Abstract
Regulation of ciliary and flagellar motility requires spatial control of dynein-driven microtubule sliding. However, the mechanism for regulating the location and symmetry of dynein activity is not understood. One hypothesis is that the asymmetrically organized central apparatus, through interactions with the radial spokes, transmits a signal to regulate dynein-driven microtubule sliding between subsets of doublet microtubules. Based on this model, we hypothesized that the orientation of the central apparatus defines positions of active microtubule sliding required to control bending in the axoneme. To test this, we induced microtubule sliding in axonemes isolated from wild-type and mutant Chlamydomonas cells, and then used electron microscopy to determine the orientation of the central apparatus. Transverse sections of wild-type axonemes revealed that the C1 microtubule is predominantly oriented toward the position of active microtubule sliding. In contrast, the central apparatus is randomly oriented in axonemes isolated from radial spoke deficient mutants. For outer arm dynein mutants, the C1 microtubule is oriented toward the position of active microtubule sliding in low calcium buffer, but is randomly oriented in high calcium buffer. These results provide evidence that the central apparatus defines the position of active microtubule sliding, and may regulate the size and shape of axonemal bends through interactions with the radial spokes. In addition, our results indicate that in high calcium conditions required to generate symmetric waveforms, the outer dynein arms are potential targets of the central pair-radial spoke control system.
The mechanism of eukaryotic ciliary and flagellar motility is based on a sliding microtubule model in which the dynein arms attached to the nine doublet microtubules generate force against adjacent doublets, causing the microtubules to slide past one another (1–5). Because the doublet microtubules are anchored to the cell by their attachment to the basal bodies, microtubule sliding causes the axoneme to bend. However, dynein generates force in a single direction (6). Therefore, all of the dynein arms cannot be simultaneously active. Moreover, to generate the complex waveforms observed for beating cilia and flagella, the dynein arms must be asymmetrically regulated around the circumference and along the length of the axoneme.
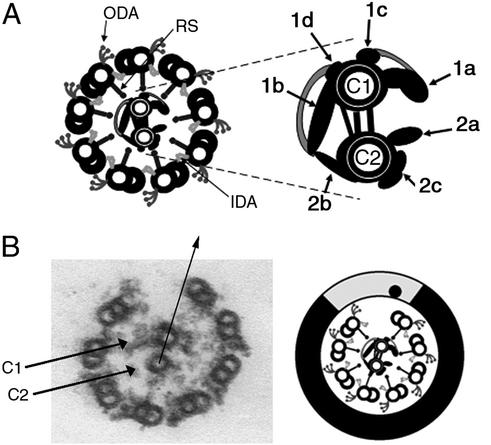
One challenge is to determine how asymmetric regulation of dynein activity is achieved. The only structure within the axoneme that exhibits obvious asymmetry is the central apparatus. The two tubules of the central apparatus are structurally and biochemically distinct (Fig. 1A) (7–10). The C1 tubule has two long projections (1a and 1b) and two short projections (1c and 1d). The C2 tubule has three short projections termed 2a, 2b, and 2c. The central apparatus is comprised of at least 23 polypeptides, subsets of which are unique to C1 and C2 (7, 8). This inherent structural and biochemical asymmetry of the central apparatus may be indicative of functional specialization.
Figure 1.
(A) Transverse section of flagellar axoneme. The outer and inner dynein arms (ODA, IDA) and radial spokes (RS) are attached to the outer doublet microtubules. The central apparatus is enlarged and offset. Central pair projections are labeled according to Mitchell and Sale (10). (B) Electron micrograph and accompanying diagram, axoneme transverse section after microtubule sliding. The C1 and C2 microtubules of the central pair are labeled. The arrow defines the orientation of the central pair. The point at which the arrow intersects the circumference of the doublets is indicated by a dot on the accompanying diagram. The position of active microtubule sliding includes the area lightly shaded. The inactive area includes the remainder of the axoneme (dark shading).
Substantial evidence has implicated the central apparatus and radial spokes in modulating dynein activity to locally control microtubule sliding (for example, ref. 11; reviewed in refs. 12–15). These studies support a model in which the central pair projections make contact with the radial spoke heads and transmit a signal to the dynein arms (Fig. 1A). The molecular mechanism of this signal transduction pathway is not well understood, but may include a combination of mechanical interactions and posttranslational modifications of axonemal components.
In many organisms, the central apparatus rotates within the nine doublet microtubules (16). In these organisms, the central pair projections may periodically interact with subsets of radial spokes, distributing an ordered and sequential signal to the dynein arms (17, 18). We hypothesized that if the asymmetry of the central apparatus specifies dynein activity on subsets of doublet microtubules, then the orientation of the central apparatus would correlate with the location of dynein-driven microtubule sliding. To test this, we induced microtubule sliding in Chlamydomonas axonemes by using an assay that uncouples sliding from flagellar beating, and then prepared the axonemes for electron microscopy. We discovered that, in wild-type axonemes, the C1 microtubule of the central pair is predominantly oriented toward the position of active sliding. These results provide evidence that the asymmetry of the central apparatus specifies the location of dynein activity, and supports a model in which the central apparatus plays a role in controlling the shape of flagellar bends.
Materials and Methods
Chlamydomonas Strains and Cell Culture.
Chlamydomonas reinhardtii strain A54-e18 (nit1-1, ac17, sr1, mt+) was obtained from Paul Lefebvre (University of Minnesota, St. Paul) and the cpc1–2 strain from David Mitchell (State University of New York, Syracuse). The strains 137c, pf6, pf14, pf17, pf28, oda1, and ida1 were obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). Motility and structural phenotypes for all strains are noted in Table 1. Cells were grown in constant light in TAP media (27).
Table 1.
Chlamydomonas strains used in this study
| Mutant | Structural/motility defect |
|---|---|
| A54-e18 | No axonemal structural defect/wild-type motility (19) |
| 137c | No axonemal structural defect/wild-type motility (20) |
| oda1, pf28 | Lack outer dynein arms/flagella beat with one-half frequency (21, 22) |
| ida1 | Lacks inner dynein arm subform I1/reduced beat frequency, altered waveform (23) |
| pf6 | Central apparatus, lacks 1a projection/flagella paralyzed or twitch (8, 24) |
| cpc1 | Central apparatus, lacks 1b projection/reduced beat frequency (10) |
| pf14 | Lacks radial spokes/paralyzed flagella (25) |
| pf17 | Lacks radial spoke heads/paralyzed flagella (26) |
References are indicated in parentheses.
Axoneme Isolation and the Microtubule Sliding Assay.
Flagella were severed from cell bodies by the dibucaine method (28) and isolated by differential centrifugation in buffer A (10 mM Hepes, pH 7.4/5 mM MgSO4/1 mM DTT/0.5 mM EDTA/50 mM potassium acetate). Axonemes were isolated by adding Nonidet P-40 (Calbiochem) for a final concentration of 0.5% (wt/vol). The axonemes were pelleted and resuspended in buffer A.
The microtubule sliding assay was based on previously described methods (2, 29). Axonemes were pelleted and resuspended in reactivation buffer (buffer A, 1 mM ATP). An equal volume of sliding buffer (buffer A, 1 mM ATP, 4 μg/ml protease) was added to the axonemes and mixed. We used either Nagarse protease (Type XXVII Protease; Sigma) or Type VIII protease (catalog no. P-5380, Sigma). Either protease produced the same results. For calcium treatments, buffer A, the reactivation buffer, and the sliding buffer included CaCl2 for a final concentration of 10−4 M free calcium according to the calculations of Wakabayashi et al. (30). Sliding was monitored by darkfield microscopy (31). Exactly 3 min after the addition of sliding buffer, 8% glutaraldehyde was added to the sample for a final concentration of 1%, and the sample was processed for electron microscopy.
Electron Microscopy.
Axonemes were pelleted and fixed in 1% glutaraldehyde and 1% tannic acid in 0.1 M sodium cacodylate, and postfixed in 1% osmium tetroxide. The samples were stained with 1% aqueous uranyl acetate, dehydrated in a graded series of ethanol, and embedded in EMbed-812 resin. Uniform gold-silver sections were mounted on copper grids, stained with uranyl acetate and Reynold's lead citrate, and examined in a JEOL 100CX transmission electron microscope. Glutaraldehyde, osmium tetroxide, tannic acid, sodium cacodylate, lead nitrate, and copper grids were purchased from EM Science. For data analysis, negatives were digitized with an optical scanner.
Statistical Analysis of Images.
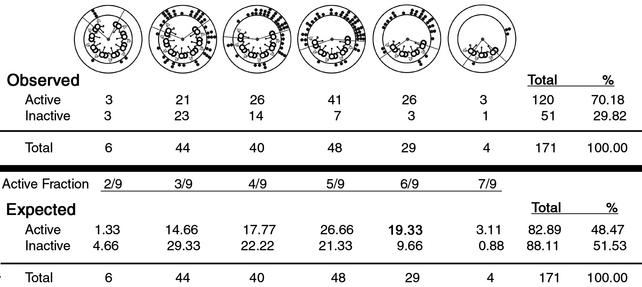
Transverse sections of axonemes lacking doublet microtubules were considered axonemes in which microtubule sliding was induced after the addition of ATP and protease. For each experiment, every transverse section in which sliding occurred and the C1 and C2 microtubules could be distinguished was analyzed for central apparatus orientation. All images were oriented with the dynein arms projecting clockwise, the axoneme viewed proximal to distal (Fig. 1B). Axoneme transverse sections corresponded to one of six different sliding patterns (Fig. 2). For each pattern, the axoneme was partitioned into active and inactive areas. The active area is the position in which microtubule sliding resulted in the loss of doublets from the remainder of the axoneme; this area included the exposed A-tubule and dynein arms and extended to the exposed B-tubule (Fig. 1B). The inactive area is the remainder of the axoneme.
Figure 2.
Quantitative analysis of central apparatus orientation. The example shown is wild-type axonemes. We calculated the expected number of events for both the active and inactive areas for each pattern of microtubule sliding, if the orientation of the central apparatus was random. Expected events = (fraction of the axoneme in the active/inactive area) × (total events for sliding pattern). The expected events in the active area were totaled for all sliding patterns (Total), expressed as a percentage of the total number of transverse sections (%), and compared with the percentage of observed events.
The orientation of the central pair was defined by an arrow drawn parallel to the axis of the central pair, from the center of the C2-tubule through the center of the C1-tubule (Fig. 1B). The point at which the arrow intersected the circumference of the doublet microtubules was indicated by a dot on the diagram for that particular sliding pattern (Fig. 1B). This method allowed us to easily view the orientation of the central apparatus for all transverse sections collected from a single experiment (Fig. 2).
To determine whether the correlation between the orientation of the central apparatus and the position of active microtubule sliding was significant, we calculated how many times we expected to observe the C1 microtubule oriented toward the active area, if central apparatus orientation was random. This calculation was made for each pattern of microtubule sliding, then totaled for all patterns, and finally expressed as a percentage of the total number of transverse sections included in the analysis (expected active percent, Fig. 2). We used the χ2 test to determine significance of differences between the number of observed and expected events (for analysis of circular arrangements see ref. 32). Probability values were calculated by using the CHIDIST function in Microsoft excel. For each strain and condition, our results represent data collected from a minimum of three independent experiments.
Results and Discussion
The Orientation of the C1 Central Tubule Correlates with the Location of Dynein-Driven Microtubule Sliding.
We hypothesized that if the asymmetry of the central apparatus is involved in modulating dynein activity, then the orientation of the central apparatus would correlate with the location of dynein-driven microtubule sliding. To test this, we analyzed the orientation of the central apparatus in flagellar axonemes isolated from Chlamydomonas cells, using a combined functional and structural approach, similar to that previously described (33, 34). The addition of ATP and protease to isolated axonemes uncouples microtubule sliding from axonemal bending. In control experiments with no ATP or protease, microtubule sliding was not detected by light microscopy, and subsequent electron microscopic examination revealed that all axonemal transverse sections contained nine doublet microtubules. Therefore, the loss of doublet microtubules from axonemes after the addition of ATP and protease most likely results from dynein-driven microtubule sliding. We defined the portion of the axoneme that lacked doublet microtubules as the location of active microtubule sliding (Fig. 1B).
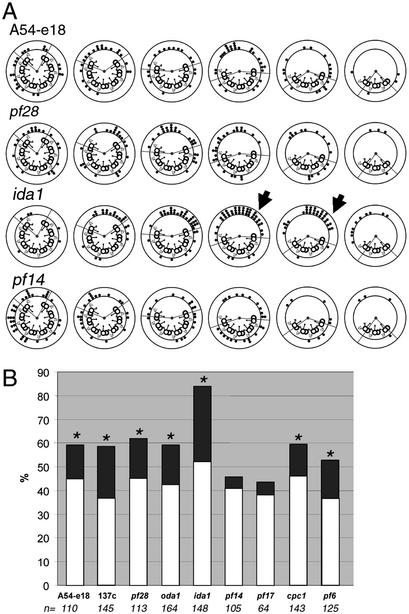
We first examined two strains, A54-e18 and 137c, that are wild-type for flagellar structure and motility. Initial inspection of axoneme transverse sections revealed that the outer edge of the C1 microtubule (including the 1c and 1d projections) was predominantly oriented toward the position of active microtubule sliding (Fig. 1B). For every transverse section examined, the orientation of the C1 microtubule was noted as a dot on a diagram for the corresponding sliding pattern (Figs. 1B and 2). To determine whether our observation was statistically significant, we devised a method for quantifying central pair orientation. For the total number of transverse sections examined, we calculated how many times we expected to observe the C1 microtubule oriented toward the position of active microtubule sliding, if the orientation of the central apparatus was random. We compared the expected value with the number of times the C1 microtubule was observed oriented toward the active position (see Materials and Methods and Figs. 1B and 2).
For wild-type axonemes, the C1 microtubule of the central apparatus was oriented toward the position of microtubule sliding in a significantly greater number of axonemes than expected, if central apparatus orientation was random (Fig. 3). We repeated the experiment using two different strains, pf28 and oda1, which lack the outer dynein arms. For both strains, the C1 microtubule of the central apparatus was predominantly oriented toward the position of active microtubule sliding (Fig. 3). Recent evidence has revealed that inner dynein arm subform I1 may play a role in controlling flagellar bending (23, 31, 35–39). Therefore, we analyzed central apparatus orientation in axonemes isolated from the I1 defective strain ida1. Remarkably, the C1 microtubule in ida1 axonemes was oriented toward the position of active microtubule sliding in >80% of transverse sections (Fig. 3). These combined results indicate that neither inner dynein arm I1 nor the outer dynein arms are exclusive targets for regulation by the central apparatus.
Figure 3.
Central apparatus orientation, low-calcium buffer. (A) Examples of data obtained from axonemes isolated from four strains after microtubule sliding. The lines drawn from the center of each cross section delineate the active and inactive areas defined in Fig. 1B. The dots represent the orientation of the C1 microtubule for individual transverse sections. Particularly striking examples of the C1 microtubule oriented toward the position of microtubule sliding are indicated by arrows. To view all data, see Fig. 6. (B) Percent of transverse sections in which the C1 microtubule was oriented toward the active area. Light bars indicate the expected percentage of events. The difference between the expected and observed percentages is darkly shaded. Strains in which this difference is significant are indicated by asterisks (χ2 test; P < 0.03). The total number of transverse sections included is indicated below each mutant.
Several studies have indicated that the C1 central microtubule plays a role in regulating dynein-driven microtubule sliding. Isolated axonemes lacking C1 cannot be reactivated to beat under physiological conditions (for reactivation under nonphysiological conditions see refs. 30 and 40–42). However, microtubule sliding velocities can be measured by using an in vitro assay (31). Sliding velocity is reduced in mutant axonemes that lack the C1 microtubule (pf16). However, sliding velocities in mutants lacking the 1a (pf6) or 1b (cpc1) projections are wild-type (31). To determine whether the 1a or 1b projections affect central apparatus orientation, we performed our analysis by using axonemes isolated from the pf6 and cpc1 mutants. In both cases, the C1 microtubule was predominantly oriented toward the position of active microtubule sliding (Fig. 3). Therefore, the remaining 1c and 1d projections appear to play an important role in regulating dynein activity to modulate flagellar motility. Interestingly, despite wild-type microtubule sliding velocity (31) and apparent wild-type orientation of the C1 microtubule toward the position of microtubule sliding (this study), pf6 cells have paralyzed flagella. The defect in pf6 appears to be in coordinating dynein activity to produce waveforms characteristic of wild-type motility. One possibility is that the central apparatus does not effectively rotate in pf6 axonemes. An assessment of central apparatus rotation in this mutant has not been reported.
Given the spatial separation of the central apparatus and outer doublet microtubules, mechanical coupling of the C1 microtubule with the dynein arms most likely requires the radial spokes. Therefore, we analyzed central apparatus orientation in axonemes isolated from mutants with radial spoke defects. For axonemes isolated from both pf14 and pf17, we found no correlation between the orientation of the central apparatus and regions of active microtubule sliding (Fig. 3). As predicted, the central apparatus is apparently uncoupled from dynein activity in radial spoke defective mutants. Based on these observations and our previous results (31), our data support a model in which the 1c and 1d projections of the C1 microtubule modulate dynein activity on subsets of doublet microtubules through interactions with the radial spokes.
It should be noted that for each strain, the number of doublet microtubules which slide away from the axoneme is not entirely random. For example, for axonemes isolated from both 137c and pf6 cells, ≈64% of transverse sections lacked only one or two microtubules. In both cases, the central apparatus was predominantly oriented toward the position of active microtubule sliding. In comparison, 42% of transverse sections of pf14 axonemes lack 3, 4, or 5 microtubules (similar to A54-e18, 46%), but in pf14 axonemes, the orientation of the central apparatus is random. Therefore, the correlation of central apparatus position with active microtubule sliding is not affected by the distribution of the number of microtubules that have slid away from the axoneme. For our complete data set and statistical analyses, see Fig. 6 and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org.
In High Calcium, the Central Apparatus Is Randomly Oriented in Mutants Lacking Outer Dynein Arms.
Chlamydomonas flagella beat with either of two distinct waveforms. The cells typically swim forward by using an asymmetric waveform. However, in response to bright light, the flagella switch to a symmetric waveform, allowing the cells to swim in reverse (reviewed, ref. 43). This change in waveform is caused by an influx of calcium ions and is mediated by calcium binding proteins that are components of the axoneme. For example, isolated axonemes can be reactivated to beat with an asymmetric waveform in buffers of pCa>8 or with a symmetric waveform in buffers of pCa4 (44–47). We recently demonstrated that the central apparatus and radial spokes are involved in regulating dynein activity in response to changes in free calcium (48). Therefore, we induced microtubule sliding in pCa4 buffer and analyzed the orientation of the central apparatus in wild-type and mutant axonemes.
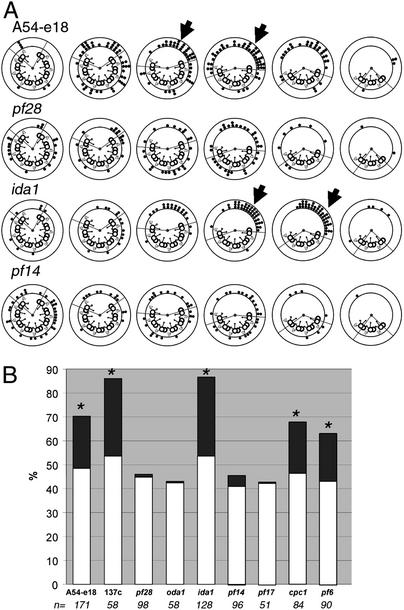
In high calcium buffer, the C1 microtubule was oriented toward the area of microtubule sliding in axonemes isolated from wild-type, ida1, pf6, and cpc1 cells; the central apparatus remained randomly oriented in pf14 and pf17 axonemes (compare Figs. 3 and 4). Importantly, however, the central apparatus was randomly oriented in pf28 and oda1 in pCa4 buffer (Fig. 4). This result is in sharp contrast to our previous observation that the C1 central tubule in pf28 and oda1 axonemes was significantly oriented toward the area of microtubule sliding in low calcium conditions (Fig. 3). Evidently, in the absence of the outer dynein arms, regulatory cues produced by the central apparatus are uncoupled from microtubule sliding under high calcium conditions. This result suggests a role for the outer dynein arms in calcium-induced changes in waveform.
Figure 4.
Central apparatus orientation, high calcium buffer. (A) Examples of data obtained from axonemes isolated from four strains after microtubule sliding in pCa4 buffer. The lines drawn from the center of each cross section delineate the active and inactive areas defined in Fig. 1B. The dots represent the orientation of the C1 microtubule for individual transverse sections. Particularly striking examples of the C1 microtubule oriented toward the position of microtubule sliding are indicated by arrows. To view all data, see Fig. 6. (B) Percentage of transverse sections in which the C1 microtubule was oriented toward the active area as shown in A. Light bars indicate the expected percentage of events. The difference between the expected and observed values is darkly shaded. Strains in which this difference is significant are indicated by asterisks (χ2 test; P < 0.01). The total number of transverse sections included is indicated below each mutant.
This conclusion is supported by the observation that outer dynein armless mutants do not exhibit normal waveform changes in response to increasing calcium concentration (22). Additional support is provided by the analysis of suppressor mutations that restore motility to both radial spoke and central apparatus defective mutants without restoring the missing structures (49). These mutations occur in genes encoding heavy chains of outer dynein arms (49–51). One mutation, sup-pf-2, is in the gene encoding the γ heavy chain subunit (51) and results in the asymmetric assembly of the outer dynein arms on subsets of doublet microtubules. Although the sup-pf-2 mutation restores motility to mutants lacking the radial spokes or central apparatus, flagella of these double mutants beat with a predominantly symmetric waveform (49, 52, 53). These observations not only implicate the outer dynein arms in calcium-induced changes in waveform, but also further illustrate the importance of establishing asymmetric regulation of dynein activity. This study does not exclude a role for the inner dynein arms or dynein regulatory complex in calcium mediated changes in motility (see refs. 35 and 54).
Precision of Central Apparatus Orientation in High and Low Calcium Buffer.
To more precisely define central apparatus orientation, we divided the area of active microtubule sliding into two equal halves, clockwise and counterclockwise (Fig. 7, which is published as supporting information on the PNAS web site). The C1 microtubule was significantly oriented toward the clockwise half of the active area in axonemes isolated from 137c, ida1, cpc1, and pf6 in buffers of pCa>8 and in axonemes isolated from 137c, A54-e18, ida1, and cpc1 in pCa4 buffer. For 137c, ida1, and cpc1 axonemes, the orientation in the clockwise direction was more pronounced in high-calcium buffer. The central apparatus is reported to rotate in buffers of both low and high calcium in Chlamydomonas (55). Although the results of our assay do not demonstrate that the central apparatus rotates, the observed orientation of the central pair is consistent with rotation in the clockwise direction.
Central Apparatus Twist and Control of Flagellar Beating.
The central apparatus in Chlamydomonas, as well as in other organisms, is reported to twist (9, 18, 55). In each case, the twist is left-handed, though the pitch of the twist is variable. If the central apparatus maintains a shallow twist in our assay, the C1 microtubule would only affect dynein activity along a fraction of the length of any particular doublet. However, in analyses of the most extended longitudinal sections viewed in our axoneme preparations, the central pair of microtubules does not appear to twist. The conditions of the sliding assay that uncouple microtubule sliding from flagellar beating may eliminate central apparatus twist. Therefore, we predict that the C1 microtubule uniformly affects dynein activity along the entire length of a particular doublet microtubule in our assay. Our data do not address the role of central apparatus twist during beating. Most likely, twist allows for regulation of dynein activity along the length of the axoneme as predicted by Omoto and Kung (18).
Similar analyses of echinoderm sperm flagella (33, 56) and mussel gill cilia (34) have been performed. The central pair does not appear to rotate in these organisms (for imposed rotation in sea urchin sperm, see refs. 57 and 58). Although sperm cells and gill cilia beat with different waveforms, the induction of microtubule sliding results in breaks between specific doublets that correlate with dynein activity (59). In both cases, the central apparatus maintains a specific orientation relative to the position of active sliding. These results imply that a single microtubule of the central apparatus may provide either positive or negative regulatory cues for microtubule sliding (60). Therefore, in these cell types, specific interactions of the radial spokes with the C1 microtubule may positively or negatively control dynein activity by a switching mechanism that does not involve central apparatus rotation. The C1 and C2 microtubules were not distinguished in these experiments, though the central apparatus is clearly an asymmetric structure in these organisms.
Our data are consistent with the hypothesis of Omoto and Kung (18) that central pair orientation specifies active sliding between peripheral doublet microtubules. If central apparatus rotation is maintained in the sliding assay, one prediction is that subsequent activation of dynein arms on specific doublet microtubules results in the progressive loss of doublets from the axoneme (Fig. 5). This prediction is supported by our observation that for all axonemes examined, >50% of the doublet microtubules that have slid away from the axoneme appear in transverse section as single doublet microtubules rather than clusters of doublets. Regulation of dynein activity may be due to mechanical forces (for example, see refs. 61 and 62) or enzymatic reactions, such as those catalyzed by kinases and phosphatases (reviewed in ref. 15). In addition, coordinating dynein activity for subsets of microtubules necessarily involves negative regulation as well (12). The C2 microtubule may play a role in modulating dynein activity; however, mutants lacking only components of the C2 microtubule have not been described. To fully understand the contribution of the central apparatus to flagellar motility will require a biophysical assessment of central pair orientation or rotation during beating as well as a complete characterization of central apparatus components, including analysis of their structural organization.
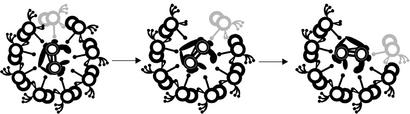
Figure 5.
As the central apparatus rotates clockwise, the C1 microtubule contacts specific radial spokes, which in turn relay a regulatory signal to the dynein arms on specific subsets of doublet microtubules. The doublets and associated dynein arms actively engaged in microtubule sliding are indicated by light shading. In each subsequent transverse section (left to right), microtubule sliding is visualized as progressive loss of doublet microtubules from the axoneme.
Supplementary Material
Acknowledgments
We thank Duane Compton for critical reading of the manuscript. This work was supported by National Institute of Health Grant GM51379 as a consortium agreement (Paul Lefebvre, University of Minnesota, St. Paul), and was supported, in part, by March of Dimes Birth Defects Foundation Research Grant 5-FY99-766 (to E.F.S.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Satir P. J Cell Biol. 1968;26:805–834. doi: 10.1083/jcb.26.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers K, Gibbons I R. Proc Natl Acad Sci USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brokaw C J. Science. 1972;178:455–462. doi: 10.1126/science.178.4060.455. [DOI] [PubMed] [Google Scholar]
- 4.Shingyoji C, Murakami A, Takahashi K. Nature. 1977;265:269–270. doi: 10.1038/265269a0. [DOI] [PubMed] [Google Scholar]
- 5.Brokaw C J. Science. 1989;243:1593–1596. doi: 10.1126/science.2928796. [DOI] [PubMed] [Google Scholar]
- 6.Sale W, Satir P. Proc Natl Acad Sci USA. 1977;74:2045–2049. doi: 10.1073/pnas.74.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams G M W, Huang B, Piperno G, Luck D J L. J Cell Biol. 1981;91:69–76. doi: 10.1083/jcb.91.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutcher S K, Huang B, Luck D J L. J Cell Biol. 1984;98:229–236. doi: 10.1083/jcb.98.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodenough U W, Heuser J E. J Cell Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell D R, Sale W S. J Cell Biol. 1999;144:293–304. doi: 10.1083/jcb.144.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner F D, Satir P. J Cell Biol. 1974;63:35–63. doi: 10.1083/jcb.63.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satir P. Mod Cell Biol. 1985;4:1–46. [Google Scholar]
- 13.Huang B. Int Rev Cytol. 1986;99:181–215. [Google Scholar]
- 14.Smith E F, Sale W S. In: Microtubules. Hyams J S, Lloyd C W, editors. New York: Wiley; 1994. pp. 381–392. [Google Scholar]
- 15.Porter M E, Sale W S. J Cell Biol. 2000;151:F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omoto C K, Gibbons I R, Kamiya R, Shingyoji C, Takahashi K, Witman G B. Mol Biol Cell. 1999;10:1–4. doi: 10.1091/mbc.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omoto C, Kung C. Nature. 1979;279:532–534. doi: 10.1038/279532a0. [DOI] [PubMed] [Google Scholar]
- 18.Omoto C, Kung C. J Cell Biol. 1980;87:33–46. doi: 10.1083/jcb.87.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith E F, Lefebvre P A. J Cell Biol. 1996;132:359–370. doi: 10.1083/jcb.132.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris E. The Chlamydomonas Sourcebook. San Diego: Academic; 1989. [Google Scholar]
- 21.Kamiya R, Okamoto M. J Cell Sci. 1985;74:181–191. doi: 10.1242/jcs.74.1.181. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell D, Rosenbaum J L. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brokaw C J, Kamiya R. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- 24.Rupp G, O'Toole E, Porter M E. Mol Biol Cell. 2001;12:739–751. doi: 10.1091/mbc.12.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piperno G, Huang B, Luck D J L. Proc Natl Acad Sci USA. 1977;74:1600–1614. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang B, Piperno G, Ramanis Z, Luck D J. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorman D S, Levine R P. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witman G B. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- 29.Okagaki T, Kamiya R. J Cell Biol. 1986;103:1895–1902. doi: 10.1083/jcb.103.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wakabayashi K, Yagi T, Kamiya R. Cell Motil Cytoskeleton. 1997;38:22–28. doi: 10.1002/(SICI)1097-0169(1997)38:1<22::AID-CM3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 31.Smith E F. Cell Motil Cytoskeleton. 2002;52:33–42. doi: 10.1002/cm.10031. [DOI] [PubMed] [Google Scholar]
- 32.Batschelet E. Circular Statistics in Biology. San Diego: Academic; 1981. [Google Scholar]
- 33.Sale W S. J Cell Biol. 1986;102:2042–2052. doi: 10.1083/jcb.102.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satir P, Matsuoka T. Cell Motil Cytoskeleton. 1989;14:345–358. doi: 10.1002/cm.970140305. [DOI] [PubMed] [Google Scholar]
- 35.Porter M E, Power J, Dutcher S K. J Cell Biol. 1992;118:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habermacher G, Sale W. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King S J, Dutcher S K. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrone C A, Myster S H, Bower R, O'Toole E T, Porter M E. Mol Biol Cell. 2000;7:2297–2313. doi: 10.1091/mbc.11.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang P, Sale W S. J Biol Chem. 2000;275:18905–18912. doi: 10.1074/jbc.M002134200. [DOI] [PubMed] [Google Scholar]
- 40.Omoto C, Yagi T, Kurimoto E, Kamyia R. Cell Motil Cytoskeleton. 1996;33:88–94. doi: 10.1002/(SICI)1097-0169(1996)33:2<88::AID-CM2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 41.Frey E, Brokaw C, Omoto C. Cell Motil Cytoskeleton. 1997;38:91–99. doi: 10.1002/(SICI)1097-0169(1997)38:1<91::AID-CM8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 42.Yagi T, Kamiya R. Cell Motil Cytoskeleton. 2000;46:190–199. doi: 10.1002/1097-0169(200007)46:3<190::AID-CM4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 43.Witman G B. Trends Cell Biol. 1993;3:403–408. doi: 10.1016/0962-8924(93)90091-e. [DOI] [PubMed] [Google Scholar]
- 44.Hyams J S, Borisy G. J Cell Sci. 1978;33:235–253. doi: 10.1242/jcs.33.1.235. [DOI] [PubMed] [Google Scholar]
- 45.Bessen M, Fay R B, Witman G B. J Cell Biol. 1980;86:446–455. doi: 10.1083/jcb.86.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamiya R, Witman G B. J Cell Biol. 1984;98:97–107. doi: 10.1083/jcb.98.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Omoto C K, Brokaw C J. Cell Motil. 1985;5:53–60. doi: 10.1002/cm.970050105. [DOI] [PubMed] [Google Scholar]
- 48.Smith E F. Mol Biol Cell. 2002;13:3344–3354. doi: 10.1091/mbc.E01-10-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang B, Ramanis Z, Luck D J L. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 50.Porter M E, Knott J A, Gardner L C, Mitchell D R, Dutcher S K. J Cell Biol. 1994;126:1495–1507. doi: 10.1083/jcb.126.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rupp G, O'Toole E, Gardner L C, Mitchell B F, Porter M E. J Cell Biol. 1996;135:1853–1865. doi: 10.1083/jcb.135.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brokaw C J, Luck D J L, Huang B. J Cell Biol. 1982;92:722–732. doi: 10.1083/jcb.92.3.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brokaw C, Luck D J L. Cell Motil. 1985;5:195–208. doi: 10.1002/cm.970050303. [DOI] [PubMed] [Google Scholar]
- 54.Piperno G, Mead K, Shestak W. J Cell Biol. 1992;118:1455–1463. doi: 10.1083/jcb.118.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamiya R, Nagai R, Nakamura S. In: Biological Functions of Microtubules and Related Structures. Sakai H, Mohri H, Borisy G, editors. New York: Academic; 1982. pp. 189–198. [Google Scholar]
- 56.Mohri H, Mohri T, Okuno M. Cell Motil Cytoskeleton. 1987;8:76–84. doi: 10.1002/cm.970080111. [DOI] [PubMed] [Google Scholar]
- 57.Shingyoji C, Katada J, Takahashi K, Gibbons I R. J Cell Sci. 1991;98:175–181. doi: 10.1242/jcs.98.2.175. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi K, Shingyoji C, Katada J, Eshel D, Gibbons I R. J Cell Sci. 1991;98:183–189. doi: 10.1242/jcs.98.2.183. [DOI] [PubMed] [Google Scholar]
- 59.Holwill M E, Satir P. Cell Motil Cytoskeleton. 1994;27:287–298. doi: 10.1002/cm.970270402. [DOI] [PubMed] [Google Scholar]
- 60.Yoshimura M, Shingyoji C. Cell Struct Funct. 1999;24:43–54. doi: 10.1247/csf.24.43. [DOI] [PubMed] [Google Scholar]
- 61.Brokaw C J. Cell Motil Cytoskeleton. 1999;42:134–148. doi: 10.1002/(SICI)1097-0169(1999)42:2<134::AID-CM5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 62.Brokaw C J. Cell Motil Cytoskeleton. 2002;53:103–124. doi: 10.1002/cm.10067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.