Abstract
The envelope glycoprotein E2 of hepatitis C virus (HCV) is the target of neutralizing antibodies and is presently being evaluated as an HCV vaccine candidate. HCV binds to human cells through the interaction of E2 with the tetraspanin CD81, a putative viral receptor component. We have analyzed four different E2 proteins from 1a and 1b viral isolates for their ability to bind to recombinant CD81 in vitro and to the native receptor displayed on the surface of Molt-4 cells. A substantial difference in binding efficiency between these E2 variants was observed, with proteins derived from 1b subtypes showing significantly lower binding than the 1a protein. To elucidate the mechanism of E2-CD81 interaction and to identify critical regions responsible for the different binding efficiencies of the E2 variants, several mutants were generated in E2 protein regions predicted by computer modeling to be exposed on the protein surface. Functional analysis of these E2 derivatives revealed that at least two distinct domains are responsible for interaction with CD81. A first segment centered around amino acid residues 613 to 618 is essential for recognition, while a second element including the two hypervariable regions (HVRs) modulates E2 receptor binding. Binding inhibition experiments with anti-HVR monoclonal antibodies confirmed this mapping and supported the hypothesis that a complex interplay between the two HVRs of E2 is responsible for modulating receptor binding, possibly through intramolecular interactions. Finally, E2 proteins from different isolates displayed a profile of binding to human hepatic cells different from that observed on Molt-4 cells or isolated recombinant CD81, indicating that additional factors are involved in viral recognition by target liver cells.
Hepatitis C virus (HCV) is the major etiological agent of both community-acquired and posttransfusionally acquired non-A, non-B viral hepatitis. Approximately 80% of infected patients develop chronic hepatitis, among which 20% to 30% progress to liver cirrhosis and end-stage liver disease. Chronic infection correlates with an increased risk of hepatocellular carcinoma. Currently available therapies are limited to administration of alpha interferon, on its own or in combination with ribavirin (15, 26). Such treatments are expensive, show low response rates, and carry the risk of significant side effects. Even if high response rates (98%) can be obtained by treating patients when they are still in the acute phase of infection (19), efficient implementation of this strategy would require a constant and population-wide monitoring, since the majority of infections are asymptomatic (9, 42). Consequently, the development of an HCV vaccine remains a high-priority goal.
The HCV viral genome is a single-stranded, positive-sense RNA of approximately 9.6 kb, encoding a single polyprotein of 3,010 to 3,033 amino acids which is cleaved into nine mature proteins by a combination of host and viral peptidases. The predicted structural components comprise the core (C) (∼21 kDa) and two heavily N-glycosylated envelope glycoproteins, E1 (∼31 kDa) and E2 (∼70 kDa). Both E1 and E2 are believed to be type 1 transmembrane proteins, with N-terminal ectodomains and C-terminal hydrophobic anchors. HCV envelope glycoproteins are deemed important, since chimpanzees immunized with purified recombinant E1/E2 heterodimeric proteins were protected against challenge with homologous virus (5). However, a major concern still remains as to whether the anti-E2 response elicited by one recombinant protein would be effective against heterologous viral inocula. In fact, HCV displays a high rate of mutation during replication and exists in the bloodstreams of infected patients as a quasispecies, which fluctuates during the course of the disease mainly as a result of immune pressure (20, 22, 37, 38, 40, 43). One of the targets of this immune response is the 27-amino-acid-long N-terminal segment of the E2 glycoprotein (amino acids [aa] 384 to 410) termed hypervariable region 1 (HVR1), which is the most variable region of the whole HCV polyprotein and contains a neutralization determinant. However, anti-HVR1 antibodies specific for one variant display only a limited ability to neutralize different viral variants (10, 11). Given this scenario, one of the tasks in developing an HCV vaccine is to find a solution that takes into account viral variability. Sequence analysis of the several HVR1s from different viral isolates suggested that a number of highly conserved segments exist within HVR1 (31, 33, 38). Thus, it would seem that HVR1, rather than being a truly variable segment, might actually adopt one out of a range of closely related conformations that is compatible with recognition of the virus' cellular receptor.
CD81 was recently reported to bind HCV through interaction with E2 and was hypothesized to act as a viral receptor component (32). Consistent with this view, high titers of antibodies neutralizing the binding of E2 to CD81 (NOB antibodies) have been shown to correlate with protection against HCV infection in chimpanzees (32). It was also reported that CD81 engagement by HCV may lead to autoimmunity or immune evasion (6, 39, 41). CD81 is a member of the tetraspanin family of membrane proteins, characterized by four transmembrane domains, a short intracellular region, and two extracellular loops. This cell surface protein is widely expressed and frequently found in association with other membrane-exposed factors that vary in cell types of different lineages (24). The CD81 binding site for E2 has been localized within the large extracellular loop (LEL) domain (21, 32), and amino acid residues essential for this interaction have been identified (27). By contrast, the E2 region(s) involved in CD81 interaction is still poorly defined; the only available information derives from binding inhibition experiments with monoclonal antibodies (MAbs) and polyclonal antibodies or from generating chimeric glycoproteins through swapping of large protein fragments between different isolates (2, 16, 17, 27, 29, 30, 44).
In this work we report the considerable variance shown by four E2 proteins from different viral isolates of the 1a and 1b subtypes in their interaction with CD81. Testing a number of mutants with deletion and swapping mutations in HVR1 and HVR2 revealed a complex interplay between these elements which is responsible for modulating binding to CD81. By contrast, through site-directed mutagenesis we identified a principal element responsible for CD81 recognition located around aa 613 to 618 of the HCV polyprotein. Our studies also indicate that additional factors other than CD81 may mediate viral recognition by liver cells, since E2 natural variants displayed a profile of binding to human hepatoma cells that was different from that observed on isolated recombinant CD81 or Molt-4 cells.
MATERIALS AND METHODS
Construction of E2 plasmids.
E2 proteins from genotypes 1a (strain H) and 1b (strains BK, N2, and J) were cloned in plasmid V1JnsTPA (8) as described below. The proteins were truncated at either aa 661 (H, BK, and J) or aa 662 (N2) and had an additional tag of six histidine residues at the C terminus. E2H (GenBank accession no. M67463) was amplified by using the sense primer5′-GGAGCAGTCTTCGTTTCGCCCGAAACCCACGTCACCGGGGGA-3′and antisense primer 5′-AGGCACAGCAGATCTTTAGTGGTGGTGGTGGTGGTGCTCGGACCTGTCCCTGTCTTCCAG-3′, E2BK (closely related to the sequence under GenBank accession no. M58335) was amplified by using the sense primer 5′-GGAGCAGTCTTCGTTTCGCCCGATACCCACGTGACAGGGGGG-3′ and antisense primer 5′-AGGCACAGCAGATCTTTAGTGGTGGTGGTGGTGGTGCTCCGCCCTATCCCTGTCCTCCAA-3′, E2N2 (Gen-Bank accession no. D13406) was amplified by using the sense primer 5′-GGAGCAGTCTTCGTTTCGCCCCACACCCTCACAACGGGGGGGCACGCTGCCCGCCTCACC-3′ and the antisense primer 5′-AGGCACAGCAGATCTTTAGTGGTGGTGGTGGTGGTGCTCCGATCTGTCCCTGTCGTCCAAGTT-3′, and E2J (1) (GenBank accession no. D90208) was amplified by using the sense primer 5′-GGAGCAGTCTTCGTTTCGCCCCATACCCGCGTGACGGGGGGG-3′ and the antisense primer 5′-AGGCACAGCAGATCTTTAGTGGTGGTGGTGGTGGTGCTCTGCTCTATCCCTGTCCTCCAA −3′, where the sequence encoding the C-terminal histidine tag is underlined. The tissue plasminogen activator (TPA) fragment was optimized to ensure optimal cleavage and was PCR amplified by using the sense primer 5′-CATGGGTCTTTTCTGCAGTCACCGTCCTTAGAT-3′ and antisense primer 5′-TCCCCCGGTGACGTGGGTTTCGGGCGAAACGAAGACTGCTCC-3′ for E2H, 5′-CCCCCCTGTCACGTGGGTATCGGGCGAAACGAAGACTGCTCC-3′ for E2BK, 5′-GGTGAGGCGGGCAGCGTGCCCCCCCGTTGTGAGGGTGTGGGGCGAAACGAAGACTGCTCC-3′ for E2N2, and 5′-CCCCCCCGTCACGCGGGTATGGGGCGAAACGAAGACTGCTCC-3′ for E2J. The E2 and TPA fragments were assembled by PCR, and the resulting products were purified with a QIAEX II gel extraction kit (catalog no. 20051; Qiagen), digested with BglII and PstI, and ligated into plasmid V1JnsTPA by using a rapid ligation kit (catalog no. 1635-379; Boehringer Mannheim). The resulting colonies were screened by PCR, and the positive ones were sequenced to confirm identity.
All of the mutants here described were constructed by oligomutagenesis.
Cell lines.
The Molt-4 cell line (human T-cell leukemia) and the 293 (human embryonic kidney) cell line were obtained from the American Type Culture Collection, as were the HuH7 and HepG2 human hepatoma cell lines. The HepG2-R2 subclone was obtained by infection of HepG2 cells with a recombinant retrovirus containing the human CD81 (hCD81) gene. The hCD81 gene was amplified by PCR and directly cloned into the pLIB retroviral vector (Clontech), giving rise to plasmid pLIB-hCD81. To produce the recombinant retroviral particles, pLIB-hCD81 was transfected into the PT67 amphotropic packaging cell line (Clontech). At 36 h after transfection, the supernatant was collected, filtered, and put over subconfluent HepG2 cells for 12 h. The medium was then replaced with fresh medium, and after 36 h cells were harvested and stained with fluorescein isothiocyanate (FITC)-conjugated anti-hCD81 MAb (Santa Cruz Biotechnology). The FITC-positive cells were analyzed and sorted with a FACS Vantage cell sorter (Becton Dickinson). The HepG2-R2 subclone was isolated by limiting dilution of hCD81-positive cells.
Production and characterization of E2 proteins.
E2 plasmids were transfected into 293 cells by using a calcium phosphate transfection kit (catalog no. 2-463335; 5-3 Prime). At 48 h after transfection, cell supernatant and crude cell extract were prepared as described below. Supernatants were cleared by centrifugation at 3,000 × g for 30 min at 4°C. They were then concentrated 20 times with Centricon Plus 80 filters (catalog no. UFC5LGC08; Amicon), supplemented with 5% glycerol, 25 mM HEPES, and 1× α-protease (catalog no. 1873580; Boehringer Mannheim) and stored in aliquots at −80°C. After removal of the supernatant, cells were washed once with cold TEN buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA) and then lysed with 1% Triton-TEN supplemented with 1× α-protease. After incubation for 30 min at 4°C, extracts were centrifuged at 9,000 rpm for 30 min at 4°C. The supernatant was recovered, supplemented with 10% glycerol and 0.05% NaN3, and stored in aliquots at −80°C. The expression of the recombinant proteins was assessed by enzyme-linked immunosorbent assay (ELISA) on Galanthus nivalis lectin (GNl) (catalog no. L8275; Sigma). Plates were coated with GNl diluted to 10 μg/ml in 1× phosphate-buffered saline (PBS) and incubated overnight at 4°C. Plates were washed with washing buffer (0.05% Tween 20, PBS), and nonspecific binding sites were saturated with bovine serum albumin (BSA) buffer (3% BSA, 0.05% Tween 20, and 0.05% NaN3 in 1× PBS). Serial dilution of either E2 cell extracts or E2 supernatants were added to the plates at a final concentration of 100 μl/well in BSA buffer and incubated for 2 h at room temperature (RT). After extensive washing, anti-His tag mouse MAb (catalog no. 34670; Qiagen) was diluted 1/400 in BSA buffer and added to the wells at a concentration of 100 μl/well. The plates were washed, incubated with secondary antibody (goat anti-mouse alkaline phosphatase conjugate) (catalog no. A7434; Sigma) diluted 1/2,000 in BSA buffer, and incubated for 1 h at RT. Plates were then washed, and alkaline phosphatase was revealed as described below.
Monomeric E2 was separated from aggregated forms by running serial dilutions of each protein preparation on a 10% acrylamide gel. Proteins were transferred onto nitrocellulose filters and, after blocking of nonspecific binding sites with blocking buffer (5% nonfat dry milk and 0.05% Tween 20 in 1× PBS) for 1 h at RT, revealed with anti-His tag mouse MAb diluted 1/100 in blocking buffer. After incubation overnight at 4°C, filters were washed six times for 5 min each with washing buffer and then incubated for 2 h at RT with goat anti-mouse peroxidase conjugate (catalog no. A8924; Sigma) diluted 1/1,000 in blocking buffer. The filters were washed as above, developed with SuperSignal West Pico chemiluminescent substrate (catalog no. 34080; Pierce), and exposed on Kodak films for a few minutes. The amount of monomer present in each preparation was evaluated by acquiring images with a Bio-Rad densitometer and analyzing them with Molecular Analyst software. The monomer content was evaluated with E2H as a reference.
Pull-down experiments.
Cells were harvested, washed in PBS, allowed to bind at RT for 1 h with E2 concentrated supernatant, washed twice with PBS, and lysed in PBS-1% Triton in the presence of protease inhibitor cocktail (Boehringer Mannheim) for 20 min at 37°. Lysates corresponding to 105 cells were loaded onto a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel to perform Western blotting for the detection of cell-bound E2. For detection of recombinant E2 protein of genotype 1a, the rat monoclonal antibody 6-1/a (13) was used diluted 1:50 in Tris-buffered saline-Triton X-100-5% nonfat dry milk, followed by incubation with anti-rat horseradish peroxidase (Dako) conjugate diluted 1:2,000. The chemiluminescent substrate Super Signal West Pico (Pierce) was used for detection, and immunoreactive proteins were visualized by exposure on X-ray film (Kodak Biomax ML).
Binding of E2 to recombinant GST-hCD81.
The hCD81 LEL was expressed as a fusion protein with the C terminus of glutathione S-transferase (GST-hCD81) and purified as previously described (27). The setting up and validation of an ELISA for detecting E2-GST-hCD81 interaction were previously reported (17, 27). For evaluation of the binding to E2, ELISA plates were coated with 1 μg of purified GST-hCD81 per well and, as a control, with 1 μg of GST per well, both diluted in PBS to 100 μl/well. GST fusion to the mouse CD81, which does not bind E2, was also used as negative control to validate the assay (27). The GST carrier protein was then used throughout this study as a negative control, since it behaved in the same way as recombinant mouse or African green monkey CD81 LEL expressed in Escherichia coli as fusions to the GST (27). After incubation overnight at 4°C, the plates were washed with washing buffer and unspecific binding sites were blocked by incubation for 1 h at 37°C with 300 μl of milk buffer (5% nonfat dry milk, 0.05% Tween 20, and 0.05% NaN3 in 1× PBS) per well. E2 supernatants containing equivalent amounts of monomer were diluted at 250 μl/well in milk buffer supplemented with GST at 5 μg/well and preincubated for 2 h at RT. The milk buffer was discarded from the plates, and E2 was added and incubated overnight at 4°C. The plates were extensively washed with washing buffer, and 100 μl of anti-His tag mouse MAb diluted 1/200 in milk buffer was added per well. After 3 h of incubation at 4°C, plates were extensively washed and incubated with 100 μl of goat anti-mouse alkaline phosphatase conjugate diluted 1/2000 in milk buffer per well. The plates were washed as described above, and alkaline phosphatase was revealed by adding 100 μl of a 1-mg/ml solution of p-nitrophenyl phosphate in ELISA substrate buffer (10% diethanolamine, 0.5 mM MgCl2 [pH 9.8]) per well. Results were expressed as the difference between the optical densities at 405 and 620 nm, measured with an automated ELISA reader (Labsystems Multiskan Bichromatic) after incubation in the dark at 37°C for 15, 30, 60, and 90 min.
Binding of E2 proteins to cell lines.
Binding of E2 to the cell surface was analyzed by using a fluorescence-activated cell sorting (FACS)-based assay. Cells were washed twice in PBS-1% fetal calf serum (FACS buffer), and then 2 × 105 cells were allowed to bind E2 concentrated supernatants at RT for 1 h. After one washing, an anti-His mouse MAb (Qiagen) was added at 2 μg/ml and left for 1 h at RT. Cells were washed again, and cell-bound MAb was revealed by an anti-mouse immunoglobulin G1-R-phycoerythrin conjugate (Serotec). E2 variants which bound very poorly to Molt-4 cells were detected by using an enzymatic amplification staining kit protocol (Flow-Amp) to increase the sensitivity of the FACS-based assay. Flow cytometry data acquisition was performed on a FACSCalibur (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson). Dead cells were detected by Sytox (Molecular Probes) staining and excluded from the analysis. Background fluorescence was measured by using a mock-transfected supernatant of HEK 293 cells and/or an isotypic mouse immunoglobulin G1 control. We did not observe significant differences in the background levels between different cell lines. This background was consistently low and was within the first log unit of fluorescence. Therefore, in analogy with previously published work, we used the background levels to set the threshold of positivity for each experiment by flow cytometric analysis of E2-cell binding and subtracted this value from each experimental datum (35). To determine the relative binding efficiency of each E2 protein analyzed in this study, a dose-response curve of the monomer species normalized to E2H monomer was performed. Data were plotted as relative monomeric doses against the median fluorescence intensity (MFI) with the background fluorescence subtracted (ΔMFI). The percent binding relative to that of E2H that is described for each E2 protein was derived from a best-fit analysis in the linear range of each curve and represents the ratio between the slope of each different E2 protein and the slope of E2H. For binding inhibition assays with the anti-CD81 MAb, Molt-4 cells were incubated in FACS buffer containing 20 μg of anti-CD81 clone 1.3.3.22 (Santa Cruz Biotechnology) per ml for 30 min at RT, washed twice in FACS buffer, and then incubated with E2 as described above.
RESULTS
Expression and characterization of recombinant E2 from 1a and 1b isolates.
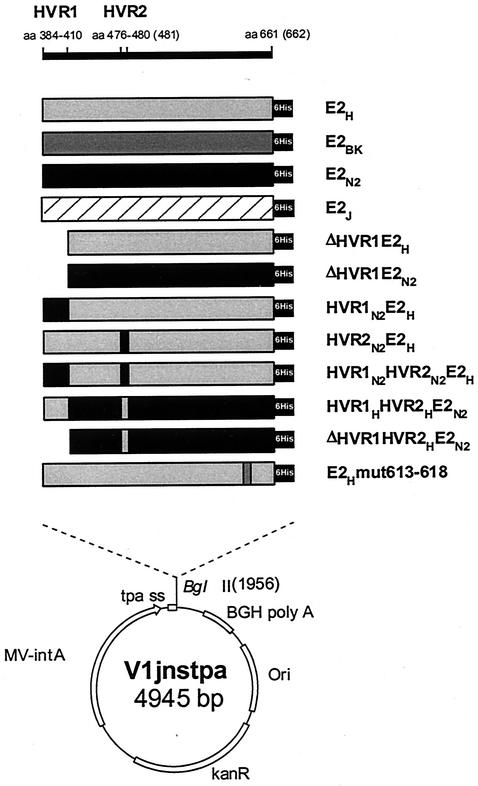
We previously reported that recombinant E2 from two different HCV strains (H and N2) belonging to the 1a and 1b subtypes bound bacterially expressed hCD81 LEL (GST-hCD81) with various efficiencies when expressed as C-terminal truncations at positions 683 and 684, respectively (44). To confirm and extend this observation, we generated expression plasmids encoding these and two additional E2 proteins of subtype 1b isolates (BK and J). The recombinant proteins were expressed as C-terminal truncations ending at position 661 for the H, BK, and J strains and at position 662 for the N2 strain variant, which contains an extra amino acid in HVR2 (Fig. 1). This choice was made on the basis of previous observations suggesting that this particular form of the E2 ectodomain has a higher tendency to fold in a correct conformation (28). In all cases the E2-coding sequence was cloned in frame downstream of the TPA to enforce secretion of the expression products, and a six-histidine tag was engineered at the C terminus to detect the various proteins with a single reagent (Fig. 2).
FIG. 1.
Sequence alignment of the H, BK, N2, and J strain E2 ectodomain sequences used in the binding assays. The two HVRs (HVR1 and HVR2) are indicated.
FIG. 2.
Expression plasmids encoding wt and mutant E2 proteins. Amino acid positions relative to the initial methionine of the HCV polyprotein are indicated at the top. The names of recombinant E2 proteins are indicated on the right. The position of the histidine tag (6His) is marked with a black box. The lower part of the figure shows the structure of the V1jnstpa vector. CMV-intA, cytomegalovirus promoter-intron A region; tpa ss, TPA signal sequence; BGH poly A: bovine growth hormone polyadenylation site; Ori, ColE1 origin of replication; kanR: kanamycin resistance gene.
Expression of recombinant proteins in transiently transfected 293 cells was assessed by GNl capture ELISA of whole-cell extracts and crude cell culture supernatants. All four proteins displayed similar expression levels, and significant amounts of recombinant products were secreted in the medium (between 30 and 50% of the total expression product [data not shown]). H and N2 strain E2 recombinants from both intracellular and secreted fractions reacted with conformation-sensitive MAbs (H33 and 166, respectively) in ELISA, confirming that a detectable proportion of these expression products is folded (data not shown). Recognition by MAb H33 is of particular relevance, since this antibody is specific for a conformation- and time-dependent epitope on non-disulfide-bridged E2 in complex with E1, which is believed to represent native prebudding forms of the HCV envelope (7, 30). We could not perform similar experiments with the BK and J proteins, since both H33 and 166 MAbs are strain specific (7, 44).
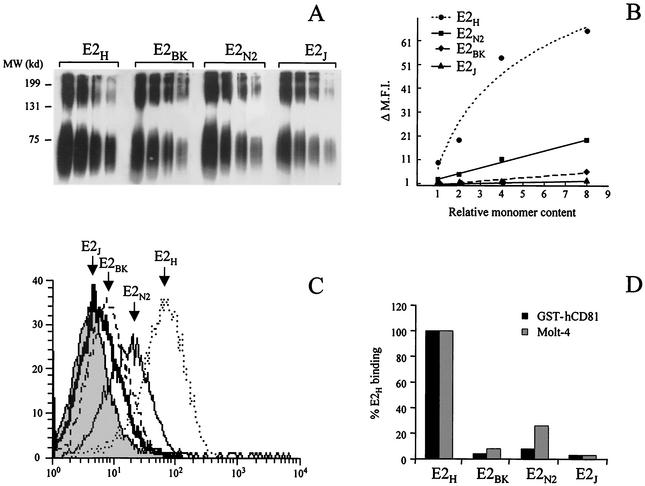
Consistent with the above observations, a significant amount of monomeric protein was detected in both the intracellular and the secreted fractions of all four expression products by nondenaturing SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (see Fig. 4A and data not shown). In general, the percentage of monomer in relation to the total amount of expression product was quite consistent between the four proteins and in different preparations of the same E2 variant (about 50% [data not shown]).
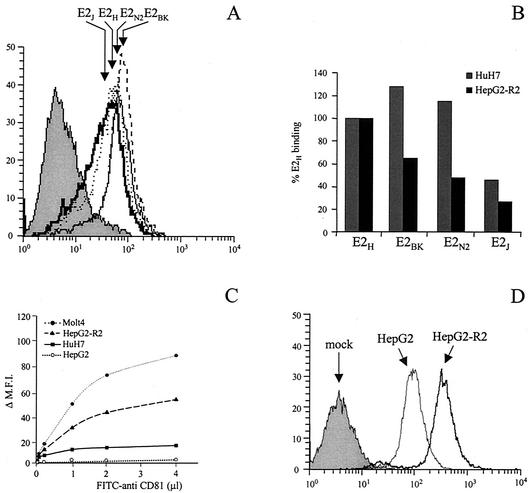
FIG. 4.
E2 binding to CD81 is strain specific. (A) Western blot analysis of serial dilutions of the secreted E2 proteins expressed from plasmids encoding E2 from different viral isolates (E2H, E2BK, E2N2, and E2J). In nonreducing SDS-PAGE, E2 aggregates are the slow-migrating species, while the faster-migrating band corresponds to the monomer and is heterogeneous in size due to glycosylation. MW, molecular mass markers. (B) Secreted E2 proteins were normalized for their content in monomeric form and tested in a dose-response FACS-based experiment for binding to Molt 4 cells. ΔMFI represents the MFI with the background fluorescence subtracted. The data show the results of a representative experiment performed in triplicate. (C) FACS data, plotted as histograms of fluorescence intensity against relative cell number, for the binding on Molt-4 cells of a normalized dose of E2 monomer from each preparation of E2 variants. E2H (dotted line), E2N2 (thin line), E2BK (dashed line), and E2J (thick line) histograms are indicated by arrows; the grey histogram represents the fluorescence intensity measured with a supernatant from mock-transfected cells. (D) Binding values for the different E2 variants are reported as percentage of E2H reactivity calculated as described in Materials and Methods. Binding to the bacterially expressed CD81 (GST-hCD81) was measured by ELISA, and the background signal observed on GST carrier protein was subtracted. Average values from two replicates were determined. Binding to Molt-4 cells was measured by FACS. The background signal measured in control reactions using equal amounts of cell culture supernatant from mock-transfected cells was subtracted from the MFI for each reaction.
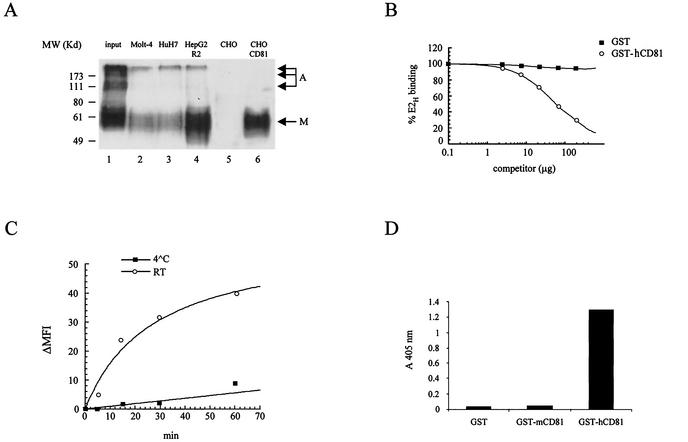
Only monomeric E2 is expected to fold in an active conformation and to bind CD81 (12). We confirmed this finding by pull-down experiments with CHO cells transfected with an expression vector encoding full-length hCD81. Mock-transfected CHO cells were used as negative control for this experiment. Soluble E2 secreted into the cell culture medium from transiently transfected 293 cells was incubated with the CD81-transfected CHO cells, and the bound material was then separated from the bulk supernatant by low-speed centrifugation and analyzed by nonreducing SDS-PAGE and Western blotting with the anti-E2 MAb 6/1a (12). As shown by the migration pattern of the recovered E2H, a significant enrichment of the monomeric form was obtained upon binding to native CD81 displayed on the surface of transfected cells, while no species reacting with the anti-E2 MAb was precipitated by mock-transfected cells (Fig. 3A, lanes 1, 5, and 6). We then performed pull-down experiments with Molt-4 cells, which display large amounts of CD81 expressed by the endogenous gene, and found essentially equivalent results (Fig. 3A, lane 2).
FIG. 3.
(A) Only monomeric E2 binds to CD81 or hepatic cell lines. Secreted E2H was used for binding and pull-down experiments with different cell lines. Cell-bound E2H was fractionated by SDS-PAGE under nonreducing conditions, and Western blot detection with anti-E2 rat MAb (61-a) followed by anti-rat horseradish peroxidase conjugate was performed. Lane 1, E2H from crude supernatant of transfected 293 cells (input); lanes 2, 3, and 4, E2H recovered after binding to Molt-4, HuH7, and HepG2-R2 cells, respectively; lanes 5 and 6, E2H recovered after pull-down with mock-transfected CHO and CD81-transfected CHO cells, respectively. Migration of monomeric (M) and aggregated (A) E2H species is indicated by arrows. Migration of molecular mass markers (MW) is shown on the left. (B) Soluble CD81 displaces E2H binding to Molt-4 cells. The extracellular domain of CD81 fused to the GST protein competes the binding of E2H to Molt-4 cells in a dose-dependent manner, while the GST protein alone has no inhibitory effect. (C) Temperature dependence of E2H binding to Molt-4 cells. (D) E2-specific binding to the extracellular domain of bacterially expressed hCD81. Binding of E2H to bacterially expressed human (GST-hCD81) and mouse (GST-mCD81) CD81 LEL fused to GST was measured by ELISA. One microgram of purified recombinant proteins was applied to the surfaces of microwell plates. Equal amounts of purified GST carrier protein were used as a negative control. Average values from two replicates are shown.
Detection of specific and saturable binding of E2 to Molt-4 cells by FACS was previously demonstrated (17, 35). We also determined the time and temperature dependence of this interaction. Binding of E2H to Molt4 cells reaches equilibrium after 40 min at RT. The kinetics of this interaction does not improve at 37°C, while E2H binds with a very slow kinetics at 4°C (Fig. 3C). To show that binding of E2 to Molt-4 cells is mediated by CD81, we performed competition assays with an anti-CD81 MAb (MAb 1.3.3.22). E2H recognition by Molt-4 cells was abolished by the anti-CD81 MAb in a dose-dependent manner but was not affected by an unrelated MAb (see Fig. 8B and data not shown). Finally, cell surface binding of E2H was efficiently competed out by an excess of purified GST-hCD81, with a 50% inhibitory concentration of about 1.5 μM (Fig. 3B). These results confirmed that Molt-4 cells specifically recognize E2 via CD81.
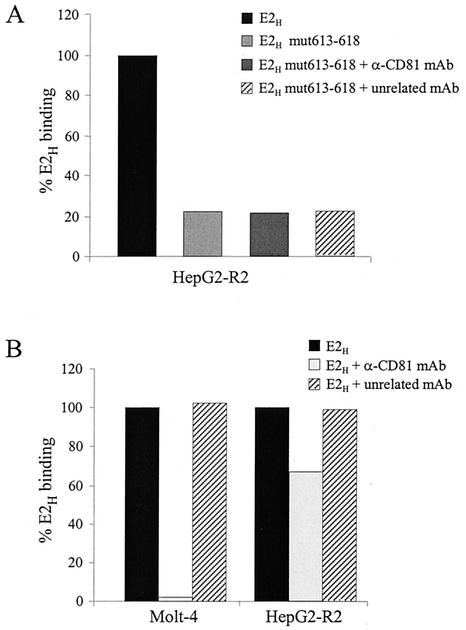
FIG. 8.
E2 can bind to hepatic cells in a CD81-independent manner. (A) Binding to HepG2-R2 cells of the complete E2H ectodomain and of mutant E2H mut613-618 in the absence of competing antibodies or in the presence of the anti-CD81 MAb 1.3.3.22 or an unrelated MAb. (B) Binding of E2H to Molt-4 or HepG2-R2 cells in the absence of competing antibodies or in the presence of the anti-CD81 MAb 1.3.3.22 or an unrelated MAb. Binding values are reported as the percentage of E2H reactivity and were calculated from dose-response curves of normalized amounts of the monomeric species of each protein, performed in duplicate.
CD81 binding of E2 is strain specific.
Based on the above observations, prior to testing the four E2 natural variants in CD81 binding assays, we determined the amount of monomeric protein present in either the intracellular or the secreted fraction of each expression product. This was done by running serial dilutions of all E2 preparations on nonreducing SDS-PAGE and by comparing the amount of monomeric forms after Western blotting and quantification by densitometric scanning of the autoradiographs. The E2H protein was used as a reference, and the normalized values of the other three proteins were expressed as relative monomer content. Only small variations in the contents of monomeric E2 were detected among different variants or in various preparations of the same protein (within a twofold difference) (Fig. 4A).
Binding of soluble E2H to bacterially expressed hCD81 LEL fused to GST (GST-hCD81) was measured by ELISA. As shown in Fig. 3D, E2H was able to recognize GST-hCD81, while no binding was observed with the corresponding recombinant fusion protein and the mouse CD81 sequence or the GST carrier protein. These data are in agreement with previous findings showing efficient and specific E2-GST-hCD81 binding (17, 27).
E2 expression products were normalized for the content in monomeric form as described in Materials and Methods (Fig. 4B). Serial dilutions of normalized protein preparations of all four E2 variants were then assayed by ELISA on microwell plates coated with purified GST-hCD81. All four E2 variants specifically interacted with the coated receptor but displayed a substantial difference in binding efficiency. In particular, the binding abilities of all three proteins of the 1b subtype were lower than that of the 1a protein (13- to 31-fold reduction) (Fig. 4D). In this assay, the binding efficiency of the secreted proteins was somewhat lower than that of the corresponding expression products from the intracellular fraction. This is probably due to the presence of modified glycans which reduce E2 recognition by CD81 (12). However, the relative binding potencies exhibited by the secreted forms of the four variants perfectly matched the reactivity pattern of the intracellular fractions (data not shown).
The four E2 variants were also tested for their ability to bind native CD81 displayed on the surface of Molt-4 cells by FACS analysis. Binding assays were performed by incubating serial dilutions of secreted E2 from expression products after normalizing them for the amount of monomeric species (Fig. 4B). Also in this case, E2H successfully bound Molt-4 cells more efficiently than the 1b variants (Fig. 4C and D). E2N2 displayed a relative binding efficiency on Molt-4 cells higher than that measured by ELISA on purified GST-hCD81 (a fourfold reduction compared to E2H); however, the overall binding profile for the four E2 isolates was unchanged (E2H > E2N2 > E2BK > E2J). Thus, the hierarchy of binding efficiency to CD81 displayed by E2 proteins from different viral isolates does not depend on their glycosylation state or on the CD81 expression system and reflects instead an intrinsic property of the E2 proteins.
Deletion of HVR1 leads to increased CD81 binding of E2.
The different CD81 binding efficiencies displayed by the four E2 variants suggested that segments containing variations in amino acid sequence between the four strains should be responsible for the observed pattern. We therefore generated expression plasmids encoding HVR1 deletion mutants of E2H and E2N2 by directly fusing the TPA leader sequence to residue Ile411 (Fig. 2). The ability of pV1jnstpaΔHVR1E2 plasmids to drive expression of the encoded mutant proteins was assayed by transient transfection of 293 cells. GNl capture ELISA using whole-cell extract and culture medium from transfected cells indicated that significant amounts of the two mutants were expressed and secreted (reference 45 and data not shown). HVR1 deletion did not affect the aggregation state of the recombinant products, since similar amounts of monomeric species were detected for the complete ectodomain (wild type [wt]) and the mutant proteins by nonreducing SDS-PAGE and Western blot analysis. Furthermore, both ΔHVR1E2H and ΔHVR1E2N2 mutants were recognized by the conformation-sensitive MAbs H33 and 166, respectively, with an efficiency comparable to that observed for the wt proteins (data not shown). Taken together, these data indicate that the folding of HVR1 deletion recombinants is very similar to that of the wt ectodomain.
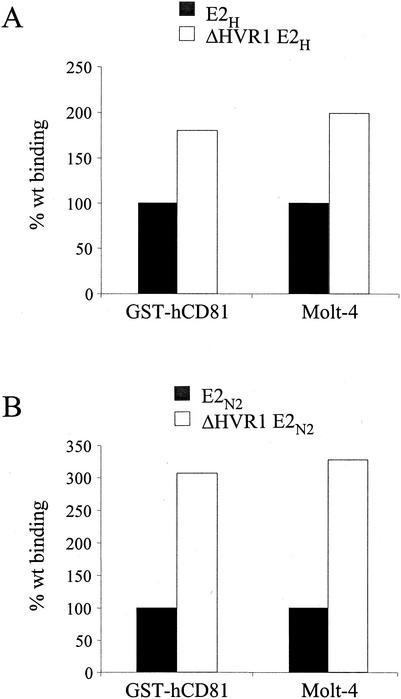
Cell culture supernatants from 293 transfected cells were again normalized for the amount of monomeric products, and serial dilutions of ΔHVR1E2 mutant preparations were tested for binding to Molt-4 cells by FACS. Both mutants showed increased binding activity on Molt-4 cells with respect to the parental proteins (Fig. 5), which was higher for the HVR1 deletion protein from the N2 strain (threefold) than for the corresponding H strain mutant (twofold). Similar results were obtained when binding experiments were performed with bacterially expressed GST-hCD81 (three- and twofold increases for the N2 and H mutants, respectively) (Fig. 5).
FIG. 5.
Deletion of the HVR1 region improves efficiency of binding of E2 to CD81. Secreted recombinant proteins expressed by plasmids encoding the complete E2 ectodomains (black bars) or E2 with HVR1 deleted (white bars) from the H strain (A) or N2 strain (B) were tested for their ability to bind to bacterially expressed CD81 (GST-hCD81) by ELISA or to Molt-4 cells by FACS. Binding values are reported as the percentage of reactivity of the complete ectodomain and were calculated from dose-response curves of normalized amounts of the monomeric species of each protein, performed in duplicate.
It is possible that HVR1 exerts a negative effect on E2-CD81 binding by masking the E2 region responsible for CD81 recognition. In agreement with this hypothesis, an anti-HVR1 MAb (MAb 9/27) (12) directed against the C-terminal half of the HVR (residues 395 to 408) was able to inhibit E2H binding to CD81 with a 50% inhibitory concentration of about 10 mM (data not shown).
The HVR1 and HVR2 regions of the N2 strain negatively modulate E2-CD81 interaction in a cooperative fashion.
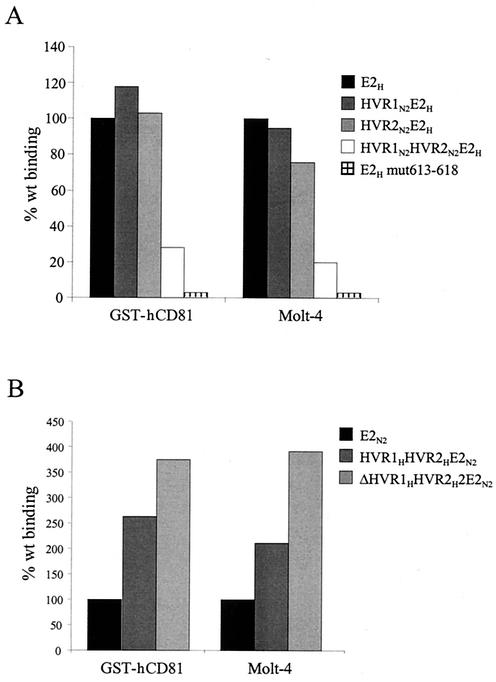
The role of the HVRs was further investigated by generating swapping mutants between the H and N2 isolates. No change in the efficiency of binding to GST-hCD81 or Molt-4 cells was observed by replacing only the HVR1 region of E2H with the corresponding sequence of the N2 strain (mutant HVR1N2E2H) (Fig. 6A). Similarly, replacement of only the E2H HVR2 region with that of the 1b isolate did not affect CD81 recognition (HVR2N2E2H mutant). However, a double E2H mutant containing both HVRs from the N2 isolate resulted in a fourfold reduction in CD81 binding (mutant HVR1N2HVR2N2E2H) (Fig. 6A).
FIG. 6.
Both the HVR1 and HVR2 regions of the N2 strain are required for maximal inhibition of E2-CD81 interaction, and mutation of residues 613 to 618 eliminates CD81 recognition. Complete E2 ectodomains and HVR-mutated E2 were tested for binding to bacterially expressed CD81 (GST-hCD81) by ELISA or to Molt-4 cells by FACS. (A) Reactivities of the E2H complete ectodomain and the HVR1N2E2H, HVR2N2E2H, HVR1N2HVR2N2E2H, and E2H smut613-618 mutants. (B) Binding of the E2N2 complete ectodomain and the HVR1HHVR2HE2N2 and ΔHVR1HVR2HE2N2 mutants. Binding values are reported as the percentage of reactivity of the complete ectodomain and were calculated from dose-response curves of normalized amounts of the monomeric species of each protein, performed in duplicate.
A different phenotype was displayed by the inverse mutation: replacement of both the HVR1 and HVR2 regions of the N2 protein with the corresponding HVRs of the H variant did not inhibit CD81 binding but rather improved receptor recognition (mutant HVR1HHVR2HE2N2) (Fig. 6B). An even better binding was displayed when the HVR1 region was deleted from the latter derivative, consistent with the hypothesis of a generally negative effect mediated by the HVR1 region (mutant ΔHVR1HVR2HE2N2) (Fig. 6B).
Thus, it appears that in both the 1a and 1b proteins HVR1 negatively affects E2-CD81 interaction but that the concomitant presence of the two HVRs from the N2 isolate is required to achieve maximal inhibition.
Identification of an E2 sequence required for CD81 recognition.
Our results are consistent with the hypothesis that the two HVRs can modulate access to a primary CD81 binding site. To identify this element, we took advantage of an E2 model recently generated by a combination of secondary-structure prediction and fold recognition methods (44). Based on this model, three protein segments were postulated to represent putative CD81 binding sites: (i) a region spanning residues 474 to 494 and including HVR2, (ii) the fragment between positions 522 and 551, and (iii) a short stretch of eight amino acids from residue 612 to 620. The last fragment appears to be spatially close to HVR2, and it is highly conserved across HCV isolates (data not shown). Therefore, we generated a substitution mutant with mutations of the six residues from position 613 to 618 (YRLWHY to SAASAS). The resulting protein variant (E2H mut613-618) showed an expression profile and monomer content similar to those of the wt protein but was no longer able to bind hCD81 either as a recombinant GST fusion or on the surface of Molt-4 cells (Fig. 6A).
The profile of E2 binding to human hepatic cells is different from that observed on Molt-4 cells.
Since HCV is a hepatotropic virus, we extended our studies to human cells of hepatic origin. Human hepatoma HuH7 cells display a highly differentiated phenotype and are the only cell line shown to support replication of subgenomic HCV replicons to date (3). They also express CD81 and, therefore, were chosen for E2 binding assays. Pull-down experiments confirmed that only the monomeric fraction of E2 can bind to HuH7 cells (Fig. 3A, lane 3). The same E2 preparations from different isolates previously tested for their binding to GST-hCD81 and Molt-4 cells were used for FACS-based quantitative assays of binding to HuH7 cells. On these cells, the subtype 1b proteins showed a binding efficiency relative to that of the 1a variant that was significantly higher than that measured on Molt-4 cells (Fig. 7A and B). To confirm and extend this observation, we also performed binding assays with human hepatoma HepG2 cells. Since this cell line expresses only low levels of or no CD81 (Fig. 7C), we generated stably CD81-transformed HepG2 cells and selected a population of cells with high levels of CD81 display by FACS. Several individual clones were isolated by limiting dilution and characterized for CD81 expression and E2 binding. A clone (HepG2-R2) with a CD81 surface display comparable to that measured on Molt-4 cells was chosen for further studies (Fig. 7C).
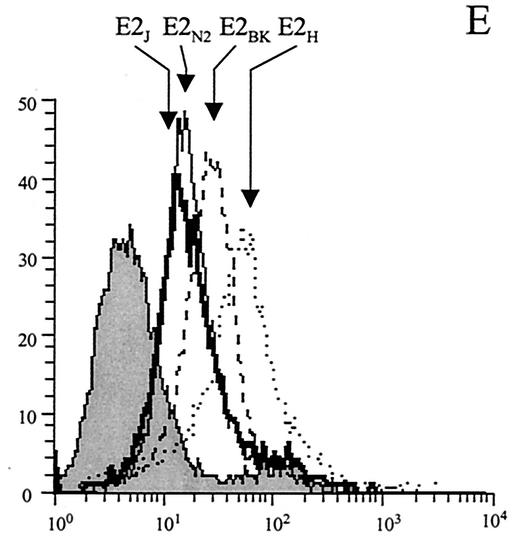
FIG. 7.
Human hepatic cells recognize E2 variants from different viral isolates with comparable efficiency. (A and E) FACS histograms of binding on HuH7 cells (A) and HepG2-R2 cells (E) of a normalized dose of E2 monomer from each preparation of E2 variants. E2H (dotted lines), E2N2 (thin lines) E2BK (dashed lines), and E2J (thick lines) histograms are indicated by arrows; the grey histograms represent the fluorescence intensity measured with a supernatant from mock-transfected cells. (B) Binding efficiencies of E2 proteins from different viral isolates (E2H, E2BK, E2 N2, and E2J) on HuH7 cells or HepG2-R2 cells. Binding values are reported as the percentage of reactivity of the E2H protein and were calculated from dose-response curves of normalized amounts of the monomeric species of each protein, performed in duplicate. (C) Cell surface expression of CD81 on the different cell lines measured by FACS analysis with an FITC-conjugated anti-CD81 MAb 1.3.3.22 (Santa Cruz) in a direct binding assay. (D) FACS histograms of the binding of the E2H protein on HepG2 (thin line) and HepG2-R2 (thick line) cells. The grey histogram represents the fluorescence intensity measured using a supernatant from mock-transfected cells.
Once again, only monomeric E2 was able to bind HepG2-R2 cells in pull-down experiments (Fig. 3A, lane 4). The amount of E2 recovered after binding to HepG2-R2 cells was higher than that observed with Molt-4 or HuH7 cells. This may reflect an intrinsically higher binding efficiency of HepG2-R2 due to E2 recognition of another E2 receptor present on HepG2 cells (the scavenger receptor type B class I) which is able to interact with soluble E2 in a CD81-independent manner (Fig. 7D) (36).
In this case also the three E2 proteins from 1b isolates showed significant binding efficiencies of between 65 and 30% of that of the E2H variant (Fig. 7B and E). Thus, while E2 interaction with CD81 appears to be an isolate-dependent phenomenon, E2 binding to cells of hepatic origin is less prone to sequence variability.
Furthermore, an E2 interaction surface distinct from that involved in CD81 recognition must contribute to the binding to hepatic cells, since mutation of residues 613 to 618, which abolished CD81 binding, did not completely impair binding of the E2H mut613-618 derivative to HepG2-R2 cells (Fig. 8A). This finding also confirms that on hepatic cells, a molecule different from CD81 is responsible for binding the E2H mut613-618. Consistently with this hypothesis, recognition of E2H mut613-618 by HepG2-R2 cells was not affected by the anti-CD81 MAb which blocks the interaction of E2H with Molt-4 cells (MAb 1.3.3.22) (Fig. 8A). To prove that such CD81-independent binding is not a specific property of the mutant protein but also contributes to the recognition of wt E2H by hepatic cells, we performed binding inhibition experiments with the anti-CD81 MAb 1.3.3.22 on HepG2-R2 cells. These results confirmed that HepG2-R2 can recognize E2H via at least two independent molecules, since only 40% of the binding was eliminated by preincubating cells with saturating amounts of MAb 1.3.3.22, while no change was observed with an unrelated isotypic MAb as a negative control (Fig. 8B and data not shown). In contrast, E2 recognition by Molt-4 cells was completely abolished by using the same amount of MAb 1.3.3.22 (Fig. 8B).
DISCUSSION
A major stumbling block in understanding the HCV infection mechanism is the lack of an efficient cell culture system to study viral attachment and entry. An alternative approach toward this end is the identification and characterization of interactions between viral and host cell components. The recent discovery that the human cell surface CD81 molecule is a putative HCV receptor component and devising of in vitro and ex vivo assays to measure interaction between CD81 and recombinant forms of the HCV E2 glycoprotein constitute a step forward in resolving this issue. Information on the E2 determinants responsible for CD81 interaction is of paramount importance in deciphering the rules that regulate viral attachment to target cells and elaborating effective strategies for prevention and therapy. As a matter of fact, antibodies capable of blocking HCV E2 interaction with CD81 (NOB antibodies) are elicited during natural or experimental infection as well as by vaccination with a recombinant E1/E2 complex (18, 32). However, NOB antibodies are difficult to induce, and the only immunogens that have been shown to elicit such a response are recombinant E1/E2 complexes produced in mammalian cells or plasmid DNA encoding E2 (14, 32). Production of pure, correctly folded E1/E2 heterodimers in sufficient amounts to immunize large numbers of individuals presents a difficult task, and while DNA vaccination is effective in small mammals, it resulted in significant paucity of immunization in larger animals or humans. Thus, the design of novel immunogens able to induce a neutralizing response is highly desirable but requires additional knowledge on the structural and functional properties of the HCV envelope proteins and the specific sites for interaction with CD81 or novel receptor components. This situation is rendered even more complicated by the fact that HCV is not a single virus but is a complex mixture of variants with significant sequence heterogeneity in protein regions deemed important for virus infection.
We previously approached the problem of identifying putative E2 regions for CD81 interaction by generating a theoretical model of its tertiary structure (44). This exercise led to three distinct protein fragments being identified as potentially important for interaction with CD81. In the present work we have generated a set of E2 substitution mutants with mutations in two of these regions and tested them for their ability to recognize isolated hCD81 as a recombinant GST fusion expressed in E. coli or in its native form as displayed on the surface of Molt-4 cells.
For our binding assays, we chose to express E2 as C-terminal truncations in mammalian cells, since soluble and correctly folded E2 can be obtained by deleting the transmembrane domain as previously reported (12, 13, 28). However, a key issue in this type of analysis is the content of active species, which can vary from one protein to another and between different preparations of the same expression product. In fact, large aggregates of cysteine-bridged products arising from misfolded E2 are generated in transfected cells, and it has been reported that the only form capable of binding to CD81 is monomeric E2 (12, 28). We confirmed this observation by pull-down experiments using paired CHO cell lines with or without hCD81, Molt-4 cells, or hepatic cells and subsequent analysis of the cell-bound fraction by nonreducing SDS-PAGE. Therefore, to allow for quantitative measurements of binding, all assays throughout this work were performed on protein preparations normalized for their content in monomeric form.
Since the E2 glycoprotein is one of the most variable HCV proteins, to increase the general relevance of our observations, we chose four different HCV isolates belonging to the most common 1a and 1b subtypes (strains H, BK, N2, and J) and examined their ability to interact with CD81. In line with our earlier observations (44), all recombinant E2 proteins from the latter three isolates were markedly less efficient than the H strain E2 in binding to bacterially expressed CD81 and to the native protein displayed on Molt-4 cells. This finding led us to hypothesize that protein fragments corresponding to less conserved E2 segments might contribute to CD81 recognition and could be responsible for the different binding levels of the four E2 variants. We focused our attention on the E2 HVRs for three reasons. First, they display the most variable sequences in the whole protein. Second, MAbs against either of the two HVRs can inhibit E2 binding to CD81, and virus recognition by Molt-4 cells is affected by anti-HVR1 MAbs (25, 35, 44). Finally, HVR2 lies in a protein segment predicted to be exposed on the surface of the virus (44).
We discovered that HVR1 deletion mutants from both the H and N2 strains increased binding efficiency. This finding together with the capability of anti-HVR1 MAbs to inhibit CD81 recognition suggested that HVR1 is not directly involved in receptor interaction but can negatively affect binding, either by steric hindrance or by blocking E2 in an unfavorable conformation. Replacement of both E2H HVRs with the corresponding sequences from the E2N2 variant generated a poor CD81 binder, whose efficiency of recognition by Molt-4 cells was comparable to that of the E2N2 protein. However, neither of the two E2N2 HVRs was able by itself to confer this phenotype. Thus, it appears that, at least in our experimental system, a particular combination of HVR1 and HVR2, such as that of the N2 strain, is required to achieve maximum inhibition. In line with this observation, in the E2N2 protein a double substitution mutant with the HVRs of the H strain protein led to an increase in binding. It should be noted that also in this chimeric protein, deletion of HVR1 improved binding, further confirming this protein region as a negative modulator.
One hypothesis to explain these results is that intramolecular interactions between the two HVRs occurs, and due to an intrinsic flexibility, the E2 protein shifts from an open to a closed conformation, with the former being more competent for CD81 recognition. Like many other viral envelope proteins, E2 also probably undergoes conformational changes during cell entry, envelope assembly, and disassembly. These changes could be blocked either by antibody binding or by intramolecular interaction between the two HVRs. According to this hypothesis, recent data indicate that the conformation of E2 changes upon binding to CD81, irrespective of the molecular context: as a soluble ectodomain, as a full-length E1/E2 complex, or as virus-like particles (16, 29). Furthermore, binding of nonneutralizing Fabs to E2 can cause conformational changes that result in reduced susceptibility to Fab-mediated NOB activity (4).
According to the previously reported E2 model, the HVR2 is located close to aa 613 to 620 (44), one of the regions proposed to be involved in the interaction with CD81. Replacement of this segment with an unrelated sequence (E2H mut613-618) led to a complete loss of CD81 interaction, while nonetheless maintaining the capacity to bind to HepG2-R2 cells. Within a second E2 region predicted to contribute to CD81 recognition (aa 522 to 551) (44), the protein segment from residue 524 to 531 has been identified as another potential site of receptor interaction (29). This study also detected a further element, residues 412 to 423, where antibody binding inhibits E2-CD81 interaction (29). The E2 protein was modeled on the structure of tick-borne encephalitis virus E envelope protein (34) on the basis of the combined results of secondary-structure prediction and threading and mapping methods (44). This model was subsequently indirectly validated by elucidating the structure of the E1 fusion glycoprotein of Semliki Forest virus (SFV) (23), since that structure was found to be remarkably similar to that of the tick-borne encephalitis virus E protein. In the HCV E2 model, HVR1 and the region spanning residues 522 to 551 are located on the opposite side of an elongated molecule with respect to the HVR2 and the region from aa 613 to 618. They could form a single binding site only if a head-to-tail dimer is postulated, and this was indeed one of the conclusions derived from our model (44), which was once again indirectly supported by the SFV E1 structure. In the present work we have shown that the E2 form capable of recognizing CD81 (or alternative receptors) migrates as a monomer in nonreducing SDS-PAGE. This is not in contradiction with the head-to-tail dimer hypothesis, and we cannot exclude the possibility that such dimers would indeed be present in solution, but their association is not mediated by disulfide bridges. Since the methods used to build the model are not as reliable as those based on detectable sequence homology, alternative hypotheses should also be considered. For example, it is in principle possible that the HCV E2 protein has two modes of binding to CD81 or that E2 is so flexible that it allows the protein to assume a completely different organization upon binding to CD81, which is not unlikely in light of the observation that a similar mechanism has to be invoked to reconcile the cryo-EM and X-ray crystallography data on SFV (23). Only further experiments will allow us to discriminate between these hypotheses, but in any case the HCV E2 model has been instrumental in designing the experiments described here and has thus fulfilled its purpose.
The mode of interaction of E2 with human hepatoma cells must be significantly different from that occurring on Molt-4 cells, since the profile of binding of E2 from the H, BK, N2, and J strains to these cell lines is remarkably different. This is unlikely to be due to differential glycosylation. In fact, hCD81 has only one potential Asn glycosylation motif; however, it resides at the very end of the intracellular C-terminal end of the protein outside of the LEL domain. Since no alternative CD81 isoform has been described to date, we do not believe that the different patterns of recognition for Molt-4 and liver-derived cell lines would result from variations in the coding sequence.
A more plausible explanation for this phenomenon could be the presence on the surface of hepatic cells of another molecule, distinct from CD81, which is capable of interacting with HCV E2. In line with this premise, binding of E2H to HepG2-R2 cells was reduced, but not completely eliminated, by a blocking anti-CD81 MAb, while binding inhibition experiments with the same MAb on Molt-4 confirmed that E2 recognition by these cells is solely due to the surface-displayed tetraspanin. Additional evidence supporting the existence of a putative HCV coreceptor is provided by the E2H mut613-618 derivative, which is unable to bind Molt-4 cells or isolated recombinant CD81 but still interacts with hepatoma cells, albeit less efficiently than the wt protein. The ability of this mutant to interact with HepG2-R2 cells was not affected by the blocking anti-CD81 MAb, confirming that E2 elements located outside the CD81 interaction surface contribute to recognition of hepatic cells, which appears to be, at least in part, CD81 independent. Finally, soluble E2 bound to HepG2 cells in spite of the fact that these cells do not display detectable amounts of CD81. During revision of this paper we were able to identify the scavenger receptor type B class I as the molecule displayed on the surface of hepatic cell lines which is able to interact with soluble E2 independently from CD81 (36). Thus, it appears that in HepG2-R2 cells as well as in liver cells that express both CD81 and scavenger receptor type B class I, binding of E2 is not restricted to a particular variant, while differential binding of E2 variants mediated by CD81 is apparently a lymphocyte-specific phenomenon and hence may have little relevance to the process of liver cell infection. However, the engagement of CD81 by E2 on the surface of T or NK cells has been recently shown to provide different signals which potentially lead to autoimmunity or immune evasion, respectively (6, 39, 41). Thus, even if no direct correlation between E2-CD81 binding efficiency and the outcome of the disease is presently available, our findings provide important information for a better understanding of the role played by HCV attachment to nonhepatic cells in the modulation of innate or adaptive immune responses.
Acknowledgments
We are grateful to Jean Dubuisson and Shoshana Levy for providing the conformation-sensitive MAb H33 and the hCD81 LEL expression plasmid. We also thank Janet Clench for critical reading of the manuscript.
Part of this work was supported by a grant from the European Community to the ENHCV Consortium.
REFERENCES
- 1.Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621-627. [DOI] [PubMed] [Google Scholar]
- 2.Allander, T., K. Drakenberg, A. Beyene, D. Rosa, S. Abrignani, M. Houghton, A. Widell, L. Grillner, and M. A. Persson. 2000. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J. Gen. Virol. 10:2451-2459. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R. 2002. In vitro models for hepatitis C. Virus Res. 82:25-32. [DOI] [PubMed] [Google Scholar]
- 4.Burioni, R., F. Bugli, N. Mancini, D. Rosa, C. Di Campli, G. Moroncini, A. Manzin, S. Abrignani, P. E. Varaldo, M. Clementi, and G. Fadda. 2001. Nonneutralizing human antibody fragments against hepatitis C virus E2 glycoprotein modulate neutralization of binding activity of human recombinant Fabs. Virology 288:29-35. [DOI] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, et al. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA 91:1294-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly, J. J., D. Martinez, K. U. Jansen, R. W. Ellis, D. L. Montgomery, and M. A. Liu. 1996. Protection against papillomavirus with a polynucleotide vaccine. J. Infect. Dis. 173:314-320. [DOI] [PubMed] [Google Scholar]
- 9.Everhart, J. E., M. Stolar, and J. H. Hoofnagle. 1997. Management of hepatitis C: a national survey of gastroenterologists and hepatologists. Hepatology 26:78S-82S. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P., H. J. Alter, D. C. Wong, R. H. Miller, S. Govindarajan, R. Engle, M. Shapiro, and R. H. Purcell. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. USA 16:7792-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farci, P. A., D. Shimoda, T. Wong, D. Cabezon, A. De Gioannis, A. Strazzera, Y. Shimizu, et al. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 96:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournillier, A., E. Depla, P. Karayiannis, O. Vidalin, G. Maertens, C. Trepo, and G. Inchauspe. 1999. Expression of noncovalent hepatitis C virus envelope E1-E2 complexes is not required for the induction of antibodies with neutralizing properties following DNA immunization. J. Virol. 73:7497-7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fried, M. W., and J. H. Hoofnagle. 1995. Therapy of hepatitis C. Semin. Liver Dis. 15:82-91. [DOI] [PubMed] [Google Scholar]
- 16.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii, K., D. Rosa, Y. Watanabe, T. Katayama, H. Harada, C. Wyatt, K. Kiyosawa, H. Aizaki, Y. Matsuura, M. Houghton, S. Abrignani, and T. Miyamura. 1998. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology 28:1117-1120. [DOI] [PubMed] [Google Scholar]
- 19.Jaeckel, E., M. Cornberg, H. Wedemeyer, T. Santantonio, J. Mayer, M. Zankel, G. Pastore, M. Dietrich, C. Trautwein, M. P. Manns, et al. 2001. Treatment of acute hepatitis C with interferon alfa-2b. N. Engl. J. Med. 345:1452-1457. [DOI] [PubMed] [Google Scholar]
- 20.Kato, N., H. Sekiya, Y. Ootsuyama, T. Nakazawa, M. Hijikata, S. Ohkoshi, and K. Shimotohno. 1993. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J. Virol. 67:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitadokoro, K., D. Bordo, G. Galli, R. Petracca, F. Falugi, S. Abrignani, G. Grandi, and M. Bolognesi. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima, M., T. Osuga, F. Tsuda, T. Tanaka, and H. Okamoto. 1994. Influence of antibodies to the hypervariable region of E2/NS1 glycoprotein on the selective replication of hepatitis C virus in chimpanzees. Virology 204:665-672. [DOI] [PubMed] [Google Scholar]
- 23.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 24.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 25.Li, C., D. Candotti, and J. P. Allain. 2001. Production and characterization of monoclonal antibodies specific for a conserved epitope within hepatitis C virus hypervariable region 1. J. Virol. 75:12412-12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 27.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 29.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 30.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 12:2873-2883. [DOI] [PubMed] [Google Scholar]
- 31.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to human CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 33.Puntoriero, G., A. Meola, A. Lahm, S. Zucchelli, B. Bruni Ercole, R. Tafi, M. Pezzanera, M. U. Mondelli, R. Cortese, A. Tramontano, G. Galfrè, and A. Nicosia. 1998. Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 17:3521-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 35.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarselli, E., H. Ansuini, R. Cerino, R. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu, Y. K., M. Hijikata, A. Iwamoto, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 65:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobolev, B. N., V. V. Poroikov, L. V. Olenina, E. F. Kolesanova, and A. L. Archakov. 2000. Comparative analysis of amino acid sequences from envelope proteins isolated from different hepatitis C virus variants: possible role of conservative and variable regions. J. Viral Hepatol. 7:368-374. [DOI] [PubMed] [Google Scholar]
- 39.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Doorn, L. J., I. Capriles, G. Maertens, R. DeLeys, K. Murray, T. Kos, H. Schellekens, and W. Quint. 1995. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J. Virol. 69:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, and S. Abrignani. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur J. Immunol. 31:166-175. [DOI] [PubMed] [Google Scholar]
- 42.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 43.Weiner, A. J., H. M. Geysen, C. Christopherson, J. E. Hall, T. J. Mason, G. Saracco, F. Bonino, et al. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. USA 89:3468-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. Bruni Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]
- 45.Zucchelli, S., R. Roccasecca, A. Meola, B. Bruni Ercole, R. Tafi, J. Dubuisson, G. Galfrè, R. Cortese, and A. Nicosia. 2001. Mimotopes of the hepatitis C virus hypervariable region 1, but not the natural sequences, induce cross-reactive antibody response by genetic immunization. Hepatology 33:692-703. [DOI] [PubMed] [Google Scholar]