Abstract
Photosystem II (PSII) is a multisubunit membrane protein complex performing light-induced electron transfer and water-splitting reactions, leading to the formation of molecular oxygen. The first crystal structure of PSII from a thermophilic cyanobacterium Thermosynechococcus elongatus was reported recently [Zouni, A., Witt, H. T., Kern, J., Fromme, P., Krauss, N., Saenger, W. & Orth, P. (2001) Nature 409, 739–743)] at 3.8-Å resolution. To analyze the PSII structure in more detail, we have obtained the crystal structure of PSII from another thermophilic cyanobacterium, Thermosynechococcus vulcanus, at 3.7-Å resolution. The present structure was built on the basis of the sequences of PSII large subunits D1, D2, CP47, and CP43; extrinsic 33- and 12-kDa proteins and cytochrome c550; and several low molecular mass subunits, among which the structure of the 12-kDa protein was not reported previously. This yielded much information concerning the molecular interactions within this large protein complex. We also show the arrangement of chlorophylls and cofactors, including two β-carotenes recently identified in a region close to the reaction center, which provided important clues to the secondary electron transfer pathways around the reaction center. Furthermore, possible ligands for the Mn-cluster were determined. In particular, the C terminus of D1 polypeptide was shown to be connected to the Mn cluster directly. The structural information obtained here provides important insights into the mechanism of PSII reactions.
The photosystem II (PSII) complex is a multisubunit membrane–protein complex consisting of >14 membrane-spanning subunits, 3 hydrophilic peripheral subunits, and >40 cofactors, including chlorophylls (chls), carotenoids, Mn, Fe, and plastoquinones, with a total molecular mass of 320 kDa for a monomer (for review, see refs. 1–3). The PSII reaction center (RC) P680 is a (pair of) chl a coordinated by RC D1-His-198 (and D2–197) and undergoes charge separation on absorption of light energy; the electrons thus generated are transferred to pheophytin, the first quinone acceptor QA, and then the second quinone acceptor QB. At the oxidizing side of PSII, a redox-active tyrosine residue D1-Tyr-161 (TyrZ) reduces P680. The oxidized TyrZ withdraws electrons from a Mn cluster consisting of four Mn atoms, which in turn withdraws electrons in a sequential way from water molecules, leading to the splitting of water and formation of molecular oxygen. This reaction provides us with the oxygen-rich atmospheric environment suitable for most of the organisms to live on the earth, whereas the protons yielded are the source for proton gradient across the thylakoid membrane required for ATP formation.
In view of its importance, PSII has received extensive studies in the past several decades regarding its protein composition, function, and dynamic regulation. As a result, our understanding of the function and reaction mechanisms of PSII has increased greatly. However, studies of the structure of PSII have appeared only in recent years. The 3D image of PSII was first studied by electron cryomicroscopy of 2D crystals of various PSII particles, and a structure at 8-Å resolution has been reported for a CP47-RC-PSII complex, which lacked oxygen-evolving activity, and its related three extrinsic proteins (4, 5). Recently, 3D crystals have been obtained for oxygen-evolving PSII complexes from two species of thermophilic cyanobacteria (6–8), and the PSII crystal structure from one of such cyanobacterium, Thermosynechococcus elongatus (formerly Synechococcus elongatus), was reported at 3.8-Å resolution by Zouni et al. (9). The reported structure was constructed as a Cα model consisting of 36 transmembrane helices (TMHs), 22 of which were assigned to the PSII large-transmembrane subunits CP47, CP43, D1, and D2. The remaining helices were assigned to low molecular mass subunits, including the α and β subunits of cytochrome (cyt) b559. In addition, two of the three extrinsic proteins, the 33-kDa protein and cyt c550, were partially assigned, and the shape and location of the Mn cluster, together with the arrangement of other cofactors, were shown. This first crystal structure of PSII provided an important basis for further studies; however, due to the limited resolution, many details need to be studied further. We report here the crystal structure of PSII from another thermophilic cyanobacterium, Thermosynechococcus vulcanus, at 3.7-Å resolution. We show the structure of major large PSII subunits, including the 12-kDa extrinsic protein, which was not identified in the previous structure; these large subunits were traced as real residues or alanines on the basis of their gene-derived amino acid sequences. We also show the structure around the Mn cluster, which provided information on ligands for the Mn cluster, and the arrangement of pigments and cofactors, including two β-carotenes (Cars) recently identified.
Materials and Methods
PSII dimers highly active in oxygen evolution were isolated (10, 11) and crystallized as in ref. 8. Diffraction experiments were performed at beamlines BL41XU and BL45XU of SPring-8, and data used for calculation in the present study were collected at BL41XU (12) at 100 K. The data obtained were processed with dps-mosflm (13) and reduced with programs in the CCP4 program suite (14). Phases were calculated with sharp (15) to a resolution of 3.7 Å and improved by the program dm (including noncrystallographic symmetry averaging with two monomers related in an asymmetric unit). The crystallographic statistics were summarized in Table 1. We used five data sets of heavy atom derivatives to obtain phase information. In addition, x-ray absorption fine structures were measured for Mn and Fe from single crystals to determine the peak and edge wavelengths of Mn and Fe, and anomalous diffraction data were collected at the peak and edge wavelengths of both Mn and Fe, respectively. The results obtained supported the number of Fe atoms and the global locations of both Mn and Fe atoms determined by the isomorphous replacement method (data not shown). Electron density map visualization and interpretation were done with the program turbo-frodo (http://afmb.cnrs-mrs.fr/TURBO_FRODO/). Real sequences of CP47, CP43, D1, D2, cyt b559 α, β subunits, PsbK, and extrinsic cyt c550 were traced into the electron density map on the basis of the large side chains of aromatic residues (His, Phe, Tyr, and Trp) present in their sequences (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). PsbI, PsbH, PsbX, and the extrinsic 33- and 12-kDa proteins were modeled as polyalanines due to the weakness of electron density assignable to the large side chains of aromatic residues. The other lower molecular mass subunits were represented as Cα, for which the direction of the polypeptide chains could not be determined. In the present study, gene-derived amino acid sequences used for D1 and cyt c550 were from T. vulcanus; the sequences of other subunits used were from T. elongatus (16), because the full sequences of T. vulcanus are not available at present. Comparisons of known sequences of PSII subunits, however, revealed that there are virtually no differences between the sequences of the two organisms as far as photosynthetic proteins are concerned. The model was subjected to rigid-body refinement with the program refmac (17) in the CCP4 suite, which resulted in an overall R factor of 0.53 and an estimated overall coordinate error of 1 Å. Figures were generated by using molscript (18) and RASTER3D (19).
Table 1.
Crystallographic statistics
| Native | Ta | Hg1 | Hg2 | Pt | Au | ||||
|---|---|---|---|---|---|---|---|---|---|
| Data collection* | |||||||||
| Resolution, Å | 25–3.7 | 25–5.5 | 25–4.5 | 25–4.4 | 25–5.5 | 25–5.0 | |||
| Unique reflections | 94,859 | 29,394 | 53,617 | 56,268 | 29,122 | 39,233 | |||
| Redundancy | 12.0 | 7.0 | 6.3 | 6.3 | 6.8 | 6.6 | |||
| Completeness, %† | 98.0 (95.0) | 97.9 (100) | 99.1 (98.4) | 99.0 (98.3) | 98.8 (99.9) | 99.1 (99.2) | |||
| Comp. of Friedels, %† | – | 98.2 (99.3) | 95.9 (89.9) | 95.8 (90.4) | 98.4 (98.2) | 97.1 (93.4) | |||
| 〈I/σ〉† | 8.0 (2.8) | 9.2 (3.5) | 5.4 (3.5) | 8.5 (4.1) | 7.7 (3.0) | 6.2 (3.5) | |||
| Rmerge, %† | 7.1 (26.1) | 6.6 (20.8) | 9.5 (19.7) | 7.1 (17.6) | 6.8 (21.4) | 8.2 (18.8) | |||
| Phasing statistics | |||||||||
| Heavy atom sites | – | 14 | 16 | 14 | 19 | 3 | |||
| RCullis (centric)‡ | – | 0.78 (0.96) | 0.78 (0.85) | 0.86 (0.87) | 0.93 (0.91) | 0.96 (0.92) | |||
| Phasing power‡ | – | 1.13/1.52 | 1.37/1.70 | 1.13/1.36 | 0.90/1.14 | 0.75/1.02 | |||
| Centric/acentric‡ | – | (0.58/0.71) | (1.09/1.36) | (0.89/1.04) | (0.92/1.04) | (0.76/1.00) | |||
| Figure of merit | |||||||||
| Resolution, Å | 9.7 | 7.2 | 5.9 | 5.2 | 4.7 | 4.3 | 4.0 | 3.7 | Total |
| Centric | 0.71 | 0.62 | 0.56 | 0.47 | 0.38 | 0.35 | 0.31 | 0.29 | 0.46 |
| Acentric | 0.84 | 0.74 | 0.60 | 0.47 | 0.38 | 0.33 | 0.29 | 0.22 | 0.41 |
All data were collected with x-rays of 1.0 Å wavelength at BL41XU of SPring-8 at a cryogenic temperature (100 K). Heavy atom derivatives used were: Ta, Ta6Br14; Hg1/Hg2, ethylmercuriphosphate; Pt, K2Pt(CN)4; Au, KAu(CN)2. Data in parentheses indicate the values in the highest-resolution shell.
Data taken from scala in the ccp4 program suite.
Data taken from sharp.
Results and Discussion
Subunit Structure.
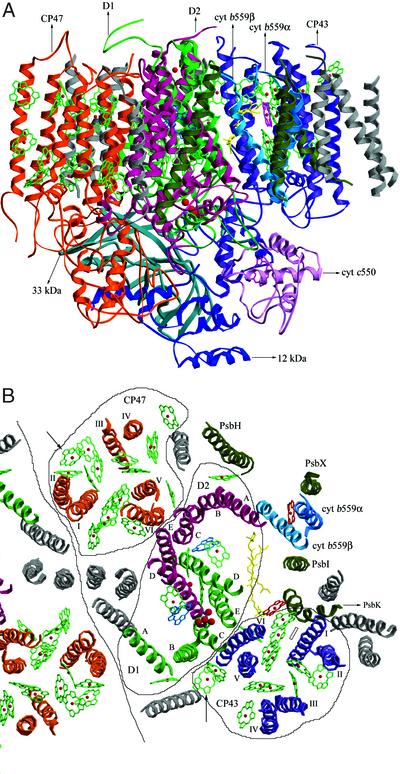
Part of the electron density map we obtained is published as Fig. 5 in supporting information on the PNAS web site, which showed enough clarity to allow the assignments of most of the protein backbones. The PSII structures thus determined for monomers in the dimer in an asymmetric unit are very similar to each other. The structure of one monomer, which contained 36 TMHs and, in overall, is similar to that reported by Zouni et al. (9), is shown in Fig. 1A. In our structure, large parts of the loop regions in the luminal side of D1, D2, CP43, and CP47 subunits, which were not assigned previously, were assigned, yielding much information concerning the possible molecular contact, not only among these molecules but also between these and other subunits (see below).
Figure 1.
Structure of PSII and the assignment of protein subunits and cofactors. (A) The structure of PSII monomer from T. vulcanus; view along the membrane plane. The red dot in the upper middle is the nonheme Fe. chl a head groups are shown in green, and the hemes and the Mn cluster are shown in purple and red. (B) The arrangement of transmembrane helices of PSII. One full PSII monomer and a part of the other monomer related by the noncrystallographic 2-fold symmetry axis were shown, together with the chls and other cofactors. Helices numbers A–E of D1 and D2 subunits and I–VI of CP47 and CP43 are indicated. Helices in gray are of subunits not assigned in the present study. Cofactors bound to CP47, CP43, and D1/D2 are circled separately. Arrowheads indicate two chls, one in CP47 and one in CP43, found in the present structure but not in the structure of T. elongatus (44), whereas a rectangle indicates a chl found in CP43 in T. elongatus's structure but not in the present structure (view from the luminal side perpendicular to the membrane plane).
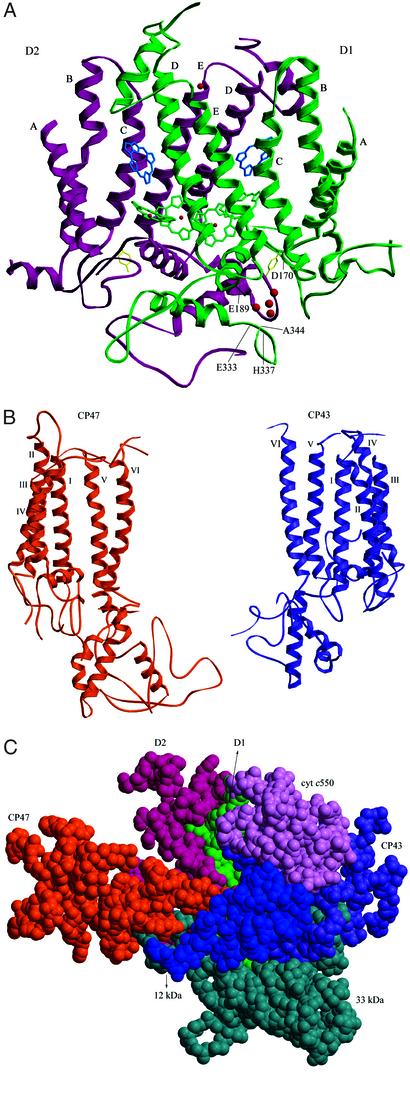
In the center of the PSII monomer, two clusters of five helices were assigned to the D1 and D2 RC subunits (Fig. 1B), which resemble those of the RC [bacterial RC (BRC)] of photosynthetic purple bacteria (20, 21) and the PSII structure determined for T. elongatus (9). All five helices of D1 and D2 are tilted against the membrane plane, with helix A and D of both D1 and D2 having the largest tilt angle (Fig. 2A). Due to this large tilting, helix D of D1 is close to helices C and E of D2 in the region near the stromal surface; similarly, helix D of D2 is close to helices C and E of D1 in the stromal part. Helix C of D1, on the other hand, is tilted toward the opposite side of helix D of D2 in the region near lumen, so that they become well separated in the luminal part (Fig. 2A). A similar situation is found for helix C of D2 and helix D of D1. These features and the total arrangement of TMHs of D1 and D2 in PSII are very similar to those of the L and M subunits in BRC (20) as well as those of the T. elongatus PSII (9), although the latter structure was represented as a Cα model with artificial numbering of the residues, which prevented a further detailed comparison with the present structure. In analog to BRC, D1-His-198 and D2-His-197 in helix D of D1 and D2 are coordinated to two RC chls arranged approximately parallel to each other in our model (see Fig. 3). D1-His-215, D1-His-272, D2-His-214, and D2-His-268 were found to be close enough to provide coordination to the nonheme iron, consistent with what has been predicted from the BRC structure. The fifth ligand found in BRC for the nonheme iron was found neither in our present model nor in our electron density map. Residues at the beginning of the DE helices of both D1 and D2 subunits in the stromal side appeared to be in the vicinity of the nonheme iron; however, their distances to the nonheme iron are >3 Å longer than the corresponding distances in BRC. This excludes the direct coordination of the nonheme iron by residues of D1 and D2 other than the His residues stated above, thus supporting the well-reported notion that bicarbonate may serve as the fifth (and sixth) ligand for the nonheme iron in PSII, which is not visible in our electron density map at the present resolution. Also not found in the BRC is the presence of two chls (ChlZD1 and ChlZD2, see below) in PSII-RC (Fig. 3A) (9, 22). Our model shows that they are coordinated by D1-His-118 and D2-His-117, respectively, consistent with results from mutagenesis studies (23–25).
Figure 2.
Structures of PSII large subunits and their interactions. (A) Structures of D1 and D2, together with four chls (green), two pheophytins (blue), TyrZ and TyrD (yellow), nonheme iron (red), and the Mn cluster (red). A–E represent helix numbers of D1 and D2. Residues of D1 determined as possible ligands for the Mn cluster are indicated (view along the membrane plane; see text for further details). (B) Structures of CP47 and CP43. Helices numbers I–VI are indicated (view from the membrane plane). (C) Interactions among CP47, CP43, D1, D2, and the three extrinsic proteins 33 kDa, 12 kDa, and cyt c550 in the luminal region. The luminal parts of CP47, CP43, D1, and D2, together with the three extrinsic proteins, are shown as a space-filling model (view perpendicular to the membrane plane from the luminal side).
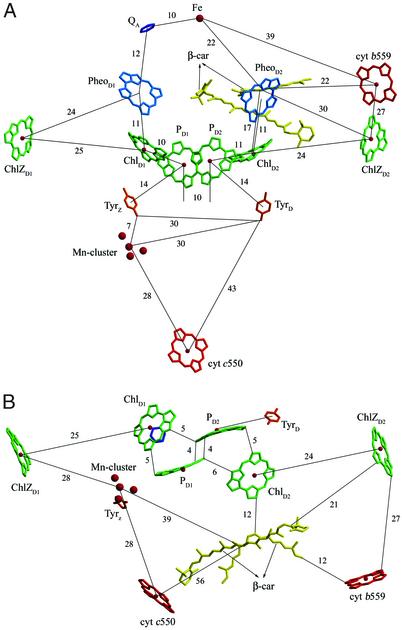
Figure 3.
Arrangements of chls and other PSII cofactors and their relative distances (Å). (A) View along the membrane plane. (B) View from the luminal side perpendicular to the membrane plane. The nonheme iron and two pheophytins are omitted for clarity.
On the luminal side, both D1 and D2 subunits contain two α-helices, one the CD helix and the other one close to the C terminus. In addition, a short helix was found in the AB loop of both D1 and D2. The presence of the CD helix is consistent with the structure of BRC (20), but the presence of the C-terminal helix is not. The structures of D1 and D2 appear very similar to each other and are related by a local pseudo-2-fold symmetry (Fig. 2A). Some of the luminal parts of D1 are close to the Mn cluster to provide coordination for it (see below); part of the luminal regions of D2 is also located near the Mn cluster but is not close enough to provide coordination.
Surrounding the D1/D2 RC are two clusters of six helices assigned to CP47 and CP43; this assignment is consistent with the fact that the former binds more chl a molecules than the latter (see below) and also agrees with the structure reported for T. elongatus (9). We determined the numbers and arrangements of the helices of both CP47 and CP43 (Fig. 1B). The results show that helix VI of CP47 is located close to helices C and E of D2, and helix VI of CP43 is close to helices C and E of D1, respectively. Our structural model is supported by the fact that almost all of the His residues in TMH regions of CP47 and CP43 are close enough to chls to provide coordination for them. The structure of CP47 appears similar to that of CP43, especially in the transmembrane region (Fig. 2B); they are also related by the local pseudo-2-fold symmetry (see also ref. 9). The structures of CP47 and CP43 are also similar to regions of PsaA and PsaB of PSI, a feature that has been noticed previously (26).
Near helix A of D2 are the α- and β-helices of cyt b559 characterized by the heme coordinated to them (Fig. 1B). In agreement with the previous report (9), we found only one heme of cyt b559 in our structure; this was confirmed directly by anomalous diffraction data collected at the peak and edge wavelengths of Fe from single crystals. Furthermore, we determined that the number of cyt b559 in our PSII complex before crystallization is close to one per RC (K. Akabori and J.-R.S., unpublished results). These results strongly suggest that the number of cyt b559 is one per PSII RC, at least in the cyanobacterial PSII. A helix adjacent to cyt b559α was assigned to PsbX, because they crosslink with each other (Fig. 1B) (27). Two helices surrounding cyt b559 were assigned to PsbH and PsbI, respectively, on the basis of the fact that the former crosslink with PsbX (28) and the latter with cyt b559α (29). PsbH is characterized by a longer TMH than PsbI, consistent with the results of secondary structure predictions (not shown). A helix close to CP43 was assigned to PsbK, on the basis of the fact that it copurifies with CP43 (30) and has two Pro residues separated by two residues in the middle of its TMH (16), which leads to a large characteristic bending of the helix as shown (Fig. 1B). The assignments of PsbH, PsbI, and PsbK are consequently different from those of Zouni et al. (9), the latter being based on the differences in composition between PSII dimer and monomer (31, 32). A possible reason for this difference can be that subunits found in the PSII dimer but not in its monomer are due to removal of these subunits by the extensive detergent treatment used to purify the monomer from the dimer, instead of their possible involvement in the dimerization of PSII. This then raises the question of placing them in the center of the PSII dimer, as did Zouni et al. (9). In fact, subunits assigned to the center of the dimer in Zouni et al.'s structure were also different from those assigned from electron microscopy of 2D crystals (33).
In the luminal side, all three extrinsic proteins were assigned (Fig. 1A). The 33-kDa protein is characterized by a large amount of β strand, as has been previously reported (9, 34, 35); in addition, a short helix was found in the region close to the C terminus. The present model was built as a polyalanine structure accounting for 83.3% of the total residues of the 33-kDa protein (Table 2), which resulted in a cylindrical structure with dimensions of 20 Å in diameter and 45 Å in length. In addition, there is a 25-Å-long loop extending from one side of the cylinder. The structure of the 33-kDa protein thus determined has some similarities to the structure of the porin family (36, 37) but is not compatible with the model predicted theoretically (38) or that analyzed by single-particle analysis of cryoelectron microscopic images from higher plants (39). The structure of cyt c550 was modeled partially with the help of the recently reported structure of this cyt from two other cyanobacteria (40, 41), generating a structure very similar to those from the other cyanobacteria. The newly assigned 12-kDa protein has an all-α architecture composed of five or more short α-helices, with no homologous structure in the database. This protein is located between the 33-kDa protein and cyt c550 but apart from the luminal surface of the membrane by ≈30 Å, in agreement with previous results that this protein has no direct contact with the membrane and cannot bind to PSII in the absence of the 33-kDa protein and cyt c550 (11). The arrangement of the three extrinsic proteins suggests that the 12-kDa protein helps to link the 33-kDa protein and cyt c550. Consequently, the 33-kDa protein and cyt c550 interact not directly but through the 12-kDa protein, which suggests a different organization of the extrinsic proteins between cyanobacteria and higher plants, because the 33- and 23-kDa proteins have been reported to interact directly in higher plant PSII (42).
The three extrinsic proteins together with the luminal regions of CP47, CP43, D1, and D2 form a large barrier to shield the Mn cluster from the bulk solution (Fig. 1A). On the basis of the present model, the luminal part of CP47 is close to those of D2, the 33- and 12-kDa proteins (Fig. 2C), suggesting possible interactions among these subunits. In particular, a close location between the large E loop of CP47 and the extrinsic 33-kDa protein was found, consistent with many reports suggesting their possible association and interaction (42, 43). Likewise, D2 is close to D1, CP47, and the 33-kDa protein; D1 is close to CP43, D2, the 33-kDa protein, and cyt c550; and CP43 is close to D1 and all three extrinsic proteins (Fig. 2C).
Arrangement of Cofactors.
Fig. 1B shows the arrangement of chls and other cofactors in PSII RC. D1/D2 binds six chls and two pheophytins, in agreement with Zouni et al. (9). We found 17 chls in CP47, which is 1 chl more than that reported for T. elongatus (44). We found 13 chls in CP43, which is the same as that reported for T. elongatus, but the position of 1 chl in our CP43 is different from that of T. elongatus (Fig. 1B) (44). These gave rise to a total number of chls of 36 in our PSII.
The relative distances of electron transfer cofactors were shown in Fig. 3A. The center-to-center distance between the two “RC chls” PD1 and PD2 is 10 Å, and those between RC chls and “accessory chls,” PD1 and ChlD1, PD2 and ChlD2, are 10 and 11 Å, respectively. These distances are not much different from each other, agreeing with the “multimer of monomer” model recently proposed for PSII RC (45, 46). However, the closest edge-to-edge distance between PD1 and PD2 is 4 Å, whereas those between PD1 and ChlD1, PD2 and ChlD2 are 5 Å (Fig. 3B) [similar values were found in Zouni et al.'s structure (9)]. Moreover, the relative orientations of these chls apparently show that the RC chls (PD1, PD2) have a larger area of their planes sitting close to each other, whereas the areas of planes located in the closest distance between the RC chls and accessory chls (ChlD1, ChlD2) are very small (Fig. 3B). These facts suggest an apparently stronger interaction between the two RC chls than those between the RC chls and accessory chls, indicating that each of the monomer chls within the tetramer chls of PSII RC is not the same. This suggests a more similar spatial organization of the PSII-RC chls with that of BRC, although the center-to-center distance between PD1 and PD2 in PSII is apparently longer than that in BRC.
In the region near the RC chls, two Cars were assigned; one was modeled as a cis- and one as an all-trans type (Fig. 3 A and B), in agreement with reports that there are two Cars in PSII-RC preparations (47, 48). The two Cars are located close to each other, with their relative distance in the closest part being 5 Å; this agrees with spectroscopic evidence reported previously that the two Cars in RC may be located closely (49). A number of experiments have been reported concerning the roles of Cars in energy and electron transfer (see ref. 48 for a recent review). The present structure showed that the center of the cis-type Car is located 22 Å to the nonheme iron, 22 Å to cyt b559 heme, 30 Å to ChlZD2, and 17 Å to ChlD2. The edge-to-edge distances between cyt b559 and the cis-type Car, between ChlZD2 and the transtype Car, are 12 and 21 Å, respectively. These results suggest that secondary electron transfer may preferentially involve the cyt b559-Car-RC pathway, and electron transfer involving ChlZD2-Car-RC may also be possible. Contrarily, a mechanism involving cyt b559-ChlZD2-Car-RC appears less preferential, because cyt b559 and ChlZD2 are located with a distance longer than their respective distances to Car. We should point out here that these discussions did not take the redox potentials of each component into consideration, which are partially unknown at present and may reasonably affect the efficiencies of the proposed electron transfer pathways. The close location of two Cars to the RC also suggests that they may be important for excess energy dissipation, a mechanism important for regulation of photosynthesis efficiency under excess light. The other distances among the electron transfer cofactors we found are similar as those reported by Zouni et al. (9).
Environment Around the Mn Cluster.
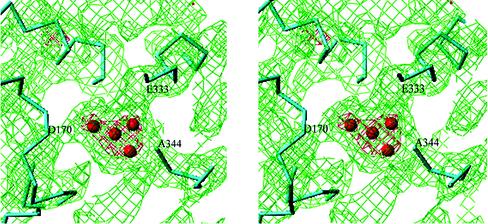
The heart of the water-splitting reaction is the Mn cluster, consisting of 4 Mn atoms, the shape of which is shown in Fig. 4 (electron density in red, countered at 5σ). This shape is similar to that reported by Zouni et al. (9), in that three Mn atoms are positioned in the corner of the density and one is in the center. The only slight difference is that in our structure, all four Mn atoms are located roughly in the same plane, whereas in Zouni et al.'s structure, the central Mn is protruded toward the luminal surface of the membrane. The Mn cluster is coordinated by the D1 polypeptide; from the electron density map, we see at least four to five connections between the Mn cluster and polypeptide backbones. One such connection has a strong electron density provided by a polypeptide main chain, which terminates at the Mn cluster. According to our assignment, this main chain is the C-terminal part of D1, indicating that the C-terminal carboxyl group of Ala-344 provides ligands directly to the Mn cluster. This is in agreement with mutagenesis studies suggesting the possible ligation of the Mn cluster by D1 C terminus (see ref. 50 for review). Other residues coordinated to the Mn cluster are Asp-170 and Glu-333 (or His-332); electron density connections between these two residues and the Mn cluster were visible at a 1σ level. His-337 and Asp-189 (or His-190) are also possibly coordinated to the Mn cluster; weak electron density connections between them and the Mn cluster can be seen at a 0.2σ level. These residues have been suggested as possible ligands from mutagenesis (reviewed in ref. 50) or chemical modification studies (51). Asp-342 has also been suggested as a possible ligand for Mn from mutagenesis studies (50), but our model does not support this, because the C-terminal part of the D1 polypeptide does not surround the Mn cluster but is oriented in such a way that residues immediately upstream of the C terminus are far from the Mn cluster. In addition, we found that Tyr-73 in the AB loop of D1 is located close to the Mn cluster. It is also clear that no polypeptide carbonyl oxygen except the C terminus is able to provide coordination to the Mn cluster, because this requires a much closer location of a corresponding polypeptide main chain with the Mn cluster, which was apparently not the case.
Figure 4.
Stereo view of the Mn cluster and the electron density surrounding it, together with the assignments of polypeptide chains. Some of the residues as possible ligands for the Mn cluster are shown. Red, electron density contoured at 5.0σ; green, electron density contoured at 1.0σ.
The present study has provided more details and additional information on the structure of proteins and arrangements of cofactors within PSII, which are useful for relevant mechanistic studies of reactions taking place in this large and important protein complex.
Supplementary Material
Acknowledgments
We thank Drs. Masahide Kawamoto (SPring-8) and Yoshiaki Kawano (RIKEN) for help with data collections; Profs. Tsuneaki Yamagata, Yuriko Yamagata, Keiichi Fukuyama, and Tomitake Tsukihara (Osaka University) for heavy atom compounds; Drs. Takakazu Kaneko and Satoshi Tabata (Kazusa DNA Research Institute) for the gene-derived amino acid sequences of T. elongatus before release; and Drs. Doug Bruce (Brock University) and Jim Barber (Imperial College) for comments. This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan (11169254, 14340257) and by a president grant from RIKEN.
Abbreviations
- PSII
photosystem II
- RC
reaction center
- chl
chlorophyll
- TMH
transmembrane helix
- cyt
cytochrome
- BRC
bacterial reaction center
- Car
β-carotene
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1IZL).
References
- 1.Hankamer B, Barber J, Boekema E J. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:641–647. doi: 10.1146/annurev.arplant.48.1.641. [DOI] [PubMed] [Google Scholar]
- 2.Debus R J. In: Metal Ions in Biological Systems. Sigel A, editor. Vol. 37. New York: Dekker; 2000. pp. 657–711. [PubMed] [Google Scholar]
- 3.Barber J. Curr Opin Struct Biol. 2002;12:523–530. doi: 10.1016/s0959-440x(02)00357-3. [DOI] [PubMed] [Google Scholar]
- 4.Rhee K-H, Morris E P, Zheleva D, Hankamer B, Kuhlbrandt W, Barber J. Nature. 1997;389:522–526. [Google Scholar]
- 5.Rhee K-H, Morris E P, Barber J, Kuhlbrandt W. Nature. 1998;396:283–286. doi: 10.1038/24421. [DOI] [PubMed] [Google Scholar]
- 6.Zouni A, Jordan R, Schlodder E, Fromme P, Witt H T. Biochim Biophys Acta. 2000;1457:103–105. doi: 10.1016/s0005-2728(00)00100-6. [DOI] [PubMed] [Google Scholar]
- 7.Kuhl H, Kruip J, Seidler A, Krieger-Liszkay A, Bünker M, Bald D, Scheidig A J, Rögner M. J Biol Chem. 2000;275:20652–20659. doi: 10.1074/jbc.M001321200. [DOI] [PubMed] [Google Scholar]
- 8.Shen J-R, Kamiya N. Biochemistry. 2000;39:14739–14744. doi: 10.1021/bi001402m. [DOI] [PubMed] [Google Scholar]
- 9.Zouni A, Witt H-T, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 10.Shen J-R, Ikeuchi M, Inoue Y. FEBS Lett. 1992;301:145–149. doi: 10.1016/0014-5793(92)81235-e. [DOI] [PubMed] [Google Scholar]
- 11.Shen J-R, Inoue Y. Biochemistry. 1993;32:1825–1832. doi: 10.1021/bi00058a017. [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto M, Kawano Y, Kamiya N. Nucl Instr Methods A. 2001;467–468:1375–1379. [Google Scholar]
- 13.Steller I, Bolotovsky B, Rossmann M G. J Appl Crystallogr D. 1997;30:1036–1040. [Google Scholar]
- 14.Collaborative Computating Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 15.de la Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S, Watanabe A, Iriguchi M, Kawashima K, Kimura T, et al. DNA Res. 2002;9:123–130. doi: 10.1093/dnares/9.4.123. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 19.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 20.Deisenhofer J, Michel H. Annu Rev Cell Biol. 1991;7:1–23. doi: 10.1146/annurev.cb.07.110191.000245. [DOI] [PubMed] [Google Scholar]
- 21.Michel H, Deisenhofer J. Biochemistry. 1988;27:1–7. [Google Scholar]
- 22.Schelvis J P M, van Noort P I, Aartsma T J, van Gorkom H J. Biochim Biophys Acta. 1994;1184:242–250. [Google Scholar]
- 23.Stewart D H, Cua A, Chisholm D A, Diner B, Bocian D F, Brudvig G W. Biochemistry. 1998;37:10040–10046. doi: 10.1021/bi980668e. [DOI] [PubMed] [Google Scholar]
- 24.Lince M T, Vermaas W F J. Eur J Biochem. 1998;256:595–602. doi: 10.1046/j.1432-1327.1998.2560595.x. [DOI] [PubMed] [Google Scholar]
- 25.Johnston H G, Wang J, Ruffle S V, Sayre R T, Gustafson T L. J Phys Chem B. 2000;104:4777–4781. [Google Scholar]
- 26.Jordan P, Fromme P, Witt H T, Klukas O, Saenger W, Krauss N. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 27.Shi L X, Kim S, J, Marchant A, Robinson C, Schroder W P. Plant Mol Biol. 1999;40:737–744. doi: 10.1023/a:1006286706708. [DOI] [PubMed] [Google Scholar]
- 28.Büchel C, Morris E, Orlova E, Barber J. J Mol Biol. 2001;312:371–379. doi: 10.1006/jmbi.2001.4951. [DOI] [PubMed] [Google Scholar]
- 29.Tomo T, Enami I, Satoh K. FEBS Lett. 1993;323:15–18. doi: 10.1016/0014-5793(93)81438-6. [DOI] [PubMed] [Google Scholar]
- 30. Sugimoto, I. & Takahashi, Y. (2001) PS2001 Proceedings, 12th International Congress on Photosynthesis, S5-033 [CSIRO Australia ( www.csiro.au/index.asp)].
- 31.Summer E J, Schmid V H, Bruns B U, Schmidt G W. Plant Physiol. 1997;113:1359–1368. doi: 10.1104/pp.113.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheleva D, Sharma J, Panico M, Morris H R, Barber J. J Biol Chem. 1998;273:16122–16127. doi: 10.1074/jbc.273.26.16122. [DOI] [PubMed] [Google Scholar]
- 33.Hankamer B, Morris E P, Nield J, Gerle C, Barber J. J Struct Biol. 2001;135:262–269. doi: 10.1006/jsbi.2001.4405. [DOI] [PubMed] [Google Scholar]
- 34.Xu Q, Nelson J, Bricker T M. Biochim Biophys Acta. 1994;1188:427–431. doi: 10.1016/0005-2728(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed A, Tajmir-Riahi H A, Carpentier R. FEBS Lett. 1995;363:65–68. doi: 10.1016/0014-5793(95)00282-e. [DOI] [PubMed] [Google Scholar]
- 36.Vogt J, Schulz G E. Struct Folding Des. 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 37.Pautsch A, Schulz G E. J Mol Biol. 2000;298:273–282. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- 38.Pazos F, Heredia P, Valencia A, de las Rivas J. Proteins Struct Funct Genet. 2001;45:372–381. doi: 10.1002/prot.10012. [DOI] [PubMed] [Google Scholar]
- 39.Nield J, Balsera M, de las Rivas J, Barber J. J Biol Chem. 2002;277:15006–15012. doi: 10.1074/jbc.M110549200. [DOI] [PubMed] [Google Scholar]
- 40.Vrettos J S, Reifler M J, Kievit O, Lakshmi K V, de Paula J C, Brudvig G W. J Biol Inorg Chem. 2001;6:708–716. doi: 10.1007/s007750100249. [DOI] [PubMed] [Google Scholar]
- 41.Sawaya M R, Krogmann D W, Serag A, Ho K K, Yeates T O, Kerfeld C A. Biochemistry. 2001;40:9215–9225. doi: 10.1021/bi002679p. [DOI] [PubMed] [Google Scholar]
- 42.Seidler A. Biochim Biophys Acta. 1996;1277:36–60. doi: 10.1016/s0005-2728(96)00102-8. [DOI] [PubMed] [Google Scholar]
- 43.Bricker T M, Frankel L K. Photosynth Res. 2002;72:131–146. doi: 10.1023/A:1016128715865. [DOI] [PubMed] [Google Scholar]
- 44.Vasil'ev S, Orth P, Zouni A, Owens T G, Bruce D. Proc Natl Acad Sci USA. 2001;98:8602–8607. doi: 10.1073/pnas.141239598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barber J, Archer M D. J Photochem Photobiol A. 2001;142:97–106. [Google Scholar]
- 46.Diner B A, Rappaport F. Annu Rev Plant Biol. 2002;53:551–580. doi: 10.1146/annurev.arplant.53.100301.135238. [DOI] [PubMed] [Google Scholar]
- 47.Bialek-Bylka G E, Tomo T, Satoh K, Koyama Y. FEBS Lett. 1995;363:137–140. doi: 10.1016/0014-5793(95)00298-n. [DOI] [PubMed] [Google Scholar]
- 48.Tracewell C A, Vrettos J S, Bautista J A, Frank H A, Brudvig G W. Arch Biochem Biophys. 2001;385:61–69. doi: 10.1006/abbi.2000.2150. [DOI] [PubMed] [Google Scholar]
- 49.Newell W R, van Amerongen H, Barber J, van Grondelle R. Biochim Biophys Acta. 1991;1057:232–238. [Google Scholar]
- 50.Debus R J. Biochim Biophys Acta. 2001;1503:164–186. doi: 10.1016/s0005-2728(00)00221-8. [DOI] [PubMed] [Google Scholar]
- 51.Preston C, Seibert M. Biochemistry. 1991;30:9625–9633. doi: 10.1021/bi00104a009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.