Abstract
The prediction of binding sites and the understanding of interfaces associated with protein complexation remains an open problem in molecular biophysics. This work shows that a crucial factor in predicting and rationalizing protein–protein interfaces can be inferred by assessing the extent of intramolecular desolvation of backbone hydrogen bonds in monomeric structures. Our statistical analysis of native structures shows that, in the majority of soluble proteins, most backbone hydrogen bonds are thoroughly wrapped intramolecularly by nonpolar groups except for a few ones. These latter underwrapped hydrogen bonds may be dramatically stabilized by removal of water. This fact implies that packing defects are “sticky” in a way that decisively contributes to determining the binding sites for proteins, as an examination of numerous complexes demonstrates.
Keywords: hydrophobic effect|protein structure|protein–ligand association| binding site
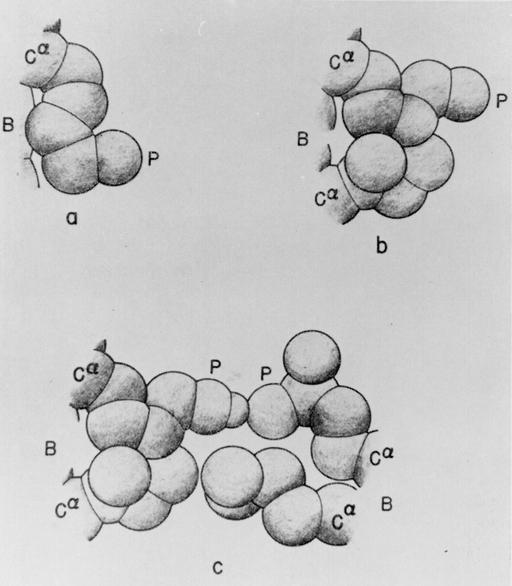
A theory of hydrophobic interactions (1) based on a statistical mechanical treatment of liquid H2O (2) and aqueous solutions of hydrocarbons (3) demonstrated how the removal of water from the neighborhood of nonpolar groups enhanced their interaction free energy in aqueous solution (4). Such dehydration-based hydrophobic interactions enhance the role of nearby intramolecular hydrogen bonds in stabilizing protein conformations (5–9) and facilitating the folding process (refs. 10–12; Fig. 1). It therefore is necessary to provide a systematic description of the nonpolar environments of hydrogen bonds, their variations among native structures, and their evolution during conformational changes. This is needed, for example, to assess the role of water removal in protein–ligand associations (13, 14), molecular disease, and aggregation (15, 16). To address such problems, we define a hydrogen-bond dehydration domain and count the number of nonpolar groups within. We show that a field must be introduced to account for spots on the protein surface where water exclusion resulting from intergroup interaction plays a key role in strengthening nearby hydrogen bonds. Such hot spots enhance the contribution of hydrophobic interactions and contribute to defining binding sites, nucleating sites for aggregation, and protein reactivity in general.
Figure 1.
Schematic representation (5) of various hydrophobic interactions of a polar side chain with its surroundings. B, backbone; P, polar head; Cα, α-carbon. (a) Interaction of a lysine side chain with the backbone. (b) Interaction of a lysine side chain with a nearby isoleucine side chain. (c) Interaction of two polar side chains (lysine and glutamic acid), engaged in hydrogen bonding, with two nonpolar side chains (isoleucine and leucine, respectively). A hydrophobic interaction is also formed between the two nonpolar side chains. [Reproduced with permission from ref. 5 (Biopolymers copyright 1963, copyright owner as specified in Biopolymers).]
The dehydration of backbone hydrogen bonds by nearby nonpolar groups makes it thermodynamically unfavorable to expose the backbone amide and carbonyl groups (Fig. 1). Similarly, as shown in Fig. 1, nearby nonpolar groups enhance the dehydration of the nonpolar parts of polar side chains as well as restricting the rotational freedom of the polar side chain, thereby increasing the stability of side-chain hydrogen bonds. Thus, the stabilization of secondary structure generally requires a higher-order organization of the chain to dehydrate the hydrogen bonds (10–12), shielding them from water attack. In view of this, we expect that most native structures of soluble proteins in their monomeric form would have most of their hydrogen bonds thoroughly dehydrated to warrant their overall stability. This is indeed the case, as can be inferred by examination (see below) of an exhaustive structural database consisting of 1,476 high-resolution (≤3 Å) entries free of sequence redundancies. The database was obtained by filtering the Protein Data Bank (PDB) with a tolerance of <40% homology in the primary sequences (17). To assess the role of dehydration of hydrogen bonds, three questions may be addressed: (i) Can we identify backbone hydrogen bonds in soluble proteins that are poorly wrapped, i.e., that are poorly dehydrated? (ii) Are most backbone hydrogen bonds thoroughly dehydrated along a folding pathway? (iii) What are the implications of individual underdehydrated hydrogen bonds (UDHBs)?
Methods
The wrapping of backbone (amide-carbonyl) hydrogen bonds by side-chain carbonaceous groups (CHn, n = 1, 2, 3) clustered around them is easily quantifiable and seems to be a straightforward way to estimate the extent of hydrophobic burial of such bonds: We define the dehydration domain of a hydrogen bond as consisting of two spheres of 6.5-Å radius centered at the α-carbons of the residues paired by the hydrogen bond. These spheres necessarily intersect, because the typical minimum distances between nonadjacent α-carbons in secondary structure are in the range of 4.8–6.1 Å (18). The choice of radius is based on the typical cutoff distance used to define pairwise interactions, but the results are qualitatively robust within the range of 6.5 ± 0.3 Å.
Thus, the extent of wrapping of hydrogen bonds is operationally defined by the number of side-chain carbonaceous groups within their dehydration domains. In the case of a complex, the dehydration shell of an intramolecular hydrogen bond near the protein surface may include carbonaceous groups from the binding partner (if they happen to lie within the desolvation domain after complexation).
Each carbonaceous group may be regarded as a third body introducing a three-body correlation (hydrophobe–hydrogen–bonded pair) (11). Thus, the extent of hydrogen-bond dehydration, ρ, averaged over all backbone hydrogen bonds of a given structure may be obtained as ρ = C3/Q, where C3 is the total number of three-body correlations and Q is the total number of backbone hydrogen bonds. A hydrogen bond is operationally defined as one satisfying the following constraints: N—O distance <3.5 Å and 45° range in the angle between the NH and CO vectors.
Significantly, we have found that the UDHBs in PDB structures are also the longest hydrogen bonds; their N—O lengths are in the range of 3.1–3.44 Å, whereas the average N—O length of a well wrapped amide-carbonyl hydrogen bond is 2.81 Å. These statistics were collected from our database and imply that, in natural proteins, the hydrogen bonds best preserved from water attack (or most stable) are also the strongest, thus providing a selective advantage; it would not be thermodynamically profitable to sacrifice conformational freedom to protect weak hydrogen bonds.
Intramolecular Wrapping of Backbone Hydrogen Bonds
The wrapping of backbone (amide-carbonyl) hydrogen bonds by side-chain carbonaceous groups (CHn, n = 1, 2, 3) clustered around them may be quantified in a straightforward way: We define the dehydration domain of a hydrogen bond as consisting of two (intersecting) dehydration spheres of 6.5-Å radius centered at the α-carbons of the residues paired by the hydrogen bond (see Methods). The choice of sphere radius is based on the cutoff distance adopted to define pairwise contributions to the internal energy and is justified a posteriori because it yields the highest regularity in the statistics of hydrogen-bond dehydration (see below). Thus, the extent of wrapping of hydrogen bonds is operationally defined by the number of carbonaceous groups within their dehydration domains. Each carbonaceous group may be regarded as a third body introducing a three-body correlation (hydrophobe–hydrogen–bonded pair) (10–12). Thus, the extent of hydrogen-bond dehydration, ρ, averaged over all backbone hydrogen bonds of a given chain conformation may be obtained as ρ = C3/Q, where C3 is the total number of three-body correlations and Q is the number of backbone hydrogen bonds.
There is a striking regularity in the average ρ value among native structures: 96% of soluble proteins in their monomeric form have ρ = 15.00 ± 2.05 (Table 1), with a maximum Gaussian dispersion of σ = 3.30 (22%) among the hydrogen bonds of a native structure. In view of these statistics, we define a UDHB as having at most nine carbonaceous groups in its dehydration domain. This definition is based on the statistics: The lowest representative ρ value (12.95) combined with the maximum dispersion (3.30) would render the probability of picking a hydrogen bond wrapped by nine or less nonpolar groups unlikely (probability <4%) for a protein chosen at random from our database.
Table 1.
Data on backbone hydrogen-bond dehydration of PDB native structures
| PDB ID code | C3 | Q | ρ | σ, % |
|---|---|---|---|---|
| 1aa2 | 771 | 52 | 14.83 | 10.18 |
| 1bz0 (α) | 1,450 | 95 | 15.26 | 12.03 |
| 1bz0 (β) | 1,472 | 99 | 14.87 | 12.00 |
| 1lou | 726 | 47 | 15.44 | 13.05 |
| 1ris | 690 | 45 | 15.33 | 12.87 |
| 1aue | 750 | 49 | 15.30 | 11.80 |
| 256b | 1,182 | 75 | 15.76 | 16.05 |
| 1ubi | 465 | 31 | 15.00 | 10.06 |
| 1gb4 | 240 | 16 | 15.00 | 10.14 |
| 1srl | 120 | 8 | 15.00 | 12.83 |
| 2ptl | 222 | 16 | 13.87 | 16.33 |
| 1crc | 408 | 28 | 14.57 | 9.60 |
| 1hhh | 1,338 | 86 | 15.56 | 12.68 |
| 1mim | 954 | 64 | 14.90 | 17.62 |
| 1ifb | 645 | 45 | 14.33 | 8.83 |
| 1hhg | 1,404 | 95 | 14.77 | 11.09 |
| 1e4j | 675 | 44 | 15.34 | 12.11 |
| 1e4k | 699 | 46 | 15.20 | 11.15 |
| 1gff-1 | 1,836 | 124 | 14.81 | 11.58 |
| 1csk-A | 333 | 23 | 14.47 | 12.01 |
| 1c3t | 315 | 21 | 15.00 | 10.78 |
| 1fas | 171 | 23 | 7.43 | 17.08 |
| 1f94 | 261 | 25 | 10.44 | 22.80 |
| 1jwi | 300 | 25 | 12.00 | 23.51 |
| 1dxo | 645 | 59 | 10.93 | 21.8 |
| 1dwz | 648 | 60 | 10.80 | 24.2 |
| 1b10 | 699 | 58 | 12.05 | 21.3 |
| 1qlx | 684 | 58 | 11.79 | 19.6 |
| 1qm0 | 648 | 57 | 11.37 | 20.2 |
| 1qm1 | 639 | 56 | 11.25 | 21.4 |
Each protein is identified by its PDB ID code, and the three groups represent good hydrogen-bond wrappers, toxins, and cellular prion proteins, respectively. Four parameters characterizing the average dehydration of hydrogen bonds are given: the total number of three-body correlations (C3), the total number of backbone hydrogen bonds (Q), the average extent of dehydration ρ = C3/Q, and the dispersion σ in the extent of dehydration over all the backbone hydrogen bonds within a native structure. The data on the worst wrapper of backbone hydrogen bonds in the PDB, the antiacetylcholinesterase toxin from green mamba venom (PDB ID code 1fas), are indicated in boldface characters.
Table 1 lists a representative group of hydrogen-bond wrapping PDB proteins with ρ = 15.00 ± 2.05 and a group of clearcut outliers. All the outliers found among soluble proteins are either cellular prion proteins (refs. 19 and 20; Table 1) or toxins (Table 1, intermediate group). The stability of the latter is determined by disulfide bonds. The worst wrapper of hydrogen bonds in the entire PDB (ρ = 7.43) is reported here to be the antiacetylcholinesterase toxin from green mamba venom (PDB ID code 1fas).
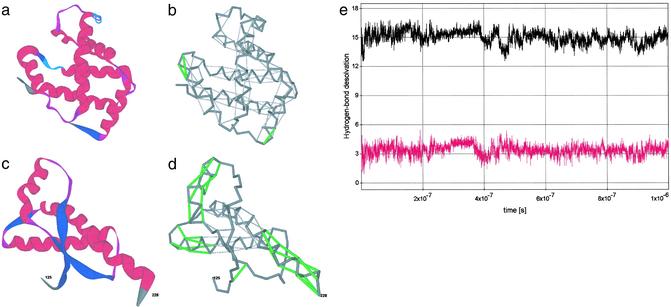
Two extreme cases in terms of average extent of hydrogen-bond wrapping among soluble proteins are displayed in Fig. 2: hemoglobin (Hb) β-subunit (18) (PDB ID code 1bz0, chain B), a good wrapper, and the human prion protein (PDB ID code 1qm0) (19). The ribbon structure of the Hb β-subunit is shown in Fig. 2a, and the 96 sufficiently wrapped hydrogen bonds (gray) and 3 UDHBs (green) are shown in Fig. 2b. Within the natural interactive context of the Hb subunit, the UDHBs signal crucial binding sites: UDHBs (with residues 90 and 94 as well as 90 and 95 paired by hydrogen bonds) are associated with the β-FG interhelical corner involved in the quaternary α1β2 interface, whereas UDHB (involving residues 5 and 9) is adjacent to Glu-6, which mutates to Val-6 in sickle-cell Hb and is located at the protein–protein Glu-6-(Phe-85, Leu-88) interface in the deoxy-HbS fiber (18).
Figure 2.
(a–d) Ribbon structure and backbone hydrogen-bond pattern for Hb β-subunit (PDB ID code 1bz0, a and b) and human prion protein (PDB ID code 1qm0, c and d). A dark-gray series of virtual bonds joining consecutive α-carbons represents the backbone in b and d. The sufficiently wrapped amide-carbonyl hydrogen bonds are represented by light-gray thin lines joining the α-carbons of paired residues, and the underwrapped hydrogen bonds are represented by green lines (b and d). Time-dependent average extent ρ = ρ(t) of backbone hydrogen-bond dehydration (black plot), and dispersion σ = σ(t) (red plot) over all backbone hydrogen bonds appearing along a time sequence of chain conformations extracted from the Duan–Kollman trajectory (21) are shown (e).
By contrast, the 30 sufficiently dehydrated hydrogen bonds and 28 UDHBs of the prion protein (PDB ID code 1qm0) are displayed in Fig. 2d, with its ribbon structure shown in Fig. 2c. The vastly higher proportion of UDHBs signals a structure vulnerable to water attack, and prone to rearrangement, especially in helix 1 (residues 143–156), where 100% of the hydrogen bonds are UDHBs. This observation agrees with current information (19, 20), which has singled out helix 1 as the probable site for rearrangement. Furthermore, helix 3 (residues 199–228) contains a significant concentration of UDHBs at the C terminus, a region assumed to define the epitope for protein-X binding (19, 20). The remaining UDHBs occur at the helix–loop junctures, which thus can be easily distorted, as required by a structural rearrangement. This follows because the UDHBs are not only the least stable but also the weakest hydrogen bonds (Methods).
Comparable low ρ values may be found in some membrane proteins, where unlike the case of soluble proteins, the underdehydration of hydrogen bonds does not imply structural defects because of the lower permittivity of the lipid medium and its inability to compete for hydrogen bonds with the protein chain. Thus, it is expected that some prion proteins with such an overall defective dehydration of their hydrogen bonds might exhibit a tendency to interact with lipid membranes (22). The comparable ρ values found for prion proteins and most membrane proteins (PDB ID code 1gl2, ρ = 11.04; PDB ID code 1i4m, ρ = 10.68; PDB ID code 1ftk, ρ = 11.01) are revealing in this regard.
Dynamics of Hydrogen-Bond Wrapping
From a dynamic perspective, our simple characterization of the hydrogen-bond environment enables us to decide whether formation of local secondary structure and large-scale structural organization are necessarily concurrent along a folding pathway. Thus, the time-dependent ρ (black plot) and σ (red plot) values are displayed in Fig. 2e for the longest all-atom explicit-solvent MD trajectory available, the Duan–Kollman 1-μs simulation of the villin headpiece (21). Strikingly, we find that not only are the constraints implied by the PDB statistics (ρ = 15.00 ± 2.05, maximum Gaussian dispersion σ = 3.30) obeyed by most native soluble proteins, but such constraints also apply as the protein explores conformation space. The results suggest that the average extent of hydrogen-bond dehydration and large-scale order are needed if secondary structure is to prevail; the constraint ρ ≈ 15.00 ± 2.05 cannot be satisfied merely by forming secondary structure alone, because in this case we would have an insufficient number of wrapping nonpolar groups surrounding the hydrogen bonds (i.e., we would invariably obtain ρ < 8).
Hydrogen-Bond Wrapping After Protein–Protein Association
To understand what individual UDHBs signal, we examined the protein–protein interface of 212 complexes from our exhaustive database, keeping in mind the difficulties in explaining and predicting binding sites on the basis of pairwise (p-p or h-h) interactions (24, 25). The overall (δ) and interface (δint) density of UDHBs on the protein surfaces were computed by calculating the total exposed surface area of the separated binding partners and the interface surface area [by subtracting the exposed surface area of the complex from that of the two separated monomers (26), as in ref. 1]. In 78 of the 212 complexes, we found a significantly higher value for the interface density of UDHBs: δint/δ > 1.5. In some cases, the density of structural defects at the interface was 7 times higher than the average density (Table 2).
Table 2.
Data on selected complexes extracted from the exhaustive and nonredundant structural database described in the main text
| Complex name (PDB ID code) | Yint | Y | C3,int | δ, 10−3 (Å−2) | δint, 10−3 (Å−2) |
|---|---|---|---|---|---|
| HLA antigen A-2 + β2-microglobulin (1i4f) | 6 | 36 | 66 | 1.58 | 3.21 |
| Ig light-chain dimer (1jvk) | 8 | 26 | 72 | 1.78 | 3.54 |
| Transthyretin dimer (1bm7) | 5 | 14 | 57 | 1.01 | 3.55 |
| Insulin dimer (6ins) | 4 | 12 | 51 | 2.80 | 4.61 |
| HIV-1 protease dimer + inhibitor (1a30) | 7 | 11 | 78 | 1.87 | 4.91 |
| SIV protease dimer (1siv) | 4 | 14 | 45 | 1.06 | 2.65 |
| CheY complex (1fqw) | 4 | 10 | 42 | 1.02 | 6.07 |
| Defensin dimer (1dfn) | 8 | 14 | 63 | 2.72 | 10.01 |
| Antitrypsin polymers (1d5s) | 14 | 22 | 165 | 1.01 | 2.76 |
| Bombyxin (1bon) | 4 | 5 | 45 | 0.60 | 3.02 |
| FcγRIII receptor + Ig (1e4k) B–C | 7 | 22 | 57 | 0.97 | 7.08 |
| Colicin + ligand (1bxi) | 6 | 12 | 57 | 0.92 | 3.97 |
| Colicin + ligand (1emv) | 5 | 11 | 60 | 0.86 | 3.60 |
| Serpin + ligand (1as4) | 14 | 31 | 183 | 1.40 | 2.02 |
| Troponin heterodimer (1pon) | 6 | 10 | 69 | 1.34 | 4.54 |
| MHC, antigen + receptor (1im9), A–D | 3 | 22 | 42 | 0.84 | 2.21 |
| MHC, antigen + ligand (1im9), A–C | 3 | 19 | 27 | 2.00 | 6.12 |
| De novo protein of α-2D (1qp6) | 8 | 12 | 96 | 1.65 | 3.67 |
Each complex is identified by its PDB ID code, and its association is defined by the following parameters: Y, number of UDHBs in the separated partners; Yint, number of UDHBs at the interface that become properly wrapped (green to gray) after binding; C3,int, number of intermolecular three-body correlations involving an overexposed hydrophobic group of one partner and a UDHB of the other partner; δ, density of UDHBs given as number of UDHBs per 1,000 Å2 averaged over the solvent-exposed surface area of both separated binding partners; and δint, density of UDHBs at the binding interface. The letters A–D label the chains in the complexes.
These results imply that the exclusion of water from structurally defective regions of the protein surface is an important factor in defining protein–protein associations. These hot spots (which involve not only nonpolar but also polar groups) should be distinguished from the exposed hydrophobic patches, although both are determined by the possibility of excluding water intermolecularly where it most counts in thermodynamic terms. In both cases, the lowering of the local degree of hydration entails a free-energy decrease: The hydrogen bond is stabilized, or the hydrophobe becomes less exposed to the solvent (5). This scenario also accounts for the stability of an alanine-based helix containing three lysine residues that deprive the backbone amide-carbonyl groups of water, thereby stabilizing the backbone hydrogen bonds (9).
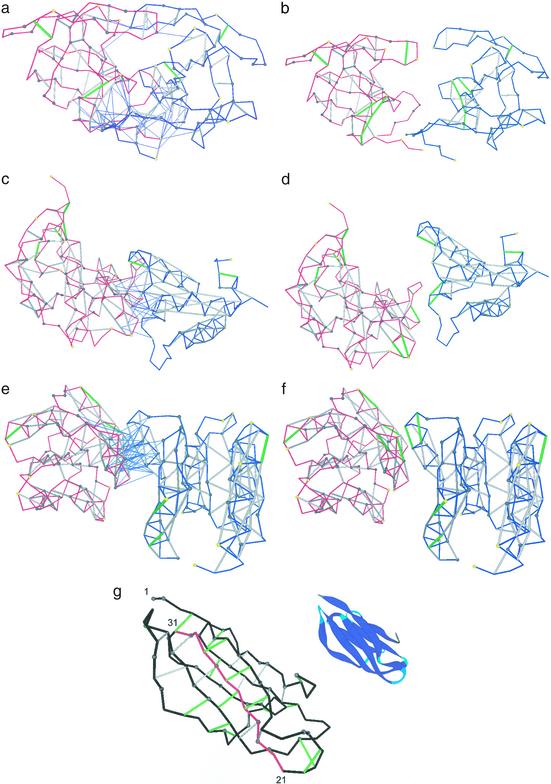
Fig. 3 displays three complexes and the separated binding partners. In the three cases, we see that the dehydration shells of the interface UDHBs are completed after binding: The overexposed aliphatic groups of the binding partner penetrates the dehydration domain of intramolecular UDHBs, thus compensating intermolecularly for defects in the monomeric structure. Induced fit lies outside the scope of this study.
Figure 3.
Three selected complexes and separated binding partners for the HIV-1 protease dimer (PDB ID code 1a30, a and b), colicin + ligand (PDB ID code 1emv, c and d), and CheY complex (PDB ID code 1fqw, e and f). The binding partners are represented by blue and red virtual-bond backbone chains; the hydrophobic residues containing >1 carbonaceous group are denoted as α-carbon spheres: gray if the residue is >60% buried and yellow otherwise. The backbone hydrogen bonds are indicated as lines joining α-carbons: gray if the bond is sufficiently dehydrated and green if it is a UDHB. A thin blue line joining an α-carbon with a hydrogen-bond center indicates that a residue in one molecule is engaged in one or several intermolecular three-body correlations contributing to the dehydration of an intramolecular hydrogen bond of the binding partner. (g) Sufficiently dehydrated hydrogen bonds and UDHBs for monomeric β2-microglobulin. The segment with the highest concentration of structural defects is highlighted in red and is part of the so-called βB–βC (residues 21–33) amyloidogenic fragment generated by treatment of β2-microglobulin with Acromobacter protease (23). The ribbon picture is an aid to the eye.
The first four complexes in Table 2 involve proteins (β2-microglobulin, Ig light chain, transthyretin, and insulin) known to be amyloidogenic under near-physiological conditions (23, 27–29). As expected, these proteins are marginally good wrappers of their hydrogen bonds in their monomeric state, with 12.5 < ρ < 13.1. Monomeric insulin (28) (ρ = 12.5) is a clear outlier vis-a-vis the statistics shown. However, after complexation, such proteins partially correct their structural defects by exclusion of water at their surface. Their ρ values (now computed by taking into account the intermolecular three-body correlations as depicted in Fig. 3 a–f) enter the “normal” range of ρ = 15.00 ± 2.05 after complexation.
These results hint at the need to introduce a new “field” to describe protein interactions: the gradient of the degree of hydration with respect to the position of a test hydrophobic moiety. Because hydrophobic moieties are solvent-structuring, the degree of hydration decreases as the hydrophobic group approaches, thus enhancing the stability of the intramolecular interaction (1, 5, 11).
This concept might prove useful in identifying the nucleation sites for amyloidogenic aggregation. To illustrate this aspect, we focus here on β2-microglobulin, although the conclusions hold for other known amyloidogenic proteins. We slid a window of a fixed number of residues along the primary sequence and identified the associated regions in the native structure; this is similar to a procedure used to identify hydrophobic nucleation sites in protein folding (30). Iterating this procedure, we identified the structural region containing the highest number of UDHBs (Fig. 3g). The associated fragment corresponds to the 21–33 window in the primary sequence. Precisely this peptide (produced by Acromobacter protease) is part of the so-called K3 fragment, which has been shown to possess high fibrillogenic propensity on its own (23). This implies that amyloid aggregation may be nucleated by an amyloidogenic region if, at the same time, the exclusion of water from that region after protein–protein association finds the highest thermodynamic benefit. We find it suggestive that the hot spot for water exclusion and the minimal amyloidogenic fragment coincide within the native structure.
To conclude, this work represents an attempt at correlating the interactive portion of a protein with inherent structural defects in its monomeric state.
Acknowledgments
We thank Prof. Y. Duan for making the Duan–Kollman trajectory (21) available for the purpose of this study and Profs. Yuji Goto, Robert Huber, Ridgeway Scott, R. Stephen Berry, Tobin R. Sosnick, and Karl F. Freed for enlightening discussions. This work was supported by National Science Foundation Grant MCB00-03722.
Abbreviation
- UDHB
underdehydrated hydrogen bond
Note Added in Proof.
The methodology and results presented here compare favorably with results from mutations of sites for association of proteins with ligands. For example, Fersht and coworkers (31) have carried out extensive mutations of the barnase site for association with the RNA substrate. They mutated the positively charged residues Lys-27, Arg-59, and His-102 to Ala. These mutations produced the expected decrease in activity, because they reduced the favorable electrostatic interactions of the protein with the negatively charged RNA. They also reported that the stability of barnase increased after mutation. We have observed that there are three underdehydrated backbone hydrogen bonds at the active site of the wild-type protein and that the mutated residues had aliphatic groups within their dehydration domains. The UDHBs at the active site are Ser-28–Ala-32, Asn-58–Gly-61, and Ser-85–His-102. Because the RNA removes surrounding water from such preexisting hydrogen bonds, which is energetically and thermodynamically favorable, we may make three conclusions. (i) The wild-type active site is operational, because this thermodynamically and energetically favorable water removal from the UDHBs acts in conjunction with the favorable electrostatics. In other words, the water removal after RNA–protein association is favored at the enzymatic site, and the water removal in turn strengthens the electrostatic interactions in the wild-type protein between the positively charged binding site and the negatively charged substrate. (ii) The mutations carried out by Fersht and coworkers not only reduced the electrostatic affinity for the substrate but also, by replacing a polar with a nonpolar residue, assured that the UDHBs of the wild-type protein are no longer underdehydrated, i.e., the UDHBs become properly dehydrated after mutation. (iii) Because the Ala residue of the mutant properly dehydrates the UDHBs of the wild-type protein, the stability of the protein itself increases as observed by Fersht and coworkers (31). This example illustrates the synergistic action of favorable water removal from the preformed hydrogen bonds in conjunction with the electrostatics at the enzymatic site.
References
- 1.Némethy G, Scheraga H A. J Phys Chem. 1962;66:1773–1789. , and erratum (1963) 67, 2888. [Google Scholar]
- 2.Némethy G, Scheraga H A. J Chem Phys. 1962;36:3382–3400. [Google Scholar]
- 3.Némethy G, Scheraga H A. J Chem Phys. 1962;36:3401–3417. [Google Scholar]
- 4.Scheraga H A. J Biomol Struct Dyn. 1998;16:447–460. doi: 10.1080/07391102.1998.10508260. [DOI] [PubMed] [Google Scholar]
- 5.Némethy G, Steinberg I Z, Scheraga H A. Biopolymers. 1963;1:43–69. [Google Scholar]
- 6.Yang A-S, Honig B. J Mol Biol. 1995;252:351–365. doi: 10.1006/jmbi.1995.0502. [DOI] [PubMed] [Google Scholar]
- 7.Myers J K, Pace N. Biophys J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avbelj F, Luo P, Baldwin R L. Proc Natl Acad Sci USA. 2000;97:10786–10791. doi: 10.1073/pnas.200343197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vila J A, Ripoll D R, Scheraga H A. Proc Natl Acad Sci USA. 2000;97:13075–13079. doi: 10.1073/pnas.240455797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández A. J Chem Phys. 2001;115:7293–7297. [Google Scholar]
- 11.Fernández A, Colubri A, Berry R S. Physica A. 2002;307:235–259. [Google Scholar]
- 12.Fernández A. Proteins Struct Funct Genet. 2002;47:447–457. doi: 10.1002/prot.10109. [DOI] [PubMed] [Google Scholar]
- 13.Ringe D. Curr Opin Struct Biol. 1995;5:825–829. doi: 10.1016/0959-440x(95)80017-4. [DOI] [PubMed] [Google Scholar]
- 14.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 15.Dobson C M. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 16.Koo E H, Lansbury P T, Jr, Kelly J W. Proc Natl Acad Sci USA. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobohm U, Scharf M, Schneider R. Protein Sci. 1993;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voet D, Voet J G. Biochemistry. New York: Wiley; 1990. [Google Scholar]
- 19.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahn R, Liu A, Luhrs T, Riek R, von Schroetter C, Lopez Garcia F, Billeter M, Calzolai L, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan Y, Kollman P A. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 22.Lin M-C, Mizabekov T, Kagan B L. J Biol Chem. 1997;272:44–47. doi: 10.1074/jbc.272.1.44. [DOI] [PubMed] [Google Scholar]
- 23.Kozhukh G V, Hagihara Y, Kawakami T, Hasegawa K, Naiki H, Goto Y. J Biol Chem. 2002;277:1310–1315. doi: 10.1074/jbc.M108753200. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sondermann P, Huber R, Oosthuizen V, Jacob U. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 26.Fraczkiewicz R, Braun W. J Comp Chem. 1998;19:319–333. [Google Scholar]
- 27.MacPhee C E, Dobson C M. J Mol Biol. 2000;297:1203–1215. doi: 10.1006/jmbi.2000.3600. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen L, Frokjaer S, Brange J, Uversky V N, Fink A L. Biochemistry. 2001;40:8397–8409. doi: 10.1021/bi0105983. [DOI] [PubMed] [Google Scholar]
- 29.Souillac P O, Uversky V N, Millett I S, Khurana R, Doniach S, Fink A L. J Biol Chem. 2002;277:12657–12665. doi: 10.1074/jbc.M109230200. [DOI] [PubMed] [Google Scholar]
- 30.Matheson R R, Jr, Scheraga H A. Macromolecules. 1978;11:819–829. [Google Scholar]
- 31.Meiering E M, Serrano L, Fersht A R. J Mol Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]