Abstract
We report our studies to probe the possible role of the host response to double-stranded RNA in cessation of alphavirus minus-strand synthesis. Mouse embryo fibroblasts (MEF) from Mx1-deficient mice that also lack either the protein kinase R (PKR) or the latent RNase L or both PKR and RNase L were screened. In RNase L-deficient but not wild-type or PKR-deficient MEF, there was continuous synthesis of minus-strand templates and the formation of new replication complexes producing viral plus strands. Inhibiting translation caused minus-strand synthesis to stop and a loss of transcription activity of the mature replication complexes. This turnover of replication complexes that were stable in cells containing RNase L suggested that RNase L plays some role, albeit possibly indirect, in the formation of stable replication complexes during alphavirus infection. In addition, confluent monolayers of RNase L-deficient murine cells readily established persistent infections and were not killed. This phenotype is contrary to what has been observed for infection in vertebrate cells with a presumably functional RNase L gene and more resembled alphavirus replication in Aedes mosquito cells, in which the activity of replication complexes making plus stands was also found to decay with inhibition of translation.
The Alphavirus genus of the togaviruses is of general interest because these viruses produce disease in a variety of animals, including humans, and most but not all replicate in invertebrates as well as mammals and birds (40). Exceptions may be recently identified alphaviruses replicating in Atlantic salmon and rainbow trout that appear not to require arthropod vectors (40). This broad replication potential is considered essential for transmission and the spread of their associated diseases. There are currently 24 known alphavirus species, of which Sindbis virus and Semliki Forest virus are the best studied.
The successful synthesis of many infectious progeny starts with the translation of the genome into the viral nonstructural proteins (nsPs). The nsPs form viral RNA-dependent RNA polymerase activities that copy the infecting genome into a complementary, or minus-strand, RNA template of genome length and then copy the minus-strand templates into genomes and a 3′-coterminal, subgenomic mRNA encoding the viral structural proteins. The crucial step after an alphavirus enters the cell is the formation of a replication complex that produces minus-strand RNA; the number of minus strands formed during the infectious cycle determines the amount or rate of viral plus-strand RNA synthesis (reviewed in references 46 and 63).
The analysis of conditionally lethal mutations of the heat-resistant strain of Sindbis virus (Sindbis virus HR) has shown that all four nsPs are essential for viral replication and transcription. The nsPs are present in membrane-associated complexes that retain transcription activity in vitro (4). Alphavirus minus-strand synthesis occurs only early in infection of vertebrate cells, and its continuation during this early phase results in the accumulation of replication complexes and increased rates of plus-strand synthesis (48). Protein synthesis is required for continued minus-strand synthesis but not for continued plus-strand synthesis. The rate of plus-strand synthesis achieves a maximum when no additional minus strands are made (48, 49).
A role for cellular factors in alphavirus RNA synthesis, and especially in minus-strand synthesis, is predicted from the sensitivity of virus replication to treatments with dactinomycin for 1.5 to 18 h (2, 11) and from the isolation of host range mutants of Sindbis virus whose lesions map to the nsP4 core polymerase subunit (30, 33). Our earlier analysis (4) of mature replication complexes identified a host protein of ∼120 to 130 kDa that coimmunoprecipitated in association with nsP1, the N-terminal protein of the nonstructural precursor polyprotein P1234, and an nsP whose alteration affected initiation of minus-strand synthesis (22, 52, 56, 68). Cellular proteins with homology to La may also be essential host factors for alphavirus replication (36).
We undertook experiments to probe whether the cell or the virus was responsible for the cessation in overall minus-strand synthesis. This loss is seen at about 4 h postinfection (p.i.) in alphavirus-infected vertebrate cells maintained at 37°C and is an event that likely contributes to superinfection exclusion. In this phenomenon, translation but not transcription of a superinfecting genome occurs (1). We suspect this is because the intracellular environment is no longer permissive for minus-strand synthesis by either the initial virus or the superinfecting virus. The cessation of minus-strand synthesis is not caused by the failure to produce the nsPs late in infection (48, 50, 51) or by the synthesis of capsid proteins and virion assembly (48).
Our recent analysis of a new class of nsP4 mutants suggested that such global suppression might be due to a host response (16). Cells infected with mutants possessing substitutions for Arg183 of the nsP4 polymerase accumulated only 1% to 50% as many minus strands as wild-type virus and yet they also shut down minus-strand synthesis at 4 h p.i. This suggested that amounts of double-stranded RNA (dsRNA) of as little as 1% of the normal 5,000 to 7,000 replicative intermediates/infected cell (68) or viral entry was sufficient to trigger the response. Furthermore, with temperature-sensitive mutants of Sindbis virus that failed to synthesize minus strands at 40°C, shifting the cells to 40°C early before a full complement of minus strands had been made and holding them at 40°C for 3 to 4 h prevented minus-strand synthesis from resuming later, after the infected cells had been returned to the permissive temperature (12).
It is commonly believed that the double-stranded structures of viral replication induce the antiviral interferon pathway. At least three classes of host defense enzymes bind to and require dsRNA for activation: the family of 2′,5′-oligoadenylate synthetases that make the 2′,5′-oligoadenylates that are required to activate a latent endonuclease, RNase L (58); the protein kinase R (PKR) (70); and an isoform of dsRNA-specific adenosine deaminase that converts adenosines to inosines (reviewed in reference 21). Basal levels of the three size classes of 2′,5′-oligoadenylate synthetases are present in many cells; each class has different dsRNA optima, and their different isozymes have different subcellular locations (38, 54). Trimers of 2′,5′-oligoadenylate activate RNase L by dimerizing its inactive monomers (10, 15); activated RNase L in turn cleaves viral and cellular mRNA and rRNA and contributes to translation inhibition (9, 59, 73). It has also been implicated in apoptotic pathways (72, 74).
Binding of dsRNA by the PKR kinase leads to its autophosphorylation, to phosphorylation of eIF2α with inhibition of translation, and to activation of the IκK complex with degradation of IκB, leading to activation of NF-κB and the family of interferon regulatory factors (IRFs) (14, 42). IRFs contain DNA-binding domains recognizing interferon response elements in interferon genes and transcriptional regulatory domains to activate or repress transcription. Some IRFs, such as IRF-3, are constitutively expressed in many cells. IRF-3 activates a cascade leading to induction of beta interferon (IFN-β) and specific IFN-α synthesis (reviewed in reference 62). The dsRNA-specific adenosine deaminase results in RNA editing and amino acid substitutions in the proteins synthesized from these templates. If this was the mechanism, nsPs would be made (as observed) late in infection but would be mutant in sequence and inactive.
Mx and other guanylate binding proteins are induced by interferons and belong to the dynamin superfamily of GTPases, involved in endocytosis and vesicle transport (37). It is unlikely these are involved, given the cessation of minus-strand synthesis under conditions preventing induction of host genes. Binding of virions or relevant viral glycoproteins to cell surface receptors and entry trigger additional stress and signaling pathways (25, 39). Therefore, it is possible that a critical amount of replicating viral RNA induces the dsRNA sensing pathways and causes alphavirus minus-strand synthesis to shut off. Alternatively, the virus may limit its own replication so as to prevent the triggering of the dsRNA pathways.
We screened mouse embryo fibroblast (MEF) cell lines deficient in Mx1 and lacking the RNase L or PKR gene (double-knockout cells) or both the RNase L and PKR genes (triple-knockout cells). These results implicated RNase L as a potential antiviral factor that may also be used by the virus to regulate aspects of its transcription.
MATERIALS AND METHODS
Cells, virus, and plasmids.
Chicken embryo fibroblast (CEF) cells were prepared from 10-day-old embryos from the eggs of leucosis-free (specific pathogen-free, Marek disease-negative) flocks (Spafas, Roanoke, Ill.). A continuous cell line of chicken embryo fibroblasts (DF-1 cells), derived from EV-O chicken embryos (24, 28, 53) was obtained from the American Type Culture Collection. CEF and DF-1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 6% (vol/vol) fetal bovine serum and 5% (vol/vol) tryptose phosphate broth at 37°C.
Mouse embryo fibroblast (MEF) cultures were from mice lacking the gene for PKR and/or RNase L (27, 73-75) and were grown as monolayers in DMEM containing 6% fetal bovine serum at 37°C. Four different MEF lines which naturally lacked the Mx1 gene were used. PKR−/− MEF (double-knockout cells lacking the Mx1 and PKR genes), wild-type MEF (parental wild-type C57BL/6 mouse cells), and RNase L−/− MEF (double-knockout cells lacking the Mx1 and RNase L genes) were from mice that had been extensively backcrossed with C57BL/6 mice. The triple-knockout (lacking the Mx1, PKR, and RNase L genes) MEF were from a mouse that was of mixed background. Additional RNase L−/− cell lines, i.e., G5 MEF, were established by limiting-dilution cloning in 96-well plates and picking cells from a well that had only one colony.
Aedes alpobictus C7/10 mosquito cells were obtained from V. Stollar (Rutgers Medical Center, Rutgers, N.J.) and adapted to grow in DMEM supplemented with 6% fetal calf serum and 5% tryptose phosphate broth at 30 or 28°C.
The heat-resistant strain of Sindbis virus (Sindbis virus HR) and the Semliki Forest virus strain were described previously (26, 49). Stocks of these alphaviruses were made in CEF and BHK-21 cells, respectively, and had titers of 4 × 109 PFU/ml and 2 × 109 PFU/ml, respectively.
Infection and RNA labeling.
The cells were infected as monolayers, usually 1 day after seeding, with virus at a multiplicity of infection (MOI) of 100 PFU/cell, as determined on CEF. When counted directly on early-passage MEF cells, Sindbis virus HR gave small plaques, which made it difficult to determine the titer. The titer appeared to be 5 to 20% of the titer determined on CEF, DF-1, and BHK-21 cells, all of which produced the same titer with Sindbis virus and Semliki Forest virus (data not shown). Cells were pulse-labeled with [3H]uridine (50 or 200 μCi/ml; Life Sciences NEN) for 1 h in complete DMEM containing 20 μg of dactinomycin/ml, 6% fetal calf serum, and 20 mM HEPES, pH 7.4. At the end of the labeling period, the cells were washed with ice-cold phosphate-buffered saline and lysed with 5% lithium dodecyl sulfate in LET buffer (0.1 M LiCl, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4) containing 200 μg of proteinase K/ml. DNA in the cell lysates was sheared by passage through a 27-gauge needle before acid-insoluble incorporation was determined.
Isolation of RF RNA and quantitation of minus strands.
Minus-strand synthesis was determined as described before (12). For accumulation experiments, all infected cultures were incubated in the presence of 200 μCi of [3H]uridine/ml plus 2 μg of dactinomycin/ml, beginning immediately after adsorption. After extraction with low-pH phenol, to remove protein and DNA, and with chloroform, the RNA was precipitated with ethanol, digested with RNase A, and chromatographed on CF-11 cellulose (Whatman, Clifton, N.J.), following the procedures of Franklin (17) for isolation of the replicative form (RF) RNAs, which are the double-stranded core of the viral replicative intermediates. Minus-strand RNA synthesis was measured with a nuclease protection assay. The [3H]uridine-labeled RF RNA was heat denatured, quickly cooled to 0°C with an ice-water bath, hybridized to a 100-fold excess of unlabeled virion 49S plus-strand RNA, and digested with RNase A or a combination of RNase A and RNase T1. The labeled RNA protected from degradation was collected on glass fiber filters after precipitation with trichloroacetic acid. The radioactivity was determined by scintillation spectroscopy.
RESULTS
Sindbis virus HR RNA synthesis in vertebrate cells.
In vertebrate cells such as CEF, Sindbis virus RNA synthesis increases rapidly during the first 4 to 5 h p.i., after which it becomes constant, as depicted in Fig. 1A. Agarose gel electrophoresis (not shown) showed that greater than 95% of the [3H]uridine-labeled viral RNA was in single-stranded 49S genomes and subgenomic 26S mRNA and only ∼5% was in the replicative intermediate RNAs, which are partially double stranded and migrate on nondenaturing 0.8 to 1.0% agarose-Tris-borate-EDTA (TBE) gels similarly to chromosomal DNA. The viral minus strands, which are genome length and are the templates for the synthesis of both genomes and subgenomic mRNA, are found in the replicative intermediates.
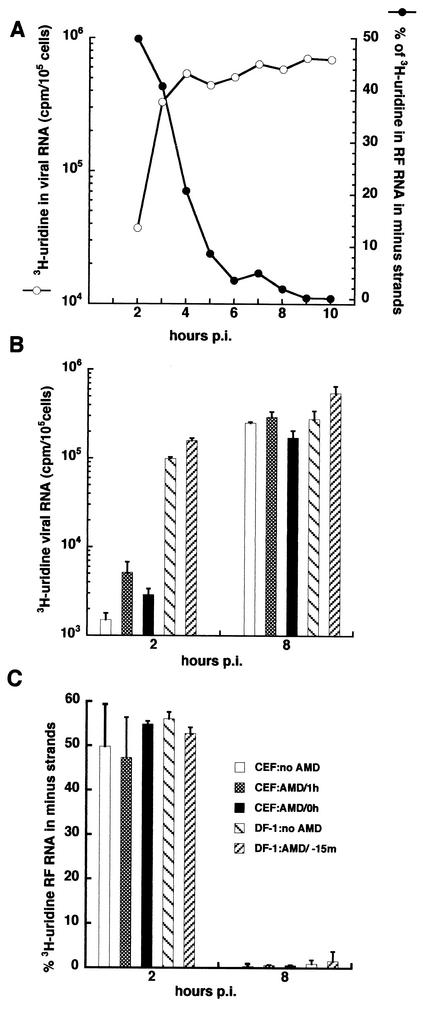
FIG. 1.
(A) Sindbis virus RNA synthesis in CEF cells. Cultures of CEF cells were infected at an MOI of 100 with Sindbis virus HR at 37°C and maintained at 37°C. At hourly intervals, beginning at 1 h p.i, cultures were pulse-labeled for 1 h with [3H]uridine (200 μCi/ml) in DMEM containing 20 μg of dactinomycin (AMD)/ml. Total acid-insoluble incorporation in 105 cells of infected CEF cultures (○) was determined. The viral RF RNA was obtained, and the percentage of the labeled RF RNA in minus-strand RNA was determined as described in Materials and Methods (•). (B) Effect of dactinomycin on plus-strand synthesis. Cultures of primary CEF cells or of the continuous DF-1 chicken cell line were infected with Sindbis virus HR at an MOI of 100 and maintained at 37°C. Triplicate cultures from each setwere pulse-labeled with [3H]uridine (200 μCi/ml) in the presence of 20 μg of dactinomycin/ml from 1 to 2 h p.i. or from 7 to 8 h p.i. and harvested at the end of the pulse period. The average total acid-insoluble incorporation of [3H]uridine into 105 cells was determined for infected CEF cultures treated with 2 μg of dactinomycin/ml at time zero or at 1 h p.i. or untreated or of DF-1 cells given 2 μg of dactinomycin/ml 15 min before infection or left untreated. (C) Effect of dactinomycin on minus-strand synthesis. The percentage of [3H]uridine in the labeled RF RNA that was in minus-strand RNA was determined for the samples in B.
Sindbis virus minus strands, also depicted in Fig. 1A as the percentage of [3H]uridine incorporated into RF RNA that is in minus strands, were synthesized only during the first 4 h p.i. at 37°C. From 1 to 2 h p.i., 50% of the [3H]uridine-labeled RF RNA, which is the RNase-resistant double-stranded cores of the replicative intermediates, was incorporated in minus strands. This indicated that essentially all (95 to 100%) of the minus strands that were present at 2 h p.i. were made during the previous 1 h. The remaining 50% of the radiolabeled RF RNA was in plus strands. With time, this ratio decreased as the minus strands accumulated, and by 5 h p.i., essentially no more minus strands were made and all of the radiolabeled RF RNA was in plus strands. Because plus-strand synthesis continued at a relatively constant rate in the absence of synthesis of new minus-strand templates, the viral replication complexes producing plus strands were very stable. The mechanism that is responsible for shutting off minus-strand synthesis after 4 h p.i. is not understood and was the focus of our studies.
We determined if the shutoff of minus-strand synthesis was caused by the loss of a short-lived host function or was the result of infection inducing a host gene(s). Earlier studies had shown that dactinomycin or α-amanitin treatment of vertebrate cells reduced viral RNA synthesis and PFU yields by 50% after 6 h of treatment and by 90% or more after 18 h of treatment (2, 3). This finding implicated host factors in viral RNA synthesis, especially in viral minus-strand synthesis, but did not address the cessation of minus-strand synthesis. As shown in Fig. 1B and C, treatment of the cell culture with 2 μg of dactinomycin/ml, which inhibited DNA-dependent RNA synthesis by more than 95%, neither significantly altered overall viral RNA synthesis (Fig. 1B) nor prevented shutoff of minus-strand synthesis in either CEF or DF-1 cultures (Fig. 1C).
Dactinomycin was added 15 min before infection of DF-1 cells, at 0 h p.i. with the virus inoculum, or at 1 h p.i., following removal of the virus inoculum, with CEF or not added until the infected cultures were radiolabeled for 1 h with both DF-1 cells and CEF. The only difference that we observed between the two types of chicken cells was a slightly faster replication cycle in DF-1 cells, in which viral RNA synthesis occurred 30 to 60 min earlier than in CEF (data not shown). Dactinomycin treatment actually appeared to enhance viral RNA synthesis slightly (Fig. 1B). The results show that if host functions contributed to the shutoff of minus-strand synthesis, they were already present in the cell, perhaps in a latent or inactive form, and did not require new transcription for their expression. If such factors existed, they would be relatively stable because they survived dactinomycin treatment. Among the possibilities would be the latent mediators of dsRNA signaling such as 2′,5′-oligoadenylate synthetase, RNase L, and the protein kinase PKR, which are involved in antiviral activities and cause translation inhibition and perhaps cell death.
We determined if minus-strand synthesis behaved any differently in CEF treated with the caspase inhibitor Z-VAD [z-Val-Ala-Asp(OMe)-CH2F; Enzyme Systems Products, Livermore, Calif.] or in BHK-21 cells overexpressing the antiapoptotic factor Bcl-2. Neither of these conditions caused a change in the pattern of viral replication or in the cytopathic effects of the virus (data not shown).
Sindbis virus replication in mouse cells lacking RNase L and/or PKR.
We screened for viral RNA synthesis and virus production by mouse fibroblast cell lines lacking Mx1 (most laboratory strains, such as the C57BL/6 mouse, lack Mx1 expression) as well as the RNase L and/or PKR gene. The wild-type C57BL/6 MEF (wild-type) cells were used as the control cell line. We also tested replication by Semliki Forest virus to determine if any phenotypic changes observed might be common to alphaviruses or specific to Sindbis virus. Cells were infected with an amount of virus equivalent to an MOI of 5 to 20 on MEF cells (this was an MOI of 100 on CEF cells). They were pulse-labeled for 1 h with [3H]uridine, beginning at 1 h p.i., and harvested at the end of the pulse period. The incorporation of [3H]uridine into total viral RNA (Fig. 2A), RF RNA (Fig. 2B), and minus strands (Fig. 2C) was monitored for each of the cell lines.
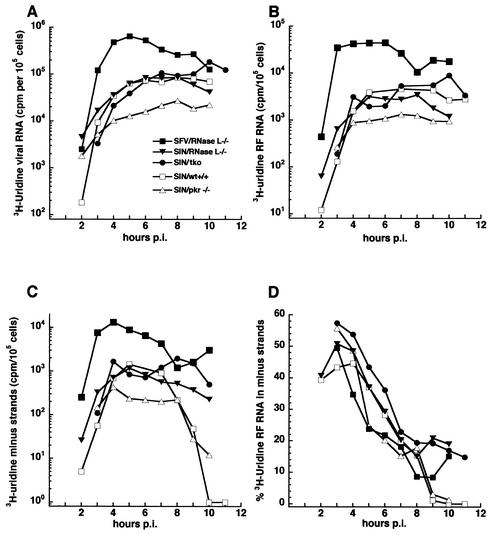
FIG. 2.
(A) RNA synthesis in MEF cells expressing or lacking PKR and/or RNase L. Cultures were infected with Sindbis virus HR (SIN) at an MOI of 100 at 37°C. Beginning at 1 h p.i., individual cultures were pulse-labeled for 1 h periods with [3H]uridine (200 μCi/ml) in DMEM containing 20 μg of dactinomycin/ml and harvested at the end of the pulse period. Wild-type (wt) MEF cells (□); MEF lacking RNase L (▾); MEF cells lacking PKR (▵); MEF cells lacking both PKR and RNase L (triple knockout [tko]; •); RNase L-deficient MEF cells infected with Semliki Forest virus (SFV) at an MOI of 100 at 37°C and similarly treated (▪). Total acid-insoluble incorporation per 105 cells is shown. (B) Incorporation of [3H]uridine into the RF cores of the replicative intermediates. RF RNA was isolated from the infected cells described in A, and the results are expressed per 105 cells. (C) Incorporation of [3H]uridine into minus strands. The RF RNA described in B was analyzed, and the results are expressed per 105 cells. (D) Percentage of [3H]uridine incorporation in the RF RNA that was in minus strands. Radioactivity in the minus strands (C) was divided by the radioactivity in the RF RNA (B).
The overall pattern of viral RNA synthesis was similar to that observed for Sindbis virus HR in CEF cells. An early, exponentially increasing phase of viral RNA synthesis was followed by a late phase in which the rate of viral RNA synthesis became constant and maximal. While similar, the kinetics of overall transcription in MEF (Fig. 2A) was delayed about 2 h compared to that in CEF (Fig. 1A), and RNA synthesis by Sindbis virus HR in wild-type and knockout MEF cells was about one-tenth of that observed in CEF cells. An exception was Semliki Forest virus, which replicated in the RNase L knockout cells at levels similar to that in CEF cells and 10 times higher than that of Sindbis virus HR (Fig. 2A).
For both viruses and in the cell lines examined, incorporation into the viral replicative intermediate cores (RF RNA) showed kinetics similar to that of overall transcription, as expected. While rates of minus-strand synthesis became maximal at 5 to 6 h p.i. in the different MEF cell lines, it decreased after this period only in the wild-type MEF and MEF lacking PKR (Fig. 2C). In wild-type MEF, only 1 to 2% of the incorporation in labeled RF cores each hour was in minus strands after 9 h p.i. A clearly different pattern was seen in both of the RNase L-deficient MEF cell lines, where minus-strand synthesis continued at high, maximal rates until at least 10 h p.i. (other experiments followed synthesis until 11 or 12 h p.i. or until development of extensive cytopathic effect prevented it; data not shown).
At late times, 11 to 20% of the [3H]uridine incorporation in RF cores was in newly made minus strands each hour (Fig. 2D). The remaining radiolabeled RNA was plus strands. A value of 50% of radiolabeled RF RNA in minus strands means that ∼100% of active templates had been made during the pulse period. Therefore, significant amounts (22 to 40% per h) of minus strands are made late in the RNase L-deficient MEF cells. Moreover, Semliki Forest virus-infected RNase L-knockout MEF cells also continued synthesis of minus strands late (Fig. 2C and D), indicating that this phenotype likely reflected alterations to common alphavirus-host interactions. We conclude that RNase L played a role in alphavirus minus-strand cessation but that Mx1 and PKR apparently did not.
Continued minus- and plus-strand synthesis in RNase L-knockout cells was due to formation of new replicases and replication complexes.
In CEF cells, minus-strand synthesis is coupled to translation of viral nonstructural polyproteins and the formation of new replication complexes. Addition of the translation inhibitor cycloheximide rapidly inhibits minus-strand synthesis but does not inhibit plus-strand synthesis by previously assembled replication complexes. We asked whether alphavirus minus-strand synthesis in the RNase L-knockout MEF cells was also sensitive to translation inhibition. RNase L-knockout MEF cells were infected with Sindbis virus HR at an MOI of at least 5 or 25 (MOI on CEF of 100 or 500), pulse-labeled with [3H]uridine for 1 h, and monitored for levels of overall transcription and of minus-strand synthesis (Fig. 3).
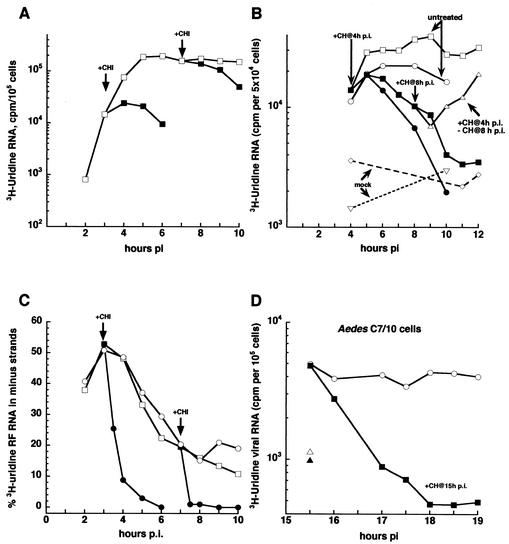
FIG. 3.
Sindbis virus replication complexes are unstable in MEF lacking RNase L and in C7/10 mosquito cells. MEF from the RNase L−/− mouse were infected at 37°C at an MOI of 100 (A) or 500 (C) PFU of Sindbis virus per ml (□; titer was determined by plaque assay on CEF monolayers) and refed fresh 37°C medium at the end of the 1-h adsorption period. Duplicate sets of cultures were refed with medium containing cycloheximide (CH, 100 μg/ml) beginning either at 3 h p.i. (▪) or at 7 h p.i. (▪). Individual dishes were pulse-labeled for 1 h with [3H]uridine at 200 μCi/ml. Labeling of cycloheximide-treated cultures was done in the continued presence of the drug; labeling of untreated cultures was done in the absence of the drug. (B) The MEF from an RNase L−/− mouse (○) or a clone (G5) of MEF that were derived from those RNase L−/− MEF (□) were given cycloheximide (• and ▪, respectively) at 4 h p.i. as described above for A. At 8 h p.i., the cycloheximide-containing medium was removed from one set of dishes, and the cells were rinsed three times and refed with medium that lacked cycloheximide (▵). Individual dishes were pulse-labeled for 1 h with [3H]uridine at 50 μCi/ml. Labeling of cycloheximide-treated cultures was done in the continued presence of the drug; labeling of untreated cultures or of cultures from which the cycloheximide was removed was done in the absence of the drug. Mock-infected cells (▿). (C) Cycloheximide stops minus-strand synthesis in MEF lacking RNase L. The RF RNA was obtained from the cells shown in A, and the [3H]uridine incorporated into minus strands was determined. Minus-strand synthesis is expressed as a percentage of [3H]uridine in the RF core RNA that was in minus-strand RNA. (D) Replication complexes in Aedes C7/10 cells are unstable. Aedes C7/10 cells were infected with Sindbis virus at 28°C at an MOI of 100 PFU/ml (titer was determined by plaque assay on CEF monolayers) and refed with fresh 28°C medium at the end of the 1-h adsorption period. At 15 h p.i., one set of dishes was refed with medium containing 100 μg of cycloheximide/ml (solid symbols) and the cells were pulse-labeled with 50 μCi of [3H]uridine/ml for 30 min in the presence (▪) or absence (○) of cycloheximide. Mock-infected (▵, ▴). Total acid-insoluble incorporation is shown.
As seen for early minus-strand synthesis in CEF cells, continued Sindbis virus minus-strand synthesis in RNase L-knockout MEF cells was sensitive to cycloheximide addition at both early and late times after infection and at both MOIs tested (Fig. 3C). Unexpectedly, and in contrast to CEF cells, viral plus-strand synthesis declined with time after cycloheximide addition (Fig. 3A). A cloned line of cells (G5 cells) derived from MEF lacking RNase L and infected with Sindbis virus HR at an MEF MOI of either 10 or 35 PFU/cell gave similar results (Fig. 3B). We tested the ability of transcription to recover in these cells and found that removal of cycloheximide at 8 h p.i. led to the full recovery of viral transcription (Fig. 3B). A similar loss of viral transcription with inhibition of translation also occurred in Aedes C7/10 mosquito cells (Fig. 3D). Taken together, these results suggest that all of the infected cells in the RNase L-deficient MEF population were infected initially and that any minus-strand synthesis observed late was not due to a second round of infection in only part of the population.
We also performed immunofluorescent staining of the infected RNase L-deficient cells with antibodies specific for Sindbis virus proteins (gift of W. B. Klimstra) and found that essentially all cells contained viral proteins compared to mock-infected monolayers, although not all cells gave an equally bright signal (data not shown). Inhibition of translation prevented the formation of minus-strand templates, and under these conditions, the old minus strands decayed and viral transcription declined coordinately. Because minus-strand synthesis failed to shut off in cells lacking RNase L, minus-strand synthesis was able to recover, and new replication complexes were created when translation restarted.
Finally, the molar ratio of 49S genome to 26S mRNA produced in RNase L double-knockout MEF cells varied slightly with the stage of the virus infection cycle, as also seen in CEF cells, and was not altered by the presence of cycloheximide (data not shown). This argued that the viral replication complexes formed in cells lacking RNase L functioned essentially the same as those assembled in cells expressing RNase L.
In RNase L double-knockout MEF cells, older Sindbis virus minus-strand templates turned over and were replaced by newly made ones.
One implication of the loss of plus-strand synthesis after cycloheximide treatment was that replication complexes became inactive in RNase L-deficient cells because the minus strands were turning over. Normally, in CEF and other vertebrate cells, alphavirus minus-strand templates are stable, i.e., they remain part of active replication complexes whose activity is not lost even if the synthesis of new polymerase proteins is prevented. If the minus strands were turning over in RNase L-deficient cells, those made late would replace those made earlier and accumulate. If, on the other hand, minus strands made late in RNase L−/− cells turned over rapidly, they would not accumulate.
We determined directly, using continuous labeling conditions, if minus strands accumulated to significant levels late in infection. CEF, wild-type MEF, and RNase L double-knockout MEF cells were infected with Sindbis virus HR and labeled with [3H]uridine, starting at 1 or 3 h p.i. or at 6 h p.i. or later. The late time period was chosen as a time after which the maximum rates of plus-strand synthesis had been reached in each cell line. As shown in Fig. 4, when [3H]uridine was added early, all three cell lines incorporated it into minus strands, and 50% of the total incorporation into the RF cores was in minus strands. When the [3H]uridine was added late in infection, new minus strands accumulated at very low levels in CEF and wild-type MEF. Radiolabeled minus strands in the RF cores accumulated at ∼0.1 to 1% per h in CEF cells and at ∼1 to 2% per h in wild-type MEF cells. This showed that a very small amount of minus-strand synthesis continued in wild-type MEF cells, which was also observed in 1-h pulse-labeling experiments, as shown in Fig. 2C. In contrast, Sindbis virus-infected RNase L-deficient MEF accumulated radiolabel into minus strands of the RF cores at ∼10% per h, and thus at a 5- to 10-fold-higher levels than wild-type MEF cells (Fig. 4).
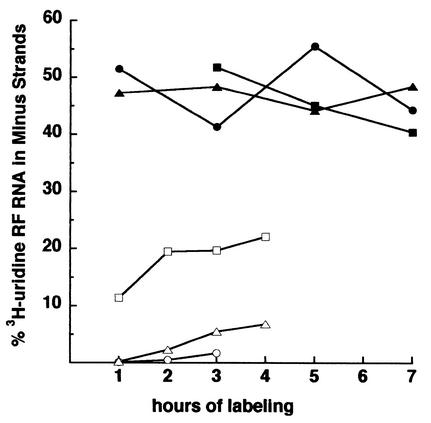
FIG. 4.
Minus strands made late in RNase L-lacking MEF cells enter replication complexes. Cultures of MEF or CEF were infected with Sindbis virus at 37°C at an MOI of 100 PFU/cell. At 1 h p.i. (▪, wild-type MEF; •, RNase L−/− MEF; ▴, CEF) or beginning only at 6 h p.i. (□, wild-type MEF; ○, RNase L−/− MEF; ▵, CEF), a set of each of the cultures was refed with medium containing 200 μCi of [3H]uridine/ml and 20 μg of dactinomycin/ml, and individual dishes were harvested at the times shown. The percentage of the [3H]uridine in the RF core RNA that was in minus strands is shown.
If these late minus strands were degraded rapidly after synthesis or released in a single-stranded state (aberrant transcription products), their presence in the RF cores would have remained at ∼10% over time. After 2 h of continuous labeling, 20% of the radiolabeled RF RNA at 8 h p.i. was in minus strands, indicating that they accumulated and that 40% of the templates active at this time were made in the preceding 2 h. To attain such a high percentage, a significant number of minus-strand templates made earlier had to have left the active replicative intermediate population. The results support the notion that older minus strands turn over and are replaced in the RNase L-deficient environment by the continuous synthesis of new ones. This finding was novel and unexpected for alphavirus replication complexes in vertebrate cells.
We also verified that the minus strands made late during infection functioned as templates for plus-strand synthesis. RFI (20S in size) is derived by RNase treatment of replicative intermediates active in the synthesis of plus or minus strands of genome size. Incorporation of [3H]uridine into RFI cannot be used to distinguish whether a minus strand or a genome is in the process of synthesis. However, [3H]uridine incorporation into minus strands in RFII (18S in size) and RFIII (15S in size) can be used. RFIII is the RNase-resistant double-stranded core of the replicative intermediate that was engaged in 26S mRNA synthesis. After limited RNase cleavage, the three RF cores were separated by velocity centrifugation with 15 to 30% sucrose gradients, from which ∼80 to 90 fractions were collected. Fractions were assayed for labeled RF RNA and pooled; samples of each pool were analyzed on a 1% agarose-TBE gel to verify the presence of RFI, -II, and/or -III cores before the RNA was used in hybridization assays (data not shown). RFII and RFIII cores obtained from Sindbis virus-infected RNase L-deficient cultures labeled between 6 and 7 h or 6 and 10 h p.i. had 11 to 13% and 20 to 22%, respectively, of their total labeled RF RNA in minus strands, amounts equal to that of the RFI cores from the same sample. We conclude that Sindbis virus minus strands made at late times in RNase L−/− MEF cells were utilized as functional templates for viral plus-strand synthesis.
Sindbis virus establishes a persistent infection in RNase L−/− MEF cells.
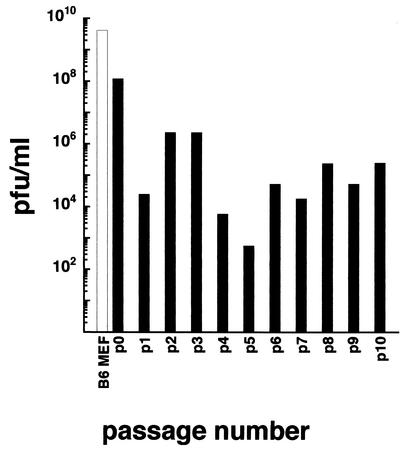
Sindbis virus established a persistent infection in RNase L−/− MEF. If RNase L−/− MEF or the G5 RNase L−/− cells were infected when the cells were a confluent cell sheet, usually 3 days after seeding, very few of the cells died from Sindbis virus infection, although >108 PFU/ml were produced in the 24 h after infection. The Sindbis virus-infected G5 MEF culture was passaged at a 1:3 dilution every 3 days. Sindbis virus in the medium after 3 days (just before passage of the culture) was counted on DF-1 cells.
As shown in Fig. 5, the G5 cloned RNase L−/− cells were persistently infected and produced ∼105 PFU/ml of culture medium over the first 10 passages. The virus produced large plaques on DF-1 cells that were indistinguishable from those of the original infecting Sindbis virus and completely killed DF-1 and BHK-21 cells, i.e., did not cause persistent infection of these cells. The number of passages and the large number of PFU recovered over time indicate that this yield is not the result of merely carrying virus from the original infection. The persistently infected G5 cultures have been maintained for over 40 passages, confirming that they are not transient persistent states and that the cells do not cure the infection. Therefore, MEF lacking RNase L appear to behave like C7/10 Aedes mosquito cells and form a persistent infection with Sindbis virus.
FIG. 5.
Virus growth and persistence. G5 RNase L-deficient MEF (solid bars) and wild-type C57BL/6 (B6) MEF (open bar) in 25-cm2 flasks were infected with Sindbis virus HR at 100 PFU/cell. After an adsorption period of 1 h at room temperature, the virus inoculum was removed, and the cells were rinsed and refed with fresh medium. At 3 h p.i., the medium was removed and the cells were refed again with 15 ml of fresh medium. One day later, the medium was harvested and the cells were refed with fresh medium. After the second day, the cells were trypsinized and split 1:3 to fresh 25-cm2 flasks with 15 ml of fresh medium. The cells were subcultured every 3 days by splitting them 1:3. The medium was collected immediately before subculturing, and virus was counted on DF-1 chicken cells.
RNase L−/− MEF cells were fully susceptible to infection and killing by Semliki Forest virus and vesicular stomatitis virus, a minus-strand RNA rhabdovirus (Table 1). With encephalomyocarditis virus used to assay susceptibility, persistently Sindbis virus infected G5 RNase L−/− cells at passage 18 formed the same number of encephalomyocarditis virus plaques as 17cl-1 cells, which are a spontaneously transformed BALB/c mouse fibroblast cell line (5.3 × 109 ± 0.7 × 109 and 4.6 × 109 ± 1.5 × 109 PFU/ml, respectively). Thus, the Sindbis virus persistently infected G5 cells remained susceptible to superinfection with heterologous virus, and their nonspecific antiviral pathways, such as interferon, were not activated or were not fully functional, as found by others (72, 74).
TABLE 1.
Cytopathic effects of Sindbis virus, Semliki Forest virus, and vesicular stomatitis virus
| Virus | Extent of cytopathic effecta in MEF:
|
||||
|---|---|---|---|---|---|
| Wild type | PKR+/+ | PKR−/− | PKR−/− RNase L−/− | RNase L−/− | |
| Sindbis virus | Full | Full | Full | Partial | Partial |
| Semliki Forest virus | Full | ND | ND | ND | Full |
| Vesicular stomatitis virus | Full | ND | ND | ND | Full |
Extent was dependent on the confluency of the monolayer; the less confluent, the more cytopathic effect was observed. ND, not determined.
Given the turnover of viral transcriptional activity, a failure to continue to produce new templates would cure the infection. This argues that RNase L may play an essential role in clearing virus from infected cells and thereby prevent persistence. The failure of the persistently infected cells to die also argues that RNase L contributes to cell killing by Sindbis virus.
DISCUSSION
In the present study, we found that fibroblast cells derived from mice that had had the RNase L gene knocked out (74) failed to shut off minus-strand synthesis when infected with either Sindbis virus or Semliki Forest virus. Unlike the MEF derived from C57BL/6 mice or from PKR knockout mice or CEF and BHK-21 cells, these infected cells continued to synthesize minus strands throughout the viral replication cycle and to use these as templates for plus-strand synthesis. Inhibiting minus-strand synthesis by inhibiting protein synthesis revealed the turnover of minus strands and the loss of replication complexes. This was also an unusual property for minus strands. Therefore, in the RNase L-deficient cellular environment, new minus strands had to be continuously assembled to maintain viral plus-strand synthesis.
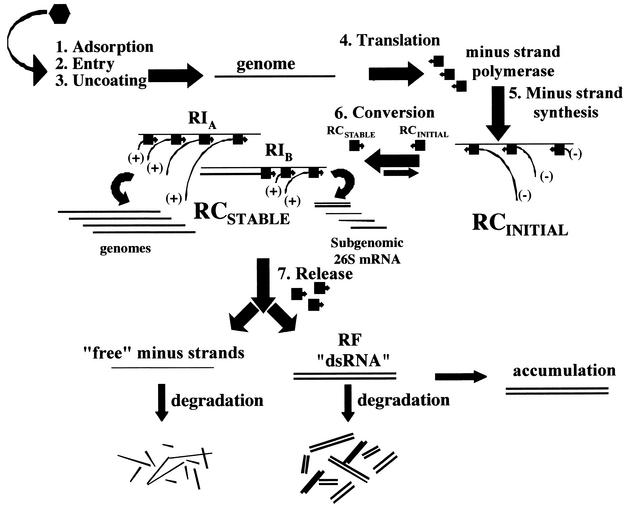
Minus-strand synthesis in RNase L knockout cells at late times p.i. resembled early minus-strand synthesis because it too required protein synthesis. This was different from minus-strand synthesis due to promoter switching observed with certain Sindbis virus HR nsP2 and nsP4 mutants that abnormally restart minus-strand synthesis late at 40°C (43, 45, 47). The model (Fig. 6) that we favor proposes that minus strands would be the preferred templates of the viral replication complexes. Once synthesized, they would be stably associated with replication complexes to produce exclusively plus strands (12, 43, 50, 51). Therefore, minus-strand synthesis occurs during the de novo formation of replication complexes. After replicative complex initiation completes the synthesis of minus strands, they mature and are converted to mature replication complexes that constitutively synthesize only plus strands (12). Treatment of cells with interferon prevented infection with Sindbis virus (41). Most likely this block occurs before the formation of mature replication complexes (step 6 in Fig. 6).
FIG. 6.
Model depicting the replication strategy of Sindbis virus and two possible fates for the inactivated minus strands in RNase L-deficient MEF cells. RI, replication intermediate; RCSTABLE, mature replication complex, active in plus-strand synthesis; RCINITIAL, initial replication complex, active in minus-strand synthesis.
Newly made templates were found in complexes late in infection, which argued that templates made earlier were lost. A similar situation was described for poliovirus replication complexes (29), in which, after about 20 rounds of plus-strand synthesis, the minus-strand templates accumulated as inactive RF RNA. As shown in Fig. 6, models for turnover of mature replication complexes include conversion of the alphavirus minus strands to dead-end RF, which would result if the polymerase were released and the nascent plus strand remained with the template minus strand. Alternatively, the minus strand might be released as single stranded, which would subsequently be degraded. With mouse hepatitis virus, which is a coronavirus and has an unstable replication complex, we found that the minus-strand templates were released as single strands and then degraded (66).
The mechanistic basis for loss of replication complex transcriptional stability is not obvious. Failure to regulate normal mRNA decay may prevent or decrease translation of other host functions needed to stably assemble replication complexes on membranes or to create permanent membrane sites. It has been found that nsP components of the complex trafficked from the endoplasmic reticulum to the plasma membrane, where assembly of the complex is proposed to occur, and then the complex is transported to a modified lysosomal compartment, where replication and transcription would occur (32). Transport may be altered in RNase L-deficient cells, preventing viral replication complexes from reaching sites that confer stability to their structure. Alternatively, in the absence of RNase L, other host functions may target part or all of the components of the replication complex to the degradation machinery. The viral nsP4 core polymerase is targeted by the ubiquitin-proteasome pathway (13).
Finally, it is likely that upstream events in the 2′,5′-oligoadenylate system are activated in RNase L-deficient cells by virus infection and they may contribute to the phenotype. In the absence of RNase L, production of 2′,5′-oligoadenylate may lead to the activation of other processes that interfere with stabilization. MEF cells lacking both RNase L and PKR behaved the same as cells lacking just RNase L, and MEF cells lacking only Mx1 or lacking both Mx1 and PKR behaved the same as CEF cells. Therefore, the lower levels of viral RNA synthesis in MEF cells compared to CEF cells were not the cause of the failure to shut off minus-strand synthesis. Also, this showed that Mx1- and PKR-mediated pathways did not play a role in these aspects of alphavirus replication.
Several mechanisms have been proposed for the temporal synthesis of minus strands, but the exact mechanism is not known (44, 46, 57, 63). If only a limited quantity of host factors were available for the formation of initial replicative complexes, minus-strand synthesis would cease when these were depleted. The fact that minus-strand synthesis ceases at the same time regardless of the number of replication and transcription complexes created does not support this hypothesis. Furthermore, studies with temperature-sensitive RNA mutants found that there was a window of time when minus-strand synthesis could resume following a shift to 40°C and a later shiftdown to 30°C; after this time, minus-strand synthesis failed to restart (12, 48, 49, 68).
The time factor, recent findings (16), and our current results (this study) support the suggestion that minus-strand shutoff is likely due to the influence of a host factor(s) that includes RNase L either directly or indirectly. The RNase L-deficient MEF cells appeared to maintain the early “more p23 permissive” environment, allowing replicase precursors to form continuously and become active in minus-strand synthesis and then in plus-strand synthesis. While this is speculative, several possibilities can be envisioned for a role of RNase L in the shutoff of minus-strand synthesis. One is that RNase L plays a role in the inactivation of essential components of the minus-strand replicase, e.g., if its actions reduced the half-life of the intermediate p23 polyprotein component of the minus-strand replicase. Alternatively, the switch from cellular to viral translation might prevent the continued synthesis of one or more essential host factors required by the minus-strand replicase, and this transition is reduced or prevented in the absence of RNase L. RNase L might play a role in the stabilization of mature replication complexes and also in inhibiting synthesis of essential host factors. In this case, stabilization would prevent recycling of such limited, essential factors. The failure to stabilize mature replication complexes may allow recycling and thus enable minus-strand synthesis to continue. Whatever the mechanism is, its role would be to prevent, when activated, the synthesis of minus strands.
The creation of a cellular environment that is nonpermissive for minus-strand synthesis would be an antiviral mechanism of the cell, especially if it were activated early enough. It might also benefit the virus by causing superinfection exclusion. It may be to the virus's advantage to allow the shutdown of minus-strand synthesis when enough templates have been made for virion production. In this way, the virus might prevent or delay provoking host defenses, e.g., interferon expression. In their normal arboviral, mosquito-bird cycles, alphaviruses persistently infect specific mosquito vectors and thus may have evolved strategies to establish and maintain persistence. Perhaps, in vertebrate hosts and in the presence of RNase L, strategies evolved to stabilize viral replication templates as compensation for a failure to prevent cessation of minus-strand synthesis and apoptosis (cytopathic effect).
In humans, the RNase L gene is located on chromosome 1q25 (61) and may be one factor predisposing to prostate cancer (8). RNase L contains an N-terminal regulatory domain with nine ankyrin repeat regions and a C-terminal part containing the endonuclease catalytic domain and a region with homology to protein kinases; the latter is thought to be nonfunctional (72). The sequence of the endonuclease domain has been proposed to show homology with the IRE1 family of endoplasmic reticulum stress response kinase/endonucleases (72). RNase L also plays a role in RNA decay during cell death. Mice and cells devoid of RNase L show reduced antiviral responses to alpha/beta interferons and are defective in apoptosis (74). This led to the proposal that RNase L is likely to play a fundamental role in the control of RNA stability distinct from its role in interferon action, and loss of RNase L might allow cell survival factors to continue to be made or to accumulate to higher levels.
In support of this possibility, alphavirus-infected, RNase L-deficient MEF cells were shown to be capable of entering a persistently infected state. RNase L may play some role in the clearance of Sindbis virus or in cell killing by the virus. Ryman et al. (41) noted that Sindbis virus did not kill dendritic cells from RNase L-negative mice. We too found that late-passage MEF, including those lacking RNase L, were killed by Sindbis virus (data not shown). In order for a cell to survive viral infection, it must not die due to the infection. RNase L might play some role in cell killing by Sindbis virus, especially since mice lacking RNase L appear to be deficient in apoptosis (74).
Virus-host interactions that block the activation of RNase L or PKR are known (20). These include viral products that degrade PKR (6) or that have dsRNA binding or other activities that compete with or prevent activation by the PKR and 2′,5′-oligoadenylate synthetase. Among the latter, the hepatitis C virus NS5A protein (19, 65), the influenza virus NS1 protein (34, 64, 67), and the vaccinia virus E3L and K3L proteins (55, 60, 69, 71) have been extensively studied. Some of these interactions also block apoptosis (20, 31). An example of a different type of interaction is the escape from interferon-mediated antiviral effects, in particular RNase L, that was suggested to affect hepatitis C and hepatitis G virus codon usage to favor fewer UA and UU dinucleotides (23). The authors proposed that such a mechanism could be used to modulate viral replication levels to avoid a vigorous host response and at the same time maintain a persistently infected state. Alphavirus genome sequences did not show a similar codon bias (D. Barton, personal communication).
Of interest, the PKR pathway also regulates the translation of IFN-γ mRNA (5) and tumor protein p23/translationally controlled tumor protein (TCTP) mRNA (7) by recognizing specific 5′ untranslated region dsRNA structures. The alphavirus 5′ untranslated region (35) and upstream regions (18) contain stem-loop structures, but the failure of PKR-knockout MEF cells to alter Sindbis virus minus-strand synthesis argues that this type of regulation is not playing a role. No mutants studied to date mimic these phenotypes, and thus the finding that viral nsPs are players or targets of this cellular response could provide new insights into alphavirus-host interactions.
Acknowledgments
We acknowledge the contribution of Han Fei Ding to the caspase inhibitor and Bcl-2 studies and the technical assistance of Chandra Mapes.
This investigation was supported by Public Health Service grants AI-15123 (D.L.S.), CA-44059 (R.H.S.), and AI-34039 (B.R.W.) from the National Institutes of Health.
REFERENCES
- 1.Adams, R. H., and D. T. Brown. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 54:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., L. J. Carlin, and R. E. Johnston. 1983. Requirement for host transcription in the replication of Sindbis virus. J. Virol. 45:200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., D. W. Lineberger, and R. E. Johnston. 1983. Reduced synthesis of Sindbis virus negative-strand RNA in cultures treated with host transcription inhibitors. J. Virol. 47:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 65:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Asouli, Y., Y. Banai, Y. Pel-Or, A. Shir, and R. Kaempfer. 2002. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 108:221-232. [DOI] [PubMed] [Google Scholar]
- 6.Black, T. L., B. Safer, A. Hovanessian, and M. G. Katze. 1989. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J. Virol. 63:2244-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bommer, U.-A., A. V. Borovjagin, M. A. Greagg, I. W. Jeffrey, P. Russell, K. G. Laing, M. Lee, and M. J. Clemens. 2002. The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA 8:478-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpten, J., N. Nupponen, S. Isaacs, R. Sood, C. Robbins, J. Xu, M. Faruque, T. Moses, C. Ewing, E. Gillanders, P. Hu, P. Bujnovszky, I. Makalowska, A. Baffoe-Bonnie, D. Faith, J. Smith, D. Stephan, K. Wiley, M. Brownstein, D. Gildea, B. Kelly, R. Jenkins, G. Hostetter, M. Matikainen, J. Schleutker, K. Klinger, T. Connors, Y. Xiang, Z. Wang, A. De Marzo, N. Papadopoulos, O. P. Kallioniemi, R. Burk, D. Meyers, H. Gronberg, P. Meltzer, R. Silverman, J. Bailey-Wilson, P. Walsh, W. Isaacs, and J. Trent. 2002. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 30:181-184. [DOI] [PubMed] [Google Scholar]
- 9.Clemens, M. J., and B. R. Williams. 1978. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell 13:565-572. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. L., S. S. Carroll, and L. C. Kuo. 1996. Stoichiometry of 2′,5′-oligoadenylate-induced dimerization of ribonuclease L. A sedimentation equilibrium study. J. Biol. Chem. 271:3979-3981. [DOI] [PubMed] [Google Scholar]
- 11.Condreay, L. D., R. H. Adams, J. Edwards, and D. T. Brown. 1988. Effect of actinomycin D and cycloheximide on replication of Sindbis virus in Aedes albopictus (mosquito) cells. J. Virol. 62:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Groot, R. J., T. Rumenapf, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 88:8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, B., and R. H. Silverman. 1997. A bipartite model of 2-5A-dependent RNase L. J. Biol. Chem. 272:22236-22242. [DOI] [PubMed] [Google Scholar]
- 16.Fata, C., S. G. Sawicki, and D. L. Sawicki. 2002. Alphavirus minus-strand RNA synthesis: role of Arg183 of the nsP4 polymerase. J. Virol. 76:8632-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin, R. M. 1966. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc. Natl. Acad. Sci. USA 55:1504-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 21.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA-temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 63:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, J. Q., and D. J. Barton. 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8:512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 25.Jan, J. T., and D. E. Griffin. 1999. Induction of apoptosis by Sindbis virus occurs at cell entry and does not require virus replication. J. Virol. 73:10296-10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaariainen, L., D. Sawicki, and P. J. Gomatos. 1978. Cleavage defect in the non-structural polyprotein of Semliki Forest virus has two separate effects on virus RNA synthesis. J. Gen. Virol. 39:463-473. [DOI] [PubMed] [Google Scholar]
- 27.Khabar, K. S., M. Dhalla, Y. Siddiqui, A. Zhou, M. N. Al-Ahdal, S. D. Der, R. H. Silverman, and B. R. Williams. 2000. Effect of deficiency of the double-stranded RNA-dependent protein kinase, PKR, on antiviral resistance in the presence or absence of ribonuclease L: HSV-1 replication is particularly sensitive to deficiency of the major IFN-mediated enzymes. J. Interferon Cytokine Res. 20:653-659. [DOI] [PubMed] [Google Scholar]
- 28.Kim, H., S. You, I. J. Kim, J. Farris, L. K. Foster, and D. N. Foster. 2001. Increased mitochondrial-encoded gene transcription in immortal DF-1 cells. Exp. Cell Res. 265:339-347. [DOI] [PubMed] [Google Scholar]
- 29.Koch, F., and G. Koch. 1985. The molecular biology of poliovirus, p. 372-420. Springer-Verlag, New York, N.Y.
- 30.Kowal, K. J., and V. Stollar. 1981. Temperature-sensitive host-dependent mutants of Sindbis virus. Virology 114:140-148. [DOI] [PubMed] [Google Scholar]
- 31.Krug, R. M. 1998. Unique functions of the NS1 protein, p. 82-92. In K. G. Nicholson, R. G. Webster, and A. H. J. Hay (ed.), Textbook of influenza. Blackwell Science, Oxford, United Kingdom.
- 32.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 35.Ou, J. H., E. G. Strauss, and J. H. Strauss. 1983. The 5′-terminal sequences of the genomic RNAs of several alphaviruses. J. Mol. Biol. 168:1-15. [DOI] [PubMed] [Google Scholar]
- 36.Pardigon, N., and J. H. Strauss. 1996. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 70:1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlovic, J., and P. Staeheli. 1991. The antiviral potentials of Mx proteins. J. Interferon Res. 11:215-219. [DOI] [PubMed] [Google Scholar]
- 38.Player, M. R., and P. F. Torrence. 1998. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther. 78:55-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popik, W., and P. M. Pitha. 2000. Exploitation of cellular signaling by HIV-1: unwelcome guests with master keys that signal their entry. Virology 276:1-6. [DOI] [PubMed] [Google Scholar]
- 40.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53-76. [DOI] [PubMed] [Google Scholar]
- 42.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 43.Sawicki, D., D. B. Barkhimer, S. G. Sawicki, C. M. Rice, and S. Schlesinger. 1990. Temperature sensitive shut-off of alphavirus minus-strand RNA synthesis maps to a nonstructural protein, nsP4. Virology 174:43-52. [DOI] [PubMed] [Google Scholar]
- 44.Sawicki, D. L., and S. G. Sawicki. 1994. Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes. Arch. Virol. Suppl. 9:393-405. [DOI] [PubMed] [Google Scholar]
- 45.Sawicki, D. L., and S. G. Sawicki. 1985. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology 144:20-34. [DOI] [PubMed] [Google Scholar]
- 46.Sawicki, D. L., and S. G. Sawicki. 1998. Role of the nonstructural polyproteins in alphavirus RNA synthesis. Adv. Exp. Med. Biol. 440:187-198. [DOI] [PubMed] [Google Scholar]
- 47.Sawicki, D. L., and S. G. Sawicki. 1993. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J. Virol. 67:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawicki, D. L., and S. G. Sawicki. 1980. Short-lived minus-strand polymerase for Semliki Forest virus. J. Virol. 34:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawicki, D. L., S. G. Sawicki, S. Keranen, and L. Kaariainen. 1981. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J. Virol. 39:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawicki, S. G., and D. L. Sawicki. 1986. The effect of loss of regulation of minus-strand RNA synthesis on Sindbis virus replication. Virology 151:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawicki, S. G., and D. L. Sawicki. 1986. The effect of overproduction of nonstructural proteins on alphavirus plus-strand and minus-strand RNA synthesis. Virology 152:507-512. [DOI] [PubMed] [Google Scholar]
- 52.Sawicki, S. G., D. L. Sawicki, L. Kaariainen, and S. Keranen. 1981. A Sindbis virus mutant temperature-sensitive in the regulation of minus-strand RNA synthesis. Virology 115:161-172. [DOI] [PubMed] [Google Scholar]
- 53.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 54.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 55.Sharp, T. V., F. Moonan, A. Romashko, B. Joshi, G. N. Barber, and R. Jagus. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302-315. [DOI] [PubMed] [Google Scholar]
- 56.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276:148-160. [DOI] [PubMed] [Google Scholar]
- 57.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silverman, R. H. 1997. 2-5A-dependent RNase L: a regulated endonuclease in the interferon system, p. 515-551. In G. D'Alessio and J. F. Riordan (ed.), Ribonucleases: structures and functions. Academic Press, Inc., New York, N.Y.
- 59.Silverman, R. H., J. J. Skehel, T. C. James, D. H. Wreschner, and I. M. Kerr. 1983. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 46:1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 61.Squire, J., A. Zhou, B. A. Hassel, H. Nie, and R. H. Silverman. 1994. Localization of the interferon-induced, 2-5A-dependent RNase gene (RNS4) to human chromosome 1q25. Genomics 19:174-175. [DOI] [PubMed] [Google Scholar]
- 62.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 63.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757-766. [DOI] [PubMed] [Google Scholar]
- 65.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 66.Wang, T., and S. G. Sawicki. 2001. Mouse hepatitis virus minus-strand templates are unstable and turnover during viral replication. Adv. Exp. Med. Biol. 494:491-497. [DOI] [PubMed] [Google Scholar]
- 67.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 65:985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watson, J. C., H. W. Chang, and B. L. Jacobs. 1991. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology 185:206-216. [DOI] [PubMed] [Google Scholar]
- 70.Williams, B. R. G. 3July2001, posting date. Signal integration via PKR. Sci. STKE. [Online.]. http://stke.sciencemag.org/cgi/content/full/sigtrans;2001/89/re2. [DOI] [PubMed]
- 71.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. G. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, A., H. Nie., and R. H. Silverman. 2000. Analysis and origins of the human and mouse RNase L genes: mediators of interferon action. Mamm. Genome 11:989-992. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 74.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]