the maximum fluorescence of the nonglycosylated form was slightly shifted to a lower wavelength. On the other hand, only GCI2(1–53) bound ANS, whereas GCI2(1–25), GCI2(1–40), and GCI2(1–64) did not show ANS binding, in agreement with what was observed for the nonglycosylated polypeptides (Fig. 2B). These results showed that GCI2(1–53) was the only species exposing substantial hydrophobic patches to the solvent. Those patches, however, must have a particular spatial distribution, because the fragment contains most of the native secondary and tertiary structure of the full-length protein.
Figure 2.
Fluorometric measurements. (A) Fluorescence spectra of CI2s and GCI2s. (B) Exposed hydrophobic regions of GCI2s monitored by ANS fluorescence. (C and D) Acrylamide quenching of CI2s and GCI2s, respectively. F and F0 stand for fluorescence in the presence and absence of acrylamide, respectively.
Table 1.
Fluorescence emission maximum wavelengths and acrylamide bimolecular quenching constants for CI2s and GCI2s
| Substrate | λmax emission, nm | Kd, M−1 |
|---|---|---|
| CI2(1–25) | 447 | 132 |
| CI2(1–40) | 447 | 132 |
| CI2(1–53) | 338 | 59 |
| CI2(1–64) | 339 | 28 |
| GCI2(1–25) | 447 | 118 |
| GCI2(1–40) | 447 | 123 |
| GCI2(1–53) | 342 | 85 |
| GCI2(1–64) | 339 | 32 |
Exposure of Trp-5 was studied by analysis of quenching with acrylamide as this reagent does not penetrate into the cores of compact folded proteins. Stern–Volmer plots of CI2s and GCI2s revealed that the two shorter forms have a highly solvent-exposed Trp-5 when compared with CI2(1–64) and GCI2(1–64), respectively (Fig. 2 C and D, Table 1). Once again, CI2(1–53) and GCI2(1–53) showed intermediate behaviors. Glycosylated and nonglycosylated forms showed similar quenching constants except for CI2(1–53) and GCI2(1–53), as the Trp of the latter was more exposed (Table 1). This observation is in agreement with the difference observed in the fluorescence emission maxima.
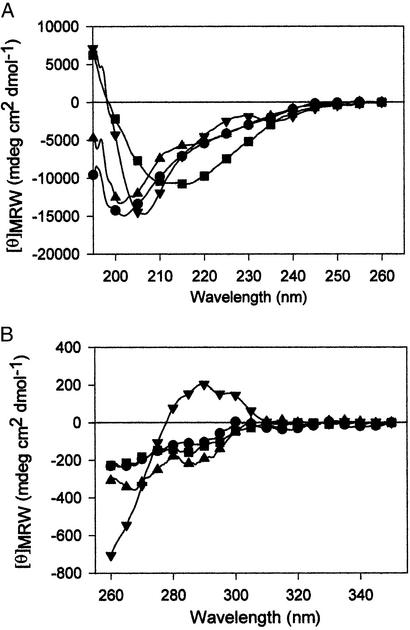
Far-UV CD spectra of GCI2s revealed the appearance of secondary structure with chain growth. GCI2(1–25) and GCI2(1–40) appeared to be disordered, both with a minimum at 200 nm (Fig. 3A). As expected, the spectrum of the full-length GCI2(1–64) was identical with that of the nonglycosylated protein (6). Fragment GCI2(1–53) was highly structured as revealed by a minimum at 211 nm. Moreover, a positive band at 190 nm in this last neoglycoprotein was also a clear indication of structure formation. The presence of secondary structure elements and ANS binding in GCI2(1–53) strongly suggested that it shared most properties with the molten globule-like structure described for the nonglycosylated conformational pathway. Near-UV CD spectra of the neoglycoprotein fragments were very similar and distinct from that of full-length GCI2(1–64), which presented a positive band at ≈290 nm, suggesting a low amount or complete absence of defined tertiary structure in the GCI2 fragments (Fig. 3B). However, NMR analysis of CI2(1–53) showed unequivocal presence of tertiary structure as evidenced from native chemical shifts and nuclear Overhauser effects (6). Acrylamide quenching experiments showing a buried tryptophan residue in GCI2(1–53) support the presence of tertiary structure, even if the near-UV CD spectrum does not suggest an asymmetric environment for the chromophore.
Figure 3.
UV-CD of GCI2s. (A) Far UV-CD spectra of GCI2s. (B) Near-UV CD spectra of GCI2s.
GT and BiP Are Selective for Substrates with Different Conformational Properties.
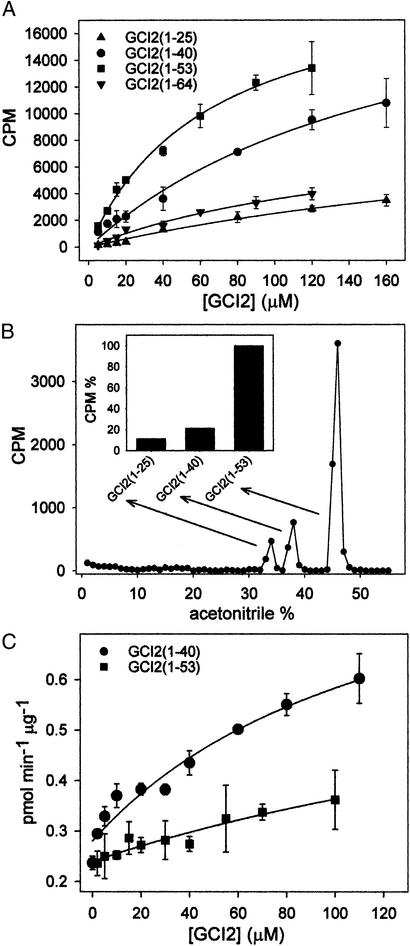
A substrate curve was performed for each member of GCI2 family of fragments, and an estimation of Km could be established in two cases. The activity of purified rat liver GT toward GCI2s varied sharply depending on the conformational properties of the neoglycoproteins (Fig. 4A). GCI2(1–25) and GCI2(1–64) were poorly glucosylated by GT and both curves did not shown saturation. The residual activity of GT toward these substrates was not surprising because even Man9GlcNAc2-Asn or fully folded glycoproteins displayed a low but reproducible glucose acceptor capacity (12). GCI2(1–53) was the best substrate (Km = 66 ± 8 μM), followed by GCI2(1–40) (Km = 180 ± 40 μM). Because none of these substrate curves reached saturation, the values of Km should be taken only as an approximation of the affinity, but clearly GT displayed a higher affinity for GCI2(1–53) than for GCI2(1–40). Aggregation of the highly hydrophobic GCI2(1–40) can be ruled out because all CI2s were monomeric in the 0–150 μM range (13, 14). Moreover, the presence of the oligosaccharide in GCI2s is expected to further enhance their solubility. In addition, aggregation of GCI2s was ruled out by the absence of light scattering in the 310–400 nm range (data not shown).
Figure 4.
Glucose acceptor capacities of GCI2s. Effect of neoglycoproteins on BiP ATPase activity. (A) Incubation of individual GCI2s with GT. (B) Radioactivity pattern of a reverse HPLC chromatography obtained on incubation of a mixture of 10 μM of GCI2(1–25), GCI2(1–40), and GCI2(1–64) as substrates of GT. (Inset) The integration of radioactivity associated with each GCI2. (C) BiP ATPase activity as function of GCI2(1–40) and GCI2(1–53) concentrations.
Next, we carried out a competition experiment between substrates for GT. In this experiment, 10 μM GCI2(1–25), GCI2(1–40), and GCI2(1–53) were incubated in the same test tube. Products were readily purified by reverse-phase HPLC and radioactivity associated to each substrate was quantified (Fig. 4B). Results confirmed those obtained from activity measurements, i.e., the acceptor capacities relative to GCI2(1–53) (100%) of GCI2(1–40), and GCI2(1–25) were 22% and 12%, respectively.
BiP, a member of the Hsp70 family of proteins, is the main ER classical molecular chaperone as it accounts for ≈5% of the ER total protein. It is required for translocation of newly synthesized peptides across the ER membrane and for their subsequent folding and assembly. This chaperone recognizes linear stretches of seven residues with large hydrophobic amino acids in alternating positions present in proteins in extended conformations (11). On the other hand, results shown in Fig. 4 A and B indicate that GT preferentially glucosylates species displaying a molten globule-like conformation, unlike to what it is known for BiP. To confirm the different recognition preferences of GT and BiP, we assayed the enhancement of purified rat liver BiP ATPase activity by GCI2(1–40) and GCI2(1–53). As depicted in Fig. 4C, GCI2(1–40) was a much more effective activator than GCI2(1–53), as opposed to what was found for GT. There are two predicted BiP recognition sites in CI2(1–64), centered around Trp-5 and L32 (Fig. 1C). The latter was apparently the site recognized by BiP in glycosylated derivatives as the oligosaccharide precluded recognition of the former one: nonglycosylated CI2(1–25) but not its glycosylated derivative [GCI2(1–25)] stimulated BiP ATPase activity (not shown).
Discussion
We have herein analyzed the structural features recognized by GT in acceptor substrates by using a family of neoglycoproteins (GCI2s) derived from CI2. As in nonglycosylated counterparts, the glycosylated derivatives modeling a nascent glycoprotein chain show the formation of substantial native structure as the polypeptide is elongated to residue 53. Intrinsic fluorescence of Trp-5 was shifted to shorter wavelengths with lower intensities, which together with parallel changes in CD spectra, are indicative of structure formation. Fluorescence spectrum of GCI2(1–53) was in between the spectra of the two shorter GCI2s and GCI2(1–64), pointing to an intermediate exposition on Trp-5 in this form. This behavior was confirmed by quenching of fluorescence with acrylamide as the quenching constant for GCI2(1–53) fell between those of GCI2(1–40) and GCI2(1–64).
Near-UV CD spectroscopy showed that all truncated forms lacked a compact tertiary structure. The three glycosylated fragments of CI2 [GCI2(1–25), GCI2(1–40), and GCI2(1–53)] studied herein presented similar near-UV CD spectra, that were very different from that of full-length GCI2(1–64). This last spectrum showed a positive band centered around 290 nm. Far-UV CD revealed that GCI2(1–53) had a high amount of secondary structure, a feature missing in GCI2(1–40) and GCI2(1–25) (Fig. 3A). These observations suggested that GCI2(1–53) displayed a molten globule-like conformation, as has been reported for the nonglycosylated CI2(1–53) species (6). The molten globule conformation of GCI2(1–53) was confirmed by its capacity for ANS binding (Fig. 2B). This molecule can be used as a probe to indicate the presence of collapsed but loosely packed intermediates, because it binds to hydrophobic amino acid patches (15). Given the overall similar behaviors between glycosylated and nonglycosylated proteins, it is likely that GCI2(1–40) displays nonnative hydrophobic clusters as revealed by NMR for CI2(1–40) (8). In summary, the family GCI2 fragments provided an ideal model for an elongating glycoprotein, with a fully folded species [GCI2(1–64)], a molten globule-like intermediate with long range interactions [GCI2(1–53)], a small fragment with local nonnative hydrophobic clusters and no secondary or tertiary structure [GCI2(1–40)], and a largely unfolded and hydrophilic [GCI2(1–25)] neoglycoprotein fragment.
The use of the GCI2 family allowed us to study substrate recognition by GT in a highly controlled system and to roughly simulate the folding and reglucosylation processes that may take place as a protein emerges into the ER lumen. The activity of purified rat liver GT (4) toward GCI2s varied substantially with the conformational properties of the acceptor neoglycoprotein (Fig. 4A). GCI2(1–53) was the best substrate followed by GCI2(1–40), whereas the glucose acceptor capacities of GCI2(1–25) and GCI2(1–64) were negligible. Preference for GCI2(1–53) over GCI2(1–40) was reflected by roughly 3-fold difference in their apparent Km values and was slightly increased when both substrates were incubated together. As the only structural feature common to both substrates [GCI2(1–40) and GCI2(1–53)] is the exposure of hydrophobic amino acid clusters, it may be concluded that this is precisely the structural determinant recognized by GT. The determinant was absent from GCI2(1–25), a hydrophilic, highly disordered fragment and from full-length folded GCI2(1–64). Hydrophobic clustering in CI2(1–40) was detected by NMR but not by ANS binding (5), suggesting that the hydrophobic residues in this fragment are not solvent accessible as in CI2(1–53) species. Alternatively, there could be other hindrances for ANS binding such as charge repulsion. A possible explanation is that GCI2(1–40) species with exposed hydrophobic clusters would not be significantly populated at the equilibrium, but GT binding to those clusters would shift the equilibrium to the exposed cluster conformation. A similar explanation may be envisaged for BiP binding of the recognition site centered around L32 in GCI2(1–40).
Recognition of the hydrophobic heptapeptide stretch by DnaK (the BiP bacterial homologue) is mediated by a cleft into which the extended polypeptide is introduced (16). Results presented here suggest that GT would require a large contact surface rather than a cleft to bind the solvent accessible structures found in molten globule-like folding intermediates. Further, the fact that GT and BiP preferentially recognize proteins displaying different conformations supports the idea that they act on different stages of folding of nascent glycoprotein chains.
A previous line of work showed that RNaseBS-protein, a short form of ribonuclease B lacking the first 20 amino acids, was a good GT substrate (17). RNaseBS-protein was better substrate than the unfolded reduced alkylated RNaseB, suggesting the preference of GT for partially folded proteins (17, 18). RNaseBS-protein has a slightly distorted conformation, shown by its greater susceptibility to PNGase F and protease digestion. Although these features were shared by RNaseBS, the full length but otherwise cleaved form of RNaseB that retained the 20 amino acids by noncovalent linkages, this latter form was poorly recognized by GT. It should be pointed out that RNaseBS is enzymatically active, whereas RNaseBS-protein is not. This is probably because three residues of the cleaved peptide (Lys-7, Gln-11, and Ala-4) are part of the substrate-binding site (19). The recognition of RNaseBS-protein by GT could arise from the exposure of residues belonging to the hydrophobic core that are located below the peptide cleaved, a point that would merit further analysis.
There is also evidence that also GT glucosylates glycoproteins in vivo at their last folding stages. It was reported that in Trypanosoma cruzi CRT only recognized cruzipain (a lysosomal glycoprotein) displaying the complete or almost complete complement of disulfide bridges (20). As in trypanosomes formation of monoglucosylated oligosaccharides (the species recognized by CNX/CRT) is exclusively dependent on GT activity, it was concluded that cruzipain molecules had to be in advanced folding stages to be glucosylated. Moreover, prevention of formation of disulfide bonds in Schizosaccharomyces pombe by the addition of DTT to live cells did not lead to generalized increase of glycoprotein glucosylation. It was proposed that the reductant prevented glycoprotein from reaching their last folding stages (21). From results with the GCI2 fragment model, it appears that the requirement for GT recognition is an accessible hydrophobic patch preferentially in species with substantial secondary structure as well as long range interactions, and not just a contiguous sequence of hydrophobic side chains.
The ER is a highly crowded environment, and several molecular chaperones have evolved to prevent protein aggregation. The substrate preference of these chaperones is expected to vary to cover a myriad of misfolded protein forms. In vivo, the affinity of GT could be modulated by the presence of other chaperones such as BiP, that recognizes hydrophobic amino acid-rich heptapeptides, preferentially in extended polypeptides. The overall folding process reflects the variety of competitive and cooperative effects exerted by ER chaperons and folding-accessory proteins. The first step toward a complete picture is to determine the selectivity of each isolated chaperone by using substrates with controlled conformations. The specificity of GT exerts its action during the last steps of protein folding, when a newly synthesized protein accumulates enough sequence information to form long-range hydrophobic clusters characteristic of molten globule conformations. It may be expected that in previous folding steps other chaperones that have evolved to preferentially recognize extended hydrophobic structures, such as BiP, could be more effective (14). There is growing evidence in several systems for a sequential intervention of BiP and CNX/CRT to assist the entire protein folding process (22–25). The herein reported different substrate recognition patterns between BiP and GT provide a molecular rationale for such sequential intervention.
Acknowledgments
We thank M. Feldman for help with GCI2s mutants construction and D. Fiore for comments on the manuscript. Financial support from the National Institutes of Health, the Howard Hughes Medical Institute, and the Agencia Nacional de Promoción Científica y Tecnológica (Argentina) is acknowledged.
Abbreviations
- ANS
8-anilinonaphthalene-1-sulfonic acid
- BiP
Ig heavy chain binding protein
- CI2
barley chymotrypsin inhibitor-2
- CNX
calnexin
- CRT
calreticulin
- GCI2
chemical glycosylated CI2
- GII
glucosidase II
- GT
UDG-Glc:glycoprotein glucosyltransferase
- ER
endoplasmic reticulum
Footnotes
This contribution is part of the special series of Inaugural articles by members of the National Academy of Sciences elected on May 2, 2000.
References
- 1.Parodi A J. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 2.Sousa M, Parodi A J. EMBO J. 1995;14:4196–4203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trombetta S E, Parodi A J. J Biol Chem. 1992;267:9236–9240. [PubMed] [Google Scholar]
- 4.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 5.de Prat Gay G, Ruiz-Sanz J, Neira J L, Itzhaki L S, Fersht A R. Proc Natl Acad Sci USA. 1995;92:3683–3686. doi: 10.1073/pnas.92.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Prat Gay G, Ruiz-Sanz J, Neira J L, Corrales F J, Otzen D E, Ladurner A G, Fersht A R. J Mol Biol. 1995;254:968–979. doi: 10.1006/jmbi.1995.0669. [DOI] [PubMed] [Google Scholar]
- 7.Itzhaki L S, Neira J L, Ruiz-Sanz J, de Prat Gay G, Fersht A R. J Mol Biol. 1995;254:289–304. doi: 10.1006/jmbi.1995.0617. [DOI] [PubMed] [Google Scholar]
- 8.de Prat Gay G, Ruiz-Sanz J, Davis B, Fersht A R. Biochemistry. 1994;91:10943–10946. doi: 10.1073/pnas.91.23.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier M, King L, Blond-Elguindi S. Methods Enzymol. 1998;290:384–409. doi: 10.1016/s0076-6879(98)90033-7. [DOI] [PubMed] [Google Scholar]
- 10.McPhalen C A, James M N G. Biochemistry. 1987;26:261–269. doi: 10.1021/bi00375a036. [DOI] [PubMed] [Google Scholar]
- 11.Blond-Elguindi S, Cwirla S E, Dower W J, Lipshutz R J, Sprang S R, Sambrook J F, Gething M J. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 12.Sousa M C, Ferrero-Garcia M A, Parodi A J. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- 13.de Prat Gay G, Fersht A R. Biochemistry. 1994;33:7957–7963. doi: 10.1021/bi00191a024. [DOI] [PubMed] [Google Scholar]
- 14.de Prat Gay G. Arch Biochem Biophys. 1996;335:1–7. doi: 10.1006/abbi.1996.0475. [DOI] [PubMed] [Google Scholar]
- 15.Semisotnov G V, Rodionova N A, Razgulyaev O I, Uversky V N, Gripas A F, Gilmanshin R I. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 16.Rüdiger S, Buchberger A, Bukau B. Nat Struct Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 17.Trombetta E S, Helenius A. J Cell Biol. 2000;148:1123–1129. doi: 10.1083/jcb.148.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritter C, Helenius A. Nat Struct Biol. 2000;7:278–280. doi: 10.1038/74035. [DOI] [PubMed] [Google Scholar]
- 19.Kim E E, Varadarajan R, Wyckoff H W, Richards F M. Biochemistry. 1992;31:12304–12314. doi: 10.1021/bi00164a004. [DOI] [PubMed] [Google Scholar]
- 20.Labriola C, Cazzulo J J, Parodi A J. Mol Biol Cell. 1999;10:1381–1394. doi: 10.1091/mbc.10.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández F, D'Alessio C, Fanchiotti S, Parodi A J. EMBO J. 1998;17:5877–5886. doi: 10.1093/emboj/17.20.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molinari M, Helenius A. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 23.Kim P S, Arvan P. J Cell Biol. 1995;128:29–38. doi: 10.1083/jcb.128.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomita Y, Yamashita T, Sato H, Taira H. J Biochem (Tokyo) 1999;126:1090–1100. doi: 10.1093/oxfordjournals.jbchem.a022554. [DOI] [PubMed] [Google Scholar]
- 25.Hammond C, Helenius A. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]