Abstract
We present fine mapping of a cis-acting nucleotide sequence found in the 5′ region of yellow fever virus genomic RNA that is required for RNA replication. There is evidence that this sequence interacts with a complementary sequence in the 3′ region of the genome to cyclize the RNA. Replicons were constructed that had various deletions in the 5′ region encoding the capsid protein and were tested for their ability to replicate. We found that a sequence of 18 nucleotides (residues 146 to 163 of the yellow fever virus genome, which encode amino acids 9 to 14 of the capsid protein) is essential for replication of the yellow fever virus replicon and that a slightly longer sequence of 21 nucleotides (residues 146 to 166, encoding amino acids 9 to 15) is required for full replication. This region is larger than the core sequence of 8 nucleotides conserved among all mosquito-borne flaviviruses and contains instead the entire sequence previously proposed to be involved in cyclization of yellow fever virus RNA.
Flaviviruses are small enveloped viruses. The virion contains a positive-strand RNA genome of about 10.7 kb encapsidated by 180 copies of the capsid (C) protein in a nucleocapsid core, which is enveloped by a lipid bilayer in which 180 copies of each of two transmembrane proteins, envelope (E) and membrane (M), are anchored (2, 9). The flaviviruses include a large number of serious human pathogens such as yellow fever virus (YFV), four serotypes of dengue virus (DENV), Japanese encephalitis virus, West Nile virus, and tick-borne encephalitis virus. Although good vaccines have been developed against some flaviviruses (YFV, Japanese encephalitis virus, and tick-borne encephalitis virus), they remain a serious health risk around the world.
The flavivirus RNA contains one large open reading frame flanked by 5′ and 3′ nontranslated regions that are required for replication and translation of the RNA (11). Analysis of the sequences of several flavivirus RNAs showed that short sequences close to the 5′ and 3′ ends are complementary, and we proposed that these sequences might function to cyclize the RNA during replication (5). The cyclization sequence at the 5′ end is located within the region encoding the N-terminal region of the capsid protein. Its 3′ counterpart is located in the 3′ nontranslated region. A core region of 8 nucleotides within the cyclization domain is conserved among all mosquito-borne flaviviruses (5). Khromykh et al. (7, 8) examined the importance of the cyclization sequences by analyzing the ability of truncated and mutated Kunjin virus (KUNV) RNA molecules to replicate in transfected cells. They found that deletion mutants lacking the sequence encoding the first 20 amino acids of KUNV capsid protein are unable to replicate (8). They further showed that a mutant RNA that had five changes within the 8-nucleotide core conserved sequence was unable to replicate. Compensating changes in the 3′ region that restored the complementarity of the cyclization sequence restored the ability of the RNA to replicate, although not to parental RNA levels (7). Thus, the ability of the RNA to cyclize is required for it to replicate. The exact sequence used in cyclization is not essential, but the wild-type sequence has a clear advantage.
Here we report the precise mapping of the 5′ sequence element required for YFV RNA replication. Of particular interest was the question of whether the core 8-nucleotide conserved sequence, whose 5′ element is UCAAUAUG, is the only required sequence or whether the longer sequence identified by Hahn et al. (5) as being involved in cyclization of YFV RNA was required. We found that the sequence required for replication of YFV RNA is identical to the entire cyclization sequence identified by Hahn et al. (5).
Construction of YFV replicons.
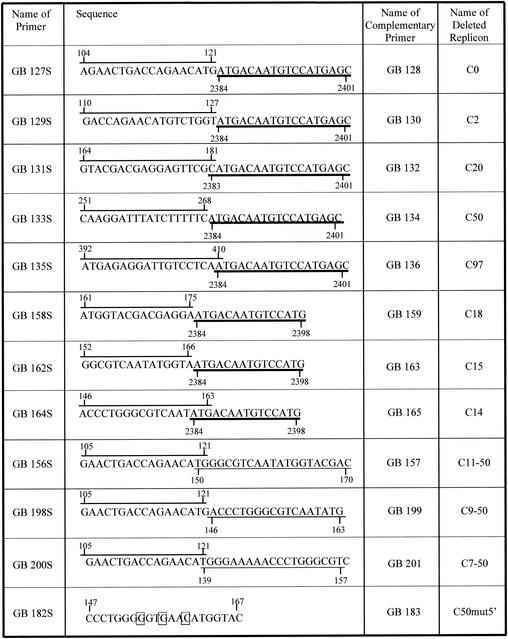
YFV replicons were constructed by using our previously described full-length YFV cDNA clone pBAC/FLYF.NI (13). Deletions were introduced into this plasmid by fusion PCR, as previously described (14). The primers used are shown in Fig. 1 and were designed to link sequences in the C protein directly to the start of the signal sequence for NS1. Thus, the entire sequence encoding prM and E, as well as various regions of C, was deleted in the replicons. Only the sequences of the plus-sense deletion primers are shown in the figure, and these are annotated to show where the sequence originates so that the reader can follow the deletions introduced. Because this sequence is coding, deletion of an integral number of triplets was required to maintain the reading frame. Also shown is a plus-sense primer used to introduce three changes into the cyclization sequence (mutant C50mut5′). Two outside primers used for PCR, one upstream of the NotI site and one downstream of the MluI site, are not illustrated. Once produced, the fusion PCR products were cloned into pBAC/FLYF.NI with restriction enzyme sites NotI and MluI. RNA was transcribed in vitro from the resulting clones and transfected into BHK cells with Lipofectamine (Invitrogen). At 24 h after transfection, the cells were treated with mouse anti-YFV serum (American Type Culture Collection) followed by a fluorescein isothiocyanate-conjugated anti-mouse antibody (Sigma) and examined for fluorescence.
FIG. 1.
Primers used to delete regions of capsid sequence. The primer sequences are annotated to show their chimeric nature, and nucleotide numbers are also shown. The overlined nucleotides are sequences in the capsid protein. The nucleotides underlined with a thick line are in the signal sequence for NS1. The nucleotides underlined with a thin line are present in the capsid sequence downstream of a deletion. Note that in some cases the origin of the sequences is ambiguous at the point of overlap. Only the plus-sense primers are illustrated; corresponding minus-sense primers are an exact complement of the plus-sense primers. Also illustrated is a primer used to introduce three mutations into the sequence of the cyclization element; the changed nucleotides are boxed.
Replicon C97.
A schematic diagram of the YFV genome with flanking sequences from the pBAC vector is shown in Fig. 2A. Below is illustrated the construction of C97, a replicon that contains essentially the entire mature capsid protein sequence fused directly to the signal sequence for NS1. Thus, prM and E have been deleted. This replicon is replication competent. When RNA is transcribed in vitro and used to transfect BHK cells, replication occurs as shown by the accumulation of YFV proteins in the transfected cells, which can be detected by an immunofluorescence assay (data not shown, but see below). Although this immunofluorescence assay is only an indirect measure of RNA replication, a positive result does prove that RNA replication has occurred because the amount of input RNA is insufficient for translation of sufficient protein to be readily detectable by our fluorescence assay. No progeny virus is produced in the assay because of the lack of prM and E.
FIG. 2.
Organization of the YFV genome and construction of replicons. (A) The first line diagrams the genome of YFV. Adjoining vector sequences are illustrated. The open boxes show the coding regions for the various proteins, which are produced as a polyprotein that is processed by viral and cellular proteases. The positions of the NotI and MluI sites used to clone PCR products into the clone are shown. The dark boxes marked CS are the cyclization sequence elements. The second line illustrates the construction of clone C97, which has all of the C protein sequence joined directly to the signal sequence for NS1. Primers GB135S and GB136 were used to delete the sequences of prM and E, together with outside primer GB4 and a primer upstream of the NotI site. The diagrams are to scale, and the third line shows a scale in kilobases. (B) Diagram of the cyclized genome with the 18 bp that stabilize it shown together with nucleotide numbers in the YFV genome. The 8-nucleotide core region is boxed. Note the unpaired G in the 5′ element.
Replicons with more extensive deletions.
Figure 2B illustrates sequences at the 5′ and 3′ ends of YFV RNA that are complementary. These sequences are capable of forming 18 bp with a single G looped out of the 5′ sequence (5). The core conserved 8-nucleotide sequence present in all mosquito-borne flaviviruses is boxed. To test the possible importance of these sequences, and to map more precisely the cis-acting sequence required for RNA replication and translation, further truncations of capsid sequence were introduced and tested for the ability of the RNA to replicate sufficiently well to produce proteins visible by immunofluorescence, as illustrated in Fig. 3. In this figure, the nucleotide sequence encoding the first 20 amino acids of the capsid protein and the encoded amino acid sequence are shown. The 19-residue sequence complementary to 3′ nucleotides and the 8-nucleotide conserved core sequence (Fig. 2B) are again indicated. Amino acids are numbered so that the initiating methionine is number 0, because this residue is removed from the mature YFV capsid protein (1). The deletion constructs are illustrated below. As for C97, in all constructs the capsid sequence is fused directly to the signal sequence for NS1, thus deleting all of prM and E. The region of the capsid protein present in each construct is indicated approximately by the name assigned to it, but because of the limitation that an integral number of codons must be deleted to maintain the reading frame, the juxtaposed sequences sometimes maintain the capsid sequence beyond the deletion point (Fig. 1).
FIG. 3.
Schematic representation of deletion constructs tested for RNA replication. The sequence encoding the first 21 amino acids of the capsid protein is shown together with the encoded amino acid sequence. The initiating Met is numbered 0 because it is removed from the mature protein. Nucleotide numbers are shown beginning at the 5′ end of the genome. The gray box encloses the cyclization sequence illustrated in Fig. 2. The 8-nucleotide core sequence is indicated by the white box, and the three nucleotides changed in construct C50mut 5′ are boxed (also Fig. 1). Below are illustrated the deletion constructs tested and the results of the replication assay. Constructs are named as in Fig. 1.
Definition of the 3′ boundary of the cis-acting replication element.
We first confirmed the results of Khromykh et al. for KUNV (8) by examining the YFV constructs C50 (which contains residues 0 to 50 of the capsid protein) and C20 (which contains residues 0 to 20 of the capsid protein). These replicons were capable of replication, as shown by immunofluorescence assay of transfected cells (results not shown). However, deletion of the entire capsid protein except for the N-terminal one or three residues (C0 and C2) gave rise to constructs that could not replicate, as evidenced by the total absence of fluorescent cells (results not shown).
In order to further delimit the RNA sequences essential for replication, we examined constructs C18, C15, and C14, and representative results for C15 and C14 are shown in Fig. 4. C18 and C15 (Fig. 4) were replication competent as evidenced by the immunofluorescent cells. C14, however, was only weakly positive (Fig. 4). Thus, the YFV RNA sequence encompassing nucleotides 1 to 163 allows some replication, but 1 to 3 additional nucleotides are required for full replication. For full replication, the 3′ boundary lies between residues 164 and 166.
FIG. 4.
Replication of YFV RNA deletion constructs. BHK cells were transfected with YFV deletion RNA C15, C14, C9-50, or C11-50, as indicated. A negative control is also shown. At 48 h posttransfection, the cells were fixed with methanol, treated with mouse anti-YFV antiserum, and stained with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G.
5′ boundary of the sequence element.
To define the 5′ boundary of the sequence element, we used C50 to construct additional deletions. In these constructs, various regions of the 5′ capsid sequence were deleted (Fig. 1 and 3). For example, construct C7-50 contains a deletion of residues 1 to 6 of the capsid protein so that the parts of the capsid sequence present are those encoding residues 0 and 7 to 50 linked to the signal sequence for NS1.
Constructs C7-50 and C9-50 were replication competent (the results for C9-50 are shown in Fig. 4). However, construct C11-50 was incapable of replication (Fig. 4). Since C9-50 has a deletion of YFV nucleotides 122 to 145 and C11-50 has a deletion of nucleotides 122 to 149, the 5′ boundary of the element lies between nucleotides 146 and 148. Together, these results define the RNA sequence needed for YFV RNA replication within narrow limits.
Mutagenesis of the sequence element.
In order to confirm that it is the nucleotide sequence that is required for RNA replication and not the encoded protein sequence, we made a mutant construct in which three nucleotides within the required sequence element were changed without disturbing the amino acid sequence (Fig. 1 and 3). This construct, C50mut5′, was unable to replicate (results not shown), confirming that it is the nucleotide sequence that is important.
Importance of the YFV cyclization elements.
Cyclization of the YFV genome is illustrated in Fig. 2B. This circular form has a calculated free energy of −34 kcal (5). The 5′ sequence element involved in cyclization spans residues 147 to 165 of the YFV genome. Thus, the 5′ and 3′ boundaries of the sequence element required for replication and translation of the RNA that we have determined in this study are in perfect agreement with the boundaries of this cyclization element, and we propose that it is this cyclization sequence that forms the required element. It is clear that the 8-nucleotide core conserved element is not sufficient for RNA replication and translation, at least in YFV. It will be of interest to determine if this is also true for other flaviviruses, as well as to examine the 3′ component of the cyclization sequence. We have found that a chimeric virus containing the YFV genome in which C, prM, and E have been replaced by the corresponding proteins of DENV-2 is defective in RNA replication (J. Corver and J. H. Strauss, unpublished observations). The cyclization sequences of YFV and DENV-2 are very similar but not identical (5), and this provides further evidence that the full sequence element is required.
Khromykh et al. (7) have provided evidence that cyclization is required for flavivirus RNA replication. Their data also indicate that, although the exact sequence of the cyclization domain is not essential for replication, the wild-type sequence has an advantage. More-extensive mutagenesis experiments would be useful in determining the extent of sequence conservation required. It is possible, for example, that changes within the core conserved region might have a larger effect than changes outside this domain, in either case assuming maintenance of complementarity between the 5′ and 3′ elements. Certainly, the precise conservation of this 8-nucleotide core among all mosquito-borne flaviviruses suggests that the exact sequence is important and recognized by some element of the RNA replication machinery. It is possible that the function of the remaining, nonconserved base pairs might be to stabilize the circular molecule so that the double-stranded 8-nucleotide core can be recognized by the replicase. Such a scenario is also consistent with the results for C14, which has the entire core sequence but lacks the two 3′ nucleotides involved in cyclization. This RNA is able to replicate but not as vigorously as a construct with the entire cyclization sequence.
It is not known which stage of RNA replication requires cyclization of the RNA. The genome of YFV is translated into one large polyprotein once it is introduced into the cytosol, by either infection or transfection. Cleavage of the polyprotein by cellular and viral proteases results in the production of the individual proteins required to form an RNA replicase responsible for minus-strand RNA synthesis, which is used in turn as a template for synthesis of more genomic RNA. Further study is required to determine whether the synthesis of minus-strand templates or of plus-strand genomes, or both, requires cyclization of the RNA. It is also possible that cyclization could be involved in the translation of the viral RNA, since bringing the two ends together could influence the interaction of the RNA with the translation machinery.
Cyclization in RNA viruses.
A number of RNA viruses, both plus and minus stranded, have been shown to have genomic RNAs that cyclize or that may potentially cyclize. One of the best-studied examples, and one that is perhaps most closely related to flavivirus cyclization, is that of alphaviruses. The plus-sense RNA genome of Sindbis virus is 11.7 kb, only slightly larger than the YFV genome. The Sindbis virus genome has been shown elsewhere to be circular in the virion (6). Although the sequences responsible for cyclization have not been identified, thermodynamic studies have shown that cyclization results from hydrogen bonding, that the circular form is stable at 37°C under physiological conditions, and that the number of base pairs involved is similar to the 11 to 19 bp predicted for different flaviviruses (3). Cyclization may have evolved so that the replicase is forced to interact with both ends of the RNA molecule in order to initiate synthesis, thus ensuring that only full-length RNAs are copied. Another possible function of cyclization in these viral RNAs is the control of the tradeoff between use of the RNA as a messenger for translation and use of it as a template for RNA synthesis, which could lead to blockades if both were to occur simultaneously. For alphaviruses, it is known that promoter and enhancer sequences near the 5′ end of the molecule are needed for efficient production of minus-strand RNA from a plus-strand template (4, 10, 12). Cyclization would bring these 5′ sequences near the 3′ promoter required for initiation of minus-strand synthesis. It is possible that similar interactions may occur in flaviviruses.
Acknowledgments
J.C. was supported by a Gosney Fellowship from Caltech. This work was supported by NIH grant AI 10793.
REFERENCES
- 1.Bell, J. R., R. W. Kinney, D. W. Trent, E. M. Lenches, L. Dalgarno, and J. H. Strauss. 1985. Amino-terminal amino acid sequences of structural proteins of three flaviviruses. Virology 143:224-229. [DOI] [PubMed] [Google Scholar]
- 2.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 3.Frey, T. K., D. L. Gard, and J. H. Strauss. 1979. Biophysical studies on circle formation by Sindbis virus 49S RNA. J. Mol. Biol. 132:1-18. [DOI] [PubMed] [Google Scholar]
- 4.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 6.Hsu, M. T., H. J. Kung, and N. Davidson. 1973. An electron microscope study of Sindbis virus RNA. Cold Spring Harbor Symp. Quant. Biol. 38:943-950. [DOI] [PubMed] [Google Scholar]
- 7.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niesters, H. G. M., and J. H. Strauss. 1990. Mutagenesis of the conserved 51-nucleotide region of Sindbis virus. J. Virol. 64:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 12.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Most, R. G., J. Corver, and J. H. Strauss. 1999. Mutagenesis of the RGD motif in the yellow fever virus 17D envelope. Virology 265:83-95. [DOI] [PubMed] [Google Scholar]
- 14.Yao, J. S., E. G. Strauss, and J. H. Strauss. 1996. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J. Virol. 70:7910-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]