Abstract
The synthesis of the Newcastle disease virus (NDV) fusion (F) protein in a cell-free protein-synthesizing system containing membranes was characterized. The membrane-associated products were in at least two different topological forms with respect to the membranes. The properties of one form were consistent with the expected membrane insertion as a classical type 1 glycoprotein. This form of the protein was fully glycosylated, and sequences amino terminal to the transmembrane domain were protected from protease digestion by the membranes. The second form of membrane-associated F protein was partially glycosylated and partially protected from protease digestion by the membranes. Protease digestion resulted in a 23-kDa protease-protected polypeptide derived from F2 sequences and sequences from the amino-terminal end of the F1 domain. Furthermore, a 10-kDa polypeptide derived from the cytoplasmic domain (CT) was also protected from protease digestion by the membranes. Protease resistance of the 23- and 10-kDa polypeptides suggested that this second form of F protein inserted in membranes in a polytopic conformation with both the amino-terminal end and the carboxyl-terminal end translocated across membranes. To determine if this second form of the fusion protein could be found in cells expressing the F protein, two different approaches were taken. A polypeptide with the size of the partially translocated F protein was detected by Western analysis of proteins in total-cell extracts of NDV strain B1 (avirulent)-infected Cos-7 cells. Using antibodies raised against a peptide with sequences from the cytoplasmic domain, CT sequences were detected on surfaces of F protein-expressing Cos-7 cells by immunofluorescence and by flow cytometry. This antibody also inhibited the fusion of red blood cells to cells expressing F and HN proteins. These results suggest that NDV F protein made both in a cell-free system and in Cos-7 cells may exist in two topological forms with respect to membranes and that the second form of the protein may be involved in cell-cell fusion.
Paramyxoviruses, such as Newcastle disease virus (NDV), encode a fusion glycoprotein (F) that directs the fusion of viral and cellular membranes, which is required for virus entry, and cell-cell fusion, which is required for syncytium formation. The NDV fusion protein, which is synthesized as a precursor, F0, has an amino-terminal signal sequence, a hydrophobic transmembrane domain near the carboxyl terminus, and a 30-amino-acid cytoplasmic tail (13, 14). The mature protein forms a homotrimer (3, 29). Fusion activity of the protein requires a proteolytic cleavage at amino acid 117 to produce disulfide-linked F2 and F1 polypeptides derived from the amino-terminal and carboxyl-terminal domains, respectively (9, 17). The F1 polypeptide has a hydrophobic sequence, the fusion peptide, located at its amino terminus (reviewed in reference 17). Shai and coworkers have proposed that a second fusion peptide exists at an internal location in the F1 polypeptide (7, 27, 28). Fusion peptides are thought to insert into target membranes on activation of fusion, disordering the lipid bilayer (reviewed in references 6 and 12). Paramyxovirus fusion proteins also have two heptad repeat (HR) regions, one located just carboxyl terminal to the fusion peptide (HR1) and the other located adjacent to the transmembrane domain (HR2) (reviewed in references 16 and 17). Results of studies in several systems have led to the hypothesis that paramyxovirus fusion proteins are folded into a metastable state in which the HR domains are not associated and the fusion peptides are masked. On fusion activation, the F protein trimer is thought to refold, inserting fusion peptides into target membranes and forming a very stable, six-stranded coiled-coil domain composed of three HR1 and three HR2 sequences (1, 6, 28).
F proteins expressed on surfaces of infected cells direct cell-cell fusion, resulting in lateral spread of the infection through an organ or through a monolayer of tissue culture cells. Surface-expressed F proteins are also assembled into virus particles. With the goal of exploring the requirements for the synthesis of the metastable form of the paramyxovirus fusion protein required for cell-cell fusion or for virus assembly, we have characterized the NDV F protein made in a cell-free protein-synthesizing system containing membranes. We have made the unexpected observation that products of this reaction contain, in nearly equimolar amounts, two different topological forms.
The topologies of membrane proteins with respect to membranes have been classed into three broad categories (reviewed in references 10 and 20). Type 1 glycoproteins have amino-terminal signal sequences and are positioned such that the carboxyl terminus is located in the cytoplasm and the amino terminus is in the ectodomain. Type 2 glycoproteins have a sequence that serves both as a signal sequence and as a membrane anchor and are positioned with the amino terminus in the cytoplasm and the carboxyl terminus in the ectodomain. Other glycoproteins are polytopic and span membranes multiple times by using alternating signal and membrane anchor sequences; depending on the protein, the amino terminus and the carboxyl terminus may be either cytoplasmic or extracellular. The paramyxovirus F proteins are assumed to be type 1 glycoproteins. Indeed, many structural studies (reviewed in reference 17) including determination of the crystal structure of the F protein (3) confirm the type 1 classification.
There are, however, an increasing number of examples of glycoproteins that are expressed in different topological forms with separate functions. For example, ductin is found in two different orientations in membranes, one of which serves as the subunit of the vacuolar H-ATPase and the other serves as a component of the connexin channel of gap junctions (5). Other examples include the prion protein (11), the human P glycoprotein (33, 39), and aquaporin-1 (19). In addition, the hepatitis B virus surface L glycoprotein assumes different conformations within the membrane, which serve in virus assembly as a matrix-like protein or in virus entry as a receptor binding protein (reference 18 and references therein). An alternative, unglycosylated form of the hepatitis C virus envelope protein that interacts with protein kinase R (PKR) has also been detected (26).
We describe results here that suggest that the NDV F protein exists in at least two different topological forms after synthesis in a cell-free system containing membranes. One form is associated with membranes typical of a type 1 glycoprotein. The second form is partially translocated with only the F2 and the amino terminus of F1 inserted into membranes. In addition, cytoplasmic tail sequences are translocated, suggesting a polytopic conformation. We have also found evidence for this second form in cells expressing the F protein and evidence that this form may be involved in cell-cell fusion.
(The initial experiments were done in partial fulfillment of the requirements for the doctoral dissertation of J.N.R.)
MATERIALS AND METHODS
Cells, virus, and plasmids.
Transcripts of wild-type and mutant NDV F genes were transcribed from pSP6 plasmids containing the inserted genes. Cos-7 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with nonessential amino acids, vitamins, penicillin-streptomycin, and 10% fetal calf serum. NDV F genes were expressed in Cos-7 cells using pSVL (Pharmacia) as previously described (32). Construction of glycosylation site mutants has been previously described (21), as has construction of cytoplasmic tail deletion mutants (31).
NDV strain AV (virulent) and strain B1 (avirulent) stocks (23, 34) were prepared by growth in eggs by standard protocols. AV stocks formed plaques in Cos cells, while B1 stocks did not, consistent with the expected phenotypes of the two strains of NDV. The F protein gene carried by purified NDV strain B1 virus was sequenced to verify the absence of a furin recognition sequence.
Cell-free translation.
Transcription and translation reactions were done as recommended by the Promega Protocols and Application Guide. Transcription reaction mixtures (50 μl) contained 5 μg of linearized DNA, transcription buffer (Promega), dithiothreitol (10 mM), GTP (0.05 mM), UTP, ATP, CTP (each at 0.5 mM), RNasin (1 μl), CAP analogue (0.5 mM), and SP6 polymerase (0.8 U/μl) and were incubated for 40 min at 37°C. RNA transcripts were isolated using RNAeasy RNA Clean Up (Qiagen) as recommended by the manufacturer.
Cell-free translations reaction mixtures (25 μl) contained rabbit reticulocyte extract (17.5 μl) (Promega), amino acids (20 μM each) minus methionine, [35S]methionine (10 μCi) (New England Nuclear), RNasin (1 U/μl), potassium acetate (96 mM), and 0.2 μg of mRNA transcript and were incubated for 1 h at 30°C. Membranes (2 μl/reaction) were a generous gift from Reid Gilmore (37, 38). Translations in wheat germ extracts were accomplished as previously described (38).
Protease digestion of reaction products was accomplished by adding 10 μg of proteinase K per ml of reaction mixture and incubating for 30 min on ice. After digestion, phenylmethylsulfonyl fluoride (0.1 M) was added. For subsequent immunoprecipitation, the reaction mixtures were made 1% with respect to Triton X-100 and 0.5% with respect to sodium deoxycholate.
All reaction products were resolved in 10 or 14% polyacrylamide gels in the with respect to presence of reducing agent. All polyacrylamide gels contained molecular weight marker proteins.
Membrane fractionation.
To separate extracts into membrane and soluble fractions, reticulocyte extracts with or without membranes were diluted in cell fractionation buffer (250 mM sucrose, 10 mM triethanolamine, 1 mM EDTA) and centrifuged for 30 min at 10,000 × g to generate a nonmembrane (soluble) and membrane (pellet) fractions. The pellet fractions were resuspended in cell fractionation buffer.
Transfections.
Transfections using Lipofectamine (Invitrogen) were done as recommended by the manufacturer. Cos-7 cells were plated at 3 × 105 per 35-mm plate. After 20 h, a mixture of DNA (0.5 μg) in 0.1 ml OptiMEM (Gibco/Invitrogen) and 10 μl of Lipofectamine in 0.2 ml of OptiMEM was incubated at room temperature for 40 min, diluted with 0.7 ml of OptiMEM, and added to a 35-mm plate previously washed with OptiMEM. The cells were incubated for 5 h, the Lipofectamine-DNA complexes were removed, and then 2 ml of supplemented DMEM was added.
Antibodies.
Anti-Ftail was raised against a synthetic peptide with the sequence of the cytoplasmic tail of the fusion protein as previously described (36) and prepared by the Peptide Core Facility of the University of Massachusetts Medical School. Anti-NDV is a polyclonal antiserum raised in rabbits against UV-inactivated virions by standard methods. Anti-HR1 was raised against a glutathione S-transferase (GST) fusion protein that contained sequences from amino acids 130 to 173 of the F protein cloned in frame to the carboxyl terminus of GST (using BamHI and EcoRI sites in the pGEX-2T vector [Amersham Biosciences]). Anti-F3b was raised against a GST fusion protein that contained sequences from amino acids 306 to 421 of the F protein similarly cloned in frame to the carboxyl terminus of GST. The GST fusion polypeptides were purified by standard methods recommended in the Novagen Applications Guide. Antibody was raised in rabbits by standard methods by Capralogics, Inc. (Hardwick, Mass.).
Immunofluorescence.
Cos-7 cells were plated in 35-mm plates containing glass coverslips and transfected as described above. After 48 h, the cells were washed twice with ice-cold IF buffer (phosphate-buffered saline [PBS] containing 1% bovine serum albumin, 0.02% sodium azide, and 0.01% CaCl2) and incubated for 1 h at 4°C in IF buffer containing antibody (diluted 1:100). The cells were washed three times with ice-cold IF buffer and incubated for 1 h on ice with IF buffer containing Alexa 488 (Molecular Probes)-labeled anti-rabbit immunoglobulin G (IgG) (Molecular Probes). They were washed with ice-cold IF buffer, fixed with 2% paraformaldehyde, and mounted for microscopy in IF buffer.
Flow cytometry.
Transfected cells, incubated overnight in DMEM without CaCl2 were removed from plates with cell dissociation buffer (Sigma Co.), washed in FACS buffer (PBS containing 1% bovine serum albumin and 0.02% azide), and incubated for 1 h at 4°C with anti-Ftail antibody diluted in FACS buffer. After being washed three times with FACS buffer, the cells were incubated for 1 h at 4°C with goat anti-rabbit IgG coupled to Alexa 488 diluted in FACS buffer. After three washes in FACS buffer, the cells were resuspended in PBS containing 2% paraformaldehye and subjected to flow cytometry (University of Massachusetts Medical School Flow Cytometry Facility).
Preparation of extracts and Western analysis.
Cos-7 cells infected with NDV for 5 to 7 h (multiplicity of infection of 10) were washed in PBS and lysed in RSB buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl) containing 1% Triton X-100, 0.5% sodium deoxycholate, 2 mg of N-ethylmaleimide per ml, and 0.2 mg of DNase per ml. Freshly made total-cell extracts, diluted in sample buffer containing β-mercaptoethanol, were loaded onto 10% polyacrylamide gels without boiling. Molecular weight markers (Amersham Biosciences) were included in each gel. After electrophoresis, the gels were equilibrated in transfer buffer (25 mM Tris, 192 mM glycine, 15% methanol [pH 8.2]) and transferred to Immobilon-P (Millipore Corp.) membranes. The membranes were blocked overnight at 4°C in PBS containing 0.5% Tween 20 and 10% nonfat dried milk. They were then washed in PBS-Tween 20 and incubated for 2 h at room temperature with primary antibody diluted in PBS-Tween 20 and 0.5% nonfat milk. They were washed and then incubated for 2 h at room temperature in secondary antibody, anti-rabbit IgG coupled to horseradish peroxidase (Amersham Biosciences) diluted in PBS-Tween 20-0.5% nonfat milk. The membranes were washed extensively and bound antibody was detected using the ECL Western blotting detection reagent system (Amersham Biosciences).
Fusion assays using R18 labeled RBC.
The method used for the fusion assays was similar to that previously described (13, 14). Avian red blood cells (RBC) (Crane Laboratories) were washed in PBS, resuspended in PBS, and incubated with 15 μg of R18 (Molecular Probes) per ml for 30 min at room temperature in the dark. Three volumes of complete medium (DMEM with 10% fetal calf serum) were added, and incubation was continued for 30 min. The RBC were washed four times in ice-cold PBS, resuspended in PBS containing CaCl2 (0.01%), and added to transfected cells, grown on coverslips, that had been washed in PBS plus CaCl2. The transfected cells were incubated with labeled RBC for 30 min on ice, washed with ice-cold PBS containing CaCl2, and then incubated at 37°C. They were washed with cold PBS containing CaCl2 and immediately visualized, counted, and photographed using a Nikon fluorescence microscope.
RESULTS
NDV F protein and F-protein mutants.
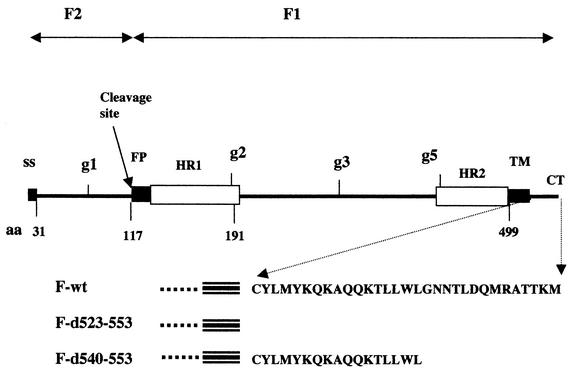
A diagram of the sequence of the NDV F protein is shown in Fig. 1, with domains of the protein indicated as well as the position of mutations utilized. Like a classical type 1 glycoprotein, the NDV F protein has an amino-terminal signal sequence of 31 amino acids (9) and a hydrophobic transmembrane domain (TM) of approximately 24 amino acids located near the carboxyl terminus of the protein (22). The entire F protein cytoplasmic domain (CT), 30 amino acids (22), was deleted in mutant d523-553, while the carboxyl-terminal half of the cytoplasmic domain was deleted in mutant d540-553 (31). The NDV F protein gene encodes a protein of 553 amino acids with a molecular mass without added carbohydrate of 59 or 56 kDa with signal sequence cleavage. There are five glycosylation addition signals within the ectodomain, one in the F2 region and four in the F1 region (21). Analysis of mutant proteins (Fg1, Fg2, Fg3, Fg4, and Fg5) altered in each of the five addition signal sites has shown that four of these sites are used in the wild-type protein, g1, g2, g3, and g5, resulting in a fully glycosylated 68-kDa protein (21).
FIG. 1.
Location of domains in the primary sequence of the NDV F protein. A diagram of the sequence of the NDV F protein (22) is presented with important domains indicated: ss, signal sequence; FP, fusion peptide; HR, heptad repeat; TM, transmembrane domain; CT, cytoplasmic tail; used glycosylation addition sites, g1, g2, g3, and g5. The position of the cleavage site used to generate F2 and F1 polypeptides is indicated by an arrow. Locations of specific amino acids (aa) are indicated below the line. Also below the line are the sequences of the CT domain of wild-type (wt) and deletion mutant F proteins, designated by the amino acids deleted.
Cell-free translation of F-protein mRNA.
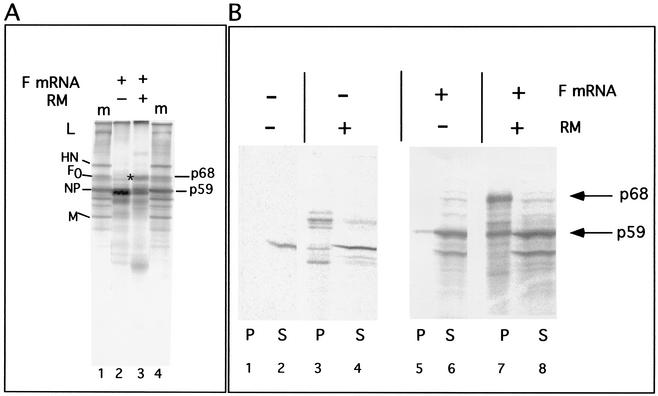
The translation of F-protein mRNA in a rabbit reticulocyte extract (Fig. 2A) in the absence of canine pancreas membranes yielded a polypeptide with a molecular mass of approximately 59 kDa (Fig. 2A, lane 2), consistent with the size of the unglycosylated F protein with an intact signal sequence. In the presence of canine pancreas membranes, F-protein mRNA directed the synthesis of the p59 product as well as a new product that comigrated with the glycosylated F0 protein found in infected cell extracts (lane 3, p68). Similar results were obtained using wheat germ extracts containing membranes (Fig. 3, lane 13). However, the fully glycosylated F protein consistently made up only 25 to 35% of the total product in reticulocyte extracts and even less in wheat germ extracts. The fraction of glycosylated F protein in the total product was not increased by using different preparations of canine pancreas membranes or increased amounts of membranes. This rather inefficient synthesis of a fully glycosylated product was in contrast to products directed by NDV HN mRNA or vesicular stomatitis virus G mRNA, in which 60 to 70% of the product was glycosylated under the same conditions (data not shown).
FIG. 2.
Translation and membrane association of F mRNA in reticulocyte cell-free protein synthesis reactions. (A) Translation of F-protein mRNA transcripts in a reticulocyte extract in the absence (lane 2) or presence (lane 3) of canine pancreas membranes (RM). *, p68 product of the reaction with membranes present. Lanes 1 and 4 contain radioactively labeled proteins present in infected-cell extracts (m, marker). L, large protein; HN, hemagglutinin-neuraminidase protein; F0, uncleaved fusion protein; NP, nucleocapsid protein; M, membrane protein. The lower-molecular-mass band in lane 3 is due to endogenous RM mRNAs (see Fig. 3). (B) Extracts incubated in the absence (lanes 1 to 4) or presence (lanes 5 to 8) of F-protein mRNA transcripts and the absence (lanes 1, 2, 5, and 6) or presence (lanes 3, 4, 7, and 8) of canine pancreas membrane were fractionated into pellet (P) and soluble (S) fractions as described in Materials and Methods.
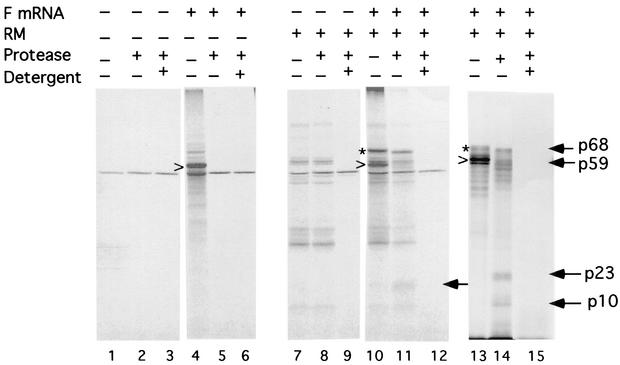
FIG. 3.
Protease protection of cell-free translation products made in the presence of membranes. Products made in the absence (lanes 1 and 4) or presence (lanes 7, 10, and 13) of membranes (RM) were digested with proteinase K in the absence (lanes 2, 5, 8, 11, and 14) or presence (lanes 3, 6, 9, 12, and 15) of Triton X-100. Products which were the result of endogenous transcripts in reticulocyte extracts or associated with the membranes are shown in lanes 1 to 3 and 7 to 9, respectively. The products of reactions in the presence of F mRNA are shown in lanes 4 to 6, 10 to 12, and 13 to 15. The fully glycosylated F protein (p68) is indicated by an asterisk. The minimally glycosylated products (p59) are indicated by the arrowhead. Smaller protected fragments are indicated by arrows. Lanes 13 to 15 contain products of wheat germ reactions carried out in the presence of membranes, while all other lanes contain products of reticulocyte extracts. The membranes used for the experiments in lanes 13 to 15 were from a different preparation from those used in the experiments in the other lanes.
Membrane association and translocation of the F protein.
One interpretation of these results was that the 59-kDa product was untranslocated polypeptide and that the membrane insertion of the F protein was unusually inefficient. To test this idea, membrane association of the products was determined by fractionation into a soluble fraction and a membrane pellet (Fig. 2B). As expected, F protein made in the absence of membranes remained largely in the supernatant or soluble fraction (Fig. 2B, lanes 5 and 6). The fully glycosylated F protein (p68) made in the presence of membranes pelleted with membranes, as expected (lane 7). However, approximately 50% of the p59 product also pelleted with membranes, suggesting some membrane association of the p59 polypeptide (lanes 7 and 8).
To explore the nature of this membrane association, the protease resistance of the products of the cell-free reaction was characterized, since translocated material should be protected from digestion. Protease digestion reduced the size of the fully glycosylated F protein by approximately 3 kDa (Fig. 3, lanes 11 and 14), a result consistent with the expected removal of the exposed cytoplasmic tail. Furthermore, the fully glycosylated molecule was completely digested on disruption of the membranes with detergent (lanes 12 and 15) showing that protease resistance was due to protection by membranes. Protease digestion of the F protein made in the absence of membranes completely digested the protein in the presence or absence of detergent (lanes 5 and 6).
The p59 product in the presence of intact membranes was also digested with protease (Fig. 3, lane 11). However, there was reproducibly a 23-kDa protected polypeptide (lanes 11 and 14) that was completely digested on membrane disruption (lanes 12 and 15). Protease digestion of the wheat germ product yielded a 23-kDa protected fragment as well as a 10-kDa protected fragment, both of which were completely digested on detergent disruption of the membranes (lanes 14 and 15). The smaller (10-kDa) protected fragment was also detected in the reticulocyte product after immunoprecipitation with antibody specific for NDV proteins and was not detected in the total product probably because of background comigrating polypeptides (see Fig. 5).
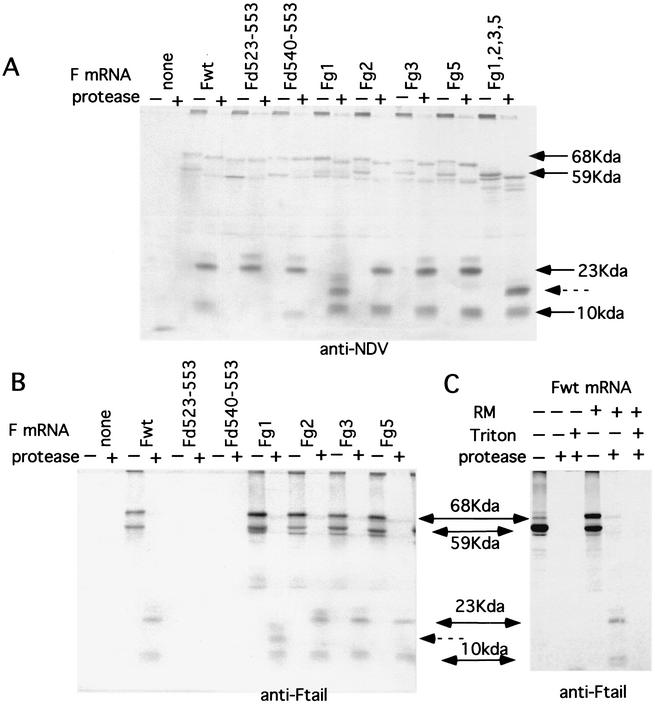
FIG. 5.
Immunoprecipitation of protease-digested products of reactions directed by mutant transcripts. (A and B) Products of reactions identical to those in Fig. 4 were digested with proteinase K and then precipitated with anti-NDV (A) or anti-Ftail (B). Precipitated material was electrophoresed on 14% polyacrylamide gels. The sizes of polypeptides indicated by arrows were determined using Rainbow markers electrophoresed on the same gel (data not shown). Fg1, Fg2, Fg3, and Fg5 are glycosylation site mutants mutated in a single-use glycosylation site. Fg1,2,3,5 encodes an F protein mutated in all four used glycosylation sites. Fd523-553 and Fd540-553 are CT deletion mutants described in Fig. 1. (C) Products of reactions directed by the wild-type transcript (Fwt) in the presence or absence of membranes (RM) and digested with protease in the presence or absence of Triton X-100. The products were then were precipitated with anti-Ftail antibody.
Glycosylation of the reaction products.
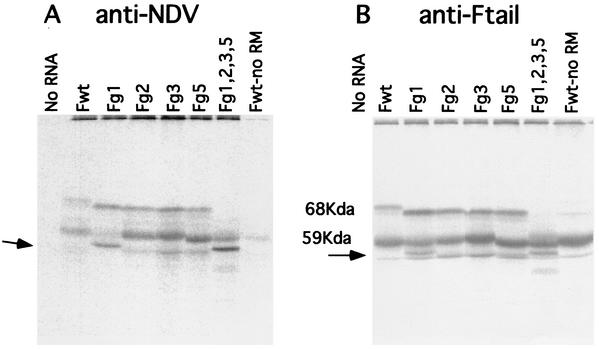
These smaller protected fragments suggested that the membrane-associated p59 product might be partially translocated. Translocation of a polypeptide can also be assessed by the glycosylation of the product. To identify glycosylation sites used in the membrane-associated forms of the F protein, mRNAs transcribed from mutant F-protein cDNAs with alterations in each of the glycosylation addition signals were translated in the presence of membranes and the products were precipitated with anti-NDV antibody or anti-Ftail antibody (Fig. 4). As expected, since each mutation resulted in the absence of a side chain on the fully glycosylated F protein, the fully glycosylated F protein derived from each mutant mRNA had a slightly lower molecular mass than did the wild-type protein. However, translation of mRNA from the Fg1 mutant gene resulted in the appearance of a new band, with a molecular mass of approximately 56 kDa, with a concomitant reduction in the amount of p59 (Fig. 4, arrow). This polypeptide was also detected in products directed by an mRNA transcribed from an F-protein cDNA missing all the glycosylation sites (Fg1, Fg2, Fg3, and Fg5). The new 56-kDa polypeptide may result from the partial translocation of the F protein, signal sequence cleavage, and absence of glycosylation due to the Fg1 mutation resulting in slightly faster migration than that of untranslocated material. Thus, the wild-type, membrane-associated p59 polypeptide may be partially translocated. Glycosylation at the g1 site would increase size of the molecule by approximately 3 kDa and signal sequence cleavage would reduce the size by 3 kDa, resulting in a molecule that comigrated with the unglycosylated product made in the absence of membranes. The fact that only the Fg1 mutation affected the size of the p59 polypeptide suggested that the amino-terminal end of the F protein was translocated but that regions of the molecule containing the other glycosylation addition sites were not translocated. Partial translocation of the F protein could account for the smaller polypeptides detected after protease digestion of membranes.
FIG. 4.
Translation of mRNA transcripts of F-protein glycosylation site mutants. Products of reticulocyte reactions in the presence of membranes and directed by mRNAs derived from glycosylation site mutant DNAs (indicated above each lane) were precipitated with anti-NDV (A) or anti-Ftail (B). Transcripts from mutant DNAs containing single-site mutations (Fg1, Fg2, Fg3, and Fg5) or containing mutations in all four used sites (Fg1,2,3,5) were used. Reaction products directed by F wild-type mRNA in the absence of membranes are shown in the right-hand lane of each panel (Fwt-no RM). The new polypeptides detected in Fg1 and in Fg1,2,3,5 products are indicated by the arrow.
Translocation of glycosylation mutants.
The notion of partial translocation of the F protein was supported by the sizes of protease-protected fragments derived from glycosylation mutant proteins. Figure 5A shows protease digestion of products made by transcripts of F glycosylation mutants immunoprecipitated with anti-NDV antibody. The migration of the 23-kDa protease-resistant polypeptide was affected by loss of the g1 glycosylation site but not by mutation of other sites (23 kDa, arrow). Interestingly, the anti-NDV antibody precipitated the protease-protected fragments more efficiently than it precipitated full-sized F protein.
On immunoprecipitation of the proteins directed by wild-type and mutant mRNAs, an additional, protease-protected polypeptide with a molecular mass of approximately 10 kDa (Fig. 5A) was also detected, a polypeptide that was masked by background bands in the unprecipitated products of the reticulocyte reaction. This species was identical in size to that seen in protease digestion products of wheat germ reactions (Fig. 3, lane 14). The size of this protected fragment was unchanged by elimination of any of the glycosylation addition sites. It was, however, completely digested in the presence of detergent (Fig. 5C).
Translocation of cytoplasmic domain mutants.
The protease-protected fragments were also characterized in products directed by mRNAs transcribed from cDNAs of two mutant F protein genes missing the entire cytoplasmic tail sequence of the protein (d523-553) or half of the cytoplasmic tail (d540-553) (Fig. 5A). As expected, protease digestion of the products did not reduce the size of the fully glycosylated F-dG523-553 and only slightly reduced the size of F-d540-553 (Fig. 5A), a result consistent with the deletion of all or part of the CT sequences in these mutant proteins. Also as expected, neither mutation affected the size of the 23-kDa protected species (Fig. 5A). However, surprisingly, deletion of the CT sequences eliminated the 10-kDa protected fragment (Fig. 5A). Furthermore, this fragment, derived from the d540-553 mutant protein, was slightly smaller, consistent with the deletion of approximately half of the cytoplasmic tail. This result suggests that the 10-kDa membrane-protected fragment may be derived from the cytoplasmic tail of the F protein.
This surprising result was confirmed by precipitating digestion products with antibody specific for the cytoplasmic tail sequences of the F protein (Fig. 5B). Using this antibody, the fully glycosylated F protein in membranes after protease digestion was not precipitated with anti-Ftail antibody, consistent with the removal of the cytoplasmic tail sequences by the protease. However, the 10-kDa fragment derived with wild-type translations was precipitated but was missing from products derived from the cytoplasmic tail deletion mutants. This result suggests that CT sequences are protected from protease digestion and may be translocated across membranes.
The 23-kDa protease-resistant fragment was also precipitated by the anti-Ftail antibody in proteins with CT domains but not in products of proteins mutated in CT sequences. This result suggests an interaction between the CT sequence and the 23-kDa polypeptide.
These combined results are consistent with a partial translocation of the amino-terminal region of the F protein as well as the carboxyl-terminal region.
Western analysis of fusion protein in infected-cell extracts.
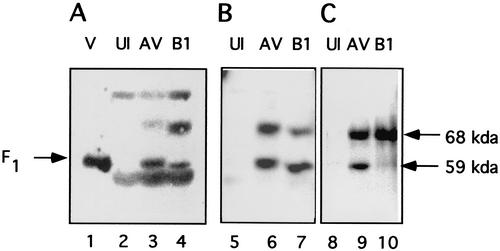
We wished to determine if the partially glycosylated form of the F protein was characteristic of only the cell-free system or if this novel form of F protein existed in NDV-infected cells. Detection of the partially glycosylated molecule in infected cells was not straightforward since the F1 protein, the result of cleavage of the fully glycosylated fusion protein, is the same size as the partially glycosylated F protein detected in the cell-free system. To eliminate complications due to the presence of F1, we used cells infected with the B1 strain of NDV. This strain encodes an F protein missing a furin recognition sequence and is therefore uncleaved in tissue culture cells (8, 35) but cleaved when grown in fertilized eggs due to the presence of factor X in the chorioallantoic fluid of eggs (reviewed in reference 17). Thus, any F1-sized polypeptide found in tissue culture cells infected with NDV strain B1 is a candidate for the partially glycosylated form of the protein. In contrast, NDV strain AV encodes an F protein with a furin sequence and the F protein is proteolytically cleaved (35). Different F-specific antibodies were screened in a Western analysis of proteins in total-cell extracts derived from NDV strain AV- or B1-infected cells (Fig. 6). Results with three different antibodies raised against different sequences in the F-protein F1 domain are shown. As expected, extracts from AV-infected cells contained two polypeptides, F0 and F1, that react with all antisera (Fig. 6, lanes 3, 6, and 9). Extracts from B1-infected cells also contained two polypeptides, F0 and an F1-sized polypeptide, that reacted with two of the antibodies (lanes 4 and 7). One of these polypeptides comigrated with F0, and the second migrated with the size of the partially glycosylated form of the F protein. This result is consistent with the presence of the partially glycosylated form of the protein, at least in NDV-B1-infected cells. It should be noted that not all antibodies would detect the p59-sized polypeptide in B1-infected cell extracts. For example, antibody specific for the cytoplasmic tail would not detect this second form or, in some experiments, would only weakly detect the polypeptide (lane 10).
FIG. 6.
Western analysis of proteins synthesized in infected cells. Cos-7 cells, infected with NDV strain AV or strain B1 or mock infected (UI) for 7 h were lysed, and aliquots of total-cell extract were subjected to Western analysis as described in Materials and Methods. Blots were incubated with anti-F3b (lanes 2 to 4), anti-HR1 (lanes 5 to 7), or anti-Ftail (lanes 8 to 10). Molecular mass markers were included in each blot and were used to determine the sizes of the bands detected. B1 virions isolated from infected, fertilized eggs (and therefore containing a cleaved F protein [17]) are shown in lane 1. Uninfected extracts are shown in lanes 2, 5, and 8; AV-infected extracts are shown in lanes 3, 6, and 9; and B1-infected extracts are shown in lanes 4, 7, and 10.
Surface expression of cytoplasmic tail sequences.
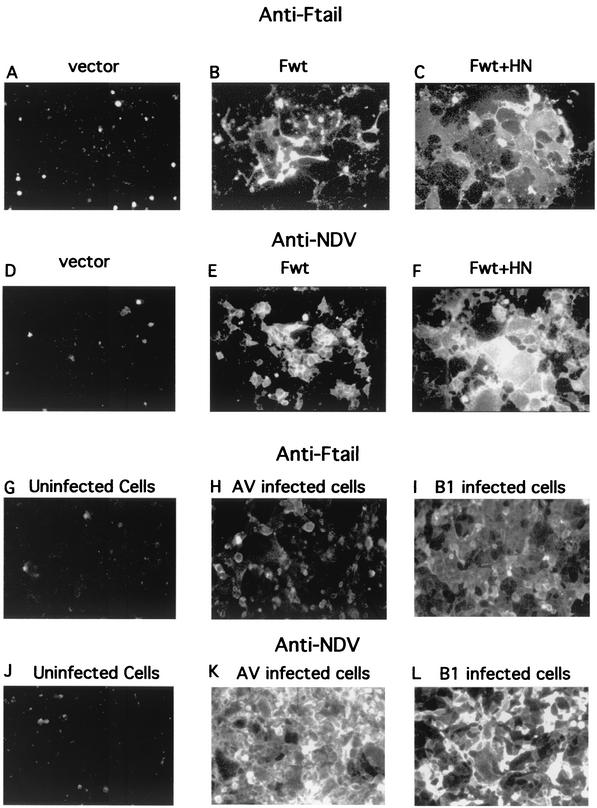
Protease resistance of CT sequences in the presence of membranes in the cell-free system raised the possibility that these sequences are translocated across the membrane. If this translocation occurs in infected cells and if this form of the F protein is transported to cell surfaces, then cytoplasmic tail sequences should be accessible to antibody on the surfaces of intact cells. To test this idea, intact cells transfected with vector alone, wild-type F-protein cDNA, or wild-type F cDNA and HN cDNA were incubated with anti-Ftail antibody and the binding of antibody was assessed by immunofluorescence of surfaces of these cells. Clearly, cells expressing F protein were positive for anti-Ftail binding (Fig. 7B). Furthermore, syncytia resulting from the coexpression of the HN and F proteins were also positive for anti-Ftail binding (Fig. 7C). Binding of anti-NDV antibody was detected in parallel cultures (Fig. 7D through F). NDV strain AV- and B1-infected cells also were positive for anti-Ftail binding (Fig. 7H and I).
FIG. 7.
Detection of CT sequences on the surfaces of transfected and infected cells by immunofluorescence. (A to F) Results obtained with transfected Cos-7 cells. (G to L) Results obtained with NDV-infected Cos-7 cells. Cos-7 cells transfected with vector alone (A and D), with pSVL-Fwt (B and E), or with pSVL-Fwt and pSVL-HN (C and F) were incubated with anti-Ftail (A to C) or anti-NDV (D to F). Panels A to C were exposed to film for 4 s, while panels D to F were exposed for 1.5 s. Uninfected Cos-7 cells (G and J) or Cos-7 cells infected with NDV strain AV (H and K) or strain B1 (I and L) were incubated with anti-Ftail (G to I) or anti-NDV (J to L). Infected cells incubated with anti-Ftail were exposed to film for 2 s, while infected cells incubated with anti-NDV were exposed to film for 1.5 s.
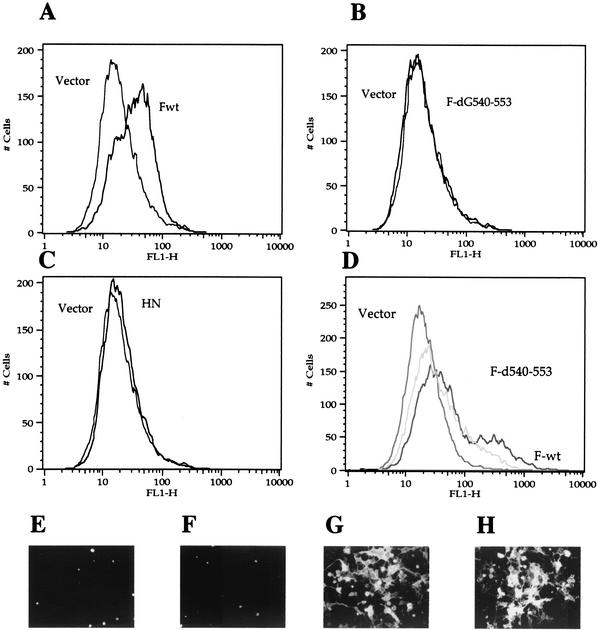
To confirm the surface expression of CT sequences, binding of anti-Ftail antibody was also analyzed by flow cytometry. Figure 8A, shows that cells expressing F protein bound anti-Ftail antibody. In addition, cells expressing only the HN protein were negative (Fig. 8C).
FIG. 8.
Detection of CT sequences on the surfaces of cells transfected with F-wt and F-d540-553 DNAs. (A-D) Results of flow cytometry. (A) pSVL-Fwt using anti-Ftail antibody; (B) pSVL-F-d540-553 using anti-Ftail antibody; (C) pSVL-HN using anti-Ftail antibody; (D) pSVL-Fwt (black line) and pSVL-F-d540-553 (gray line) using anti-NDV antibody. FL1-H, fluorescence. Each panel also shows cells transfected with vector alone and incubated with both primary and secondary antibody as a background control. (E to H) Detection of CT sequences on surfaces of transfected cells by immunofluorescence, (E) Vector incubated with anti-Ftail; (F) F-d540-553 incubated with anti-Ftail; (G) F-d540-553 incubated with anti-NDV; (H) F-wt incubated with anti-NDV. Panels E and F were exposed for 3 s, and panels G and H were exposed for 2 s.
To verify that the binding by anti-Ftail antibody was specific for the CT sequences of F protein, cells expressing F protein with a deletion of CT sequences, F-d540-553, were analyzed by flow cytometry and immunofluorescence (Fig. 8B and F). Anti-Ftail antibody did not bind to these cells in either assay, although, as previously reported (31), this mutant protein is expressed at cell surfaces, as shown by flow cytometry and immunofluorescence using anti-NDV antibody (Fig. 8D and G, respectively). These results indicated that binding of anti-Ftail to cell surfaces is specific to the CT sequences in the F protein.
Inhibition of fusion by antibody to CT sequences.
To ask if the CT sequences detected at cell surfaces played any role in cell-cell fusion, the effect of antibody to CT sequences on membrane fusion was determined. We tested the effect of antibody on the fusion of RBC with Cos-7 cells expressing the HN and F proteins. Initial steps in RBC fusion to HN- and F-protein-expressing cells can be monitored by the transfer of fluorescence-labeled lipid from RBC membranes to the membranes of cells expressing viral fusion proteins. Cos-7 cells transfected with genes encoding the HN protein and the wild-type F protein form large syncytia which bind RBC. On incubation at 37°C, these cells fused with the RBC and the fluorescence-labeled lipid in RBC spread into the syncytia (Fig. 9A). Eighty percent of syncytia were positive for fusion with RBC, showing that proteins expressed on the surfaces of the syncytia are capable of directing fusion on binding of the RBC.
FIG. 9.
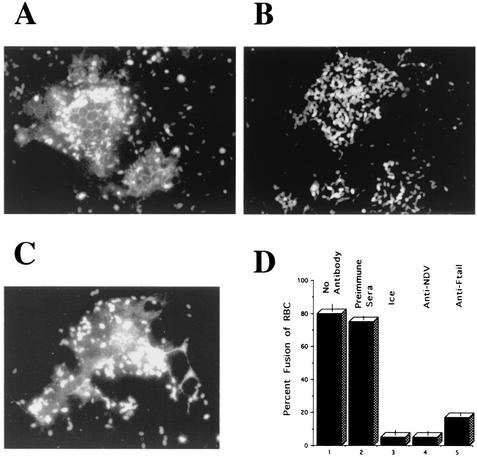
Inhibition of fusion by anti-Ftail. At 48 h after cotransfection of Cos-7 cells with pSVL-Fwt and pSVL-HN, monolayers were incubated in the presence or absence of anti-Ftail antibody for 30 min on ice. The cells were then incubated on ice with R18-labeled RBC in the presence or absence of antibody. After the unbound RBC were washed away, the cells were incubated at 37°C in the presence or absence of antibody. Each field shows a syncytium. Similar results were obtained in four separate experiments using different lots of RBC. (A) No antibody; (B) anti-Ftail; (C) preimmune sera; (D) percentage of syncytia that were positive for dye transfer (fusion): column 1, no antibody added; column 2, preimmune sera; column 3, cells incubated only on ice; column 4, anti-NDV antisera (added only after RBC binding); column 5, anti-Ftail. The results in panel D are the average of three experiments. In each experiment, 50 to 100 syncytia were counted. Bars indicate variation between experiments.
To determine if anti-Ftail antibody could inhibit this fusion, the antibody was present before, during, and after RBC binding. The presence of the antibody did not decrease the total binding of RBC to the cell monolayer (data not shown). Clearly, anti-Ftail antibody inhibited the fusion of the RBC with syncytia (Fig. 9B), since there was no transfer of the label from the RBC into syncytia. Eighty five percent of all syncytia were negative for fusion. This inhibition is not due to a general response to IgG. Preimmune sera did not inhibit RBC fusion with syncytia (Fig. 9C). These results were quantitated (Fig. 9D) by determining the percentage of syncytia that were positive for fusion of RBC. In addition, as shown in Fig. 9D, no fusion was observed after incubation on ice or after incubation at 37°C in the presence of anti-NDV added after RBC binding. That anti-Ftail inhibited fusion raises the possibility that a second form of the fusion protein participates in cell-cell fusion.
DISCUSSION
The membrane-associated products of cell-free synthesis directed by the NDV F mRNA existed in at least two different topological forms that were found in roughly equal proportions. The properties of one of these forms are consistent with the expected membrane insertion as a classical type 1 glycoprotein with a short carboxyl-terminal sequence located in the cytoplasmic side of membranes (Fig. 10, species 1). This form was glycosylated at the g1, g2, g3, and g5 sites, and sequences amino terminal to the TM domain were protected from protease digestion by membranes. Another translocated form of the protein product was detected by its membrane association, partial glycosylation, and protease digestion products. This form had properties consistent with the partial insertion of the protein into membranes, an insertion of sequences from the F2 region as well as the amino terminus of the F1. After accounting for signal sequence cleavage and glycosylation at site g1, the 23-kDa size of the protected fragment is consistent with translocation of approximately 200 amino acids of the amino terminus of F0. In addition, the predominant form of the fragment was glycosylated at only the g1 site, not the g2 site, which is at amino acid 191. Since a glycosylation addition site must be 12 to 14 amino acids from the membrane in order to be recognized by the luminal oligosaccharide transferase (24), the most distal region of the F protein that could serve as a stop transfer sequence would be from approximately amino acids 200 to 225. Thus, both the sizes and glycosylation status of the protected fragments argue that the most C-terminal sequence that could serve in translocation arrest would be somewhere between approximately residues 190 and 225. However, given the uncertainties of size determination of glycoproteins on polyacrylamide gels, it is also possible that more amino-terminal sequences in the fusion peptide or HR1 domain serve as a translocation arrest signal. It is interesting that Shai and colleagues have reported that a Sendai virus F peptide with sequences comparable to amino acids 200 to 225 in the NDV F protein inserts into membranes (7, 27).
FIG. 10.
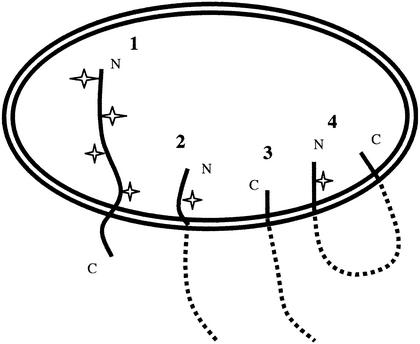
Membrane topologies of the F protein. Shown are the possible topologies of the F protein in the rough endoplasmic reticulum. Species 1 is typical of a type 1 glycoprotein, with the amino terminus (N) in the lumen and the carboxyl terminus (C) in the cytoplasm, and would account for the fully glycosylated form of the protein. Species 2 shows a partially translocated molecule with F2 sequences inserted and would account for the partially translocated form of the protein that is glycosylated at site 1. Species 3 shows the translocation of only the CT sequences, with the amino terminus of the protein untranslocated. Species 4 would occur if the F2 and CT sequences detected on the luminal side of membranes were on the same molecule. Glycosylation sites are indicated by “stars”. The precise disposition with respect to membranes of the presumed cytoplasmic domains of the protein in species 2 to 4 is uncertain and is therefore represented by dashed lines.
Protease digestion of membrane-associated products also detected another, smaller protected fragment with an approximate size of 10 kDa. That this fragment was derived from CT sequences was suggested by its precipitation with antibody specific for these sequences as well as its disappearance from the digestion products of a mutant F protein with a CT deletion. These sequences could be embedded in membranes in a way that protects them from protease digestion. Alternatively, the protease protection of these sequences could be due to their translocation. Indeed, detection of CT sequences on surfaces of cells expressing the F protein argue that the CT sequences are translocated. The predicted size of a fragment with CT sequences as well as TM sequences would be approximately 6 kDa. However, the CT sequence of the NDV F protein has a glycosylation addition signal (21), which, if used, would increase the size of the fragment by approximately 3 kDa, bringing the predicted size of the combined CT and TM sequences within the range of the detected fragment. Alternatively, to account for the size of the fragment, a portion of the HR2 domain may be protected from protease digestion.
Whether the protease-protected CT sequences and the amino-terminal protected fragment are derived from the same molecule is not clear. Potentially, these protected species are derived from two different topological forms, illustrated in Fig. 10 as forms 2 and 3. Alternatively, they could be derived from the same molecule (form 4) that was inserted as a loop. This latter form might be possible if the TM sequence serves as a signal sequence when located after a translocation arrest mediated by more amino-terminal sequences. Such a scenario is typical of membrane insertion of polytopic membrane proteins. That both protected fragments were immunoprecipitated with anti-Ftail suggests that the two sequences may interact.
Use of cell-free translation systems has been a reliable and widely used method for exploring the disposition of proteins in membranes. However, a mixed topology was potentially a characteristic of the cell-free system only, and it was important to demonstrate the phenomenon in F-protein-expressing cells. Two lines of evidence do suggest that the F protein may assume a dual topology in cells expressing this protein. First, CT sequences could be detected, both by immunofluorescence and by flow cytometry, on surfaces of cells expressing the wild-type F protein but not of cells expressing F protein with CT sequences deleted or cells expressing only the HN protein. Surfaces of infected cells were also positive for CT sequences. Second, a polypeptide with the size of the partially glycosylated form of the F protein could be detected by Western analysis of proteins in total-cell extracts prepared from NDV strain B1-infected cells. This form of the NDV F protein was probably not previously detected for several reasons. First, it comigrates with the cleaved form of the F protein, F1. Second, the protein, with minimal glycosylation, is very hydrophobic (22) and probably readily aggregates and sediments with nuclei in the preparation of cytoplasmic extracts. Indeed, we reproducibly detected this second form only in freshly made and unfrozen total-cell extracts. Third, not all antibodies detected this form of the protein.
The potential of internal sequences of a paramyxovirus F protein to serve as stop transfer signals was a question explored many years ago (25). The focus of these early experiments was the fusion peptide. Indeed, it was concluded that the fusion peptide did not serve as a stop transfer/membrane anchor when located internally in a polypeptide although, when placed at the end of a sequence, the fusion peptide could function in this way (25). In contrast, the results presented here are consistent with sequences carboxyl terminal to the fusion peptide functioning in translocation arrest. The potential translocation arrest activity of these sequences was not previously explored. Furthermore, since there were no examples of proteins with mixed topology at the time of the earlier studies, the possibility that the F protein might fold in two different ways was not considered.
Clearly, a key question is the functional significance of this second form of the F protein detected in the cell-free system and in cells, a question currently under investigation. While this second form of the F protein may play no role, this possibility seems unlikely, given the economy of viral replication mechanisms. This second form could participate directly in membrane fusion, in virus assembly, or in glycoprotein folding. Alternatively, the partially translocated form may play some regulatory role in any of these processes. For example, the partial translocation of the F protein could serve to down regulate the expression of a fully active F protein. It is also possible that its function is unrelated to membrane fusion, as suggested for the alternative form of the HCV env protein (26). However, it has been found that deletion of the cytoplasmic tail of the NDV F protein renders the protein fusion defective, for reasons that have not been clear (31), although this result may suggest some role of the CT domain in fusion. That antibody against the cytoplasmic tail sequences can block the fusion of RBC with cells expressing the HN and F proteins also suggests that these sequences, exposed on cell surfaces, may play a role in fusion. The antibody may indirectly interfere with conformational changes in the fully translocated F protein required for activation of fusion by virtue of its proximity to molecules capable of directing fusion. Alternatively, a second form of the F protein may play some direct role in membrane fusion. This second possibility would considerably complicate current models for F-protein-mediated fusion.
It is interesting that some antibodies raised against the cytoplasmic tail of human immunodeficiency virus gp41 neutralize infection (2, 15). This observation led to the suggestion that these sequences were expressed on cell surfaces, although this proposal is controversial (30). It was suggested that these surface-exposed human immunodeficiency virus gp41 CT sequences play some role in postattachment steps in virus entry (4).
Acknowledgments
This work was supported by grants GM 37745 and AI 30572 from the National Institutes of Health.
We thank Reid Gilmore for generously supplying canine pancreas membranes. We thank Anne Haywood for critically reading the manuscript.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Chanh, T. C., G. R. Dreesman, P. Kanda, G. P. Linette, J. T. Sparrow, D. D. Ho, and R. C. Kennedy. 1986. Induction of anti-HIV neutralizing antibodies by synthetic peptides. EMBO J. 5:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland, S. M., T. D. Jones, and N. J. Dimmock. 2000. Properties of a neutralizing antibody that recognizes a conformational form of epitope ERDRD in the the gp41 C-terminal tail of human immunodeficiency virus type 1. J. Gen. Virol. 81:1251-1260. [DOI] [PubMed] [Google Scholar]
- 5.Dunlop, J., P. C. Jones, and M. E. Finbow. 1995. Membrane insertion and assembly of ductin: a polytopic chanel with dual orientations. EMBO J. 14:3609-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh, J. K., S. G. Peisajovich, and Y. Shai. 2000. Sendai virus internal fusion peptide: structural and functional characterization and a plausible mode of viral entry inhibition. Biochemistry 39:11581-11592. [DOI] [PubMed] [Google Scholar]
- 8.Glickman, R. L., R. J. Syddall, R. M. Sheehan, and M. A. Bratt. 1988. Quantitative basic residue requirements in the cleavage activation site of the fusion glycoprotein as a determinant of virulence of Newcastle disease virus. J. Virol. 62:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorman, J. J., A. Nestorowicz, S. J. Mitchell, G. L. Corino, and P. W. Selleck. 1988. Charaterization of the sites of proteolytic activation of Newcastle disease virus membrane glycoprotein precursors. J. Biol. Chem. 263:12522-12531. [PubMed] [Google Scholar]
- 10.Hegde, R. S., and V. R. Lingappa. 1999. Regulation of protein biogenesis at the endoplasmic reticulum membrane. Trends Cell Biol. 9:132-137. [DOI] [PubMed] [Google Scholar]
- 11.Hegde, R. S., J. A. Mastrianni, M. R. Scott, K. DeFea, A., P. Tremblay, S. B. Torchia, and V. R. Lingappa. 1998. A transmembrane form of the prion protein in neurodegenerative disease. Science 279:827-834. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 13.Kemble, G. W., T. Danieli, and J. W. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 14.Kemble, G. W., Y. I. Henis, and J. M. White. 1993. GPI- and transmembrane-anchored influenza hemagglutinin differ in structure and receptor binding activity. J. Cell Biol. 122:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy, R. C., R. D. Henkel, D. Pauletti, J. S. Allan, T. H. Lee, M. Essex, and G. R. Dreesman. 1986. Antiserum to a synthetic peptide recongnized the HTLV-III envelope glycoprotein. Science 231:1556-1559. [DOI] [PubMed] [Google Scholar]
- 16.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis of changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 18.Lambert, C., and R. Prange. 2001. Dual topology of the hepatitis B virus large envelope protein. J. Biol. Chem. 276:22265-22272. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Y., I. R. Turnbull, A. Bragin, K. Carveth, A. S. Verkman, and W. R. Skach. 2000. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell 11:2973-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 21.McGinnes, L., T. Sergel, J. Reitter, and T. Morrison. 2001. Carbohydrate modifications of the NDV fusion protein heptad repeat domains influence maturation and fusion activity. Virology 283:332-342. [DOI] [PubMed] [Google Scholar]
- 22.McGinnes, L. W., and T. G. Morrison. 1986. Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 5:343-356. [DOI] [PubMed] [Google Scholar]
- 23.Nagai, Y., and H.-D. Klenk. 1977. Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology 77:125-134. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson, I., and G. von Heijne. 1993. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268:5798-5801. [PubMed] [Google Scholar]
- 25.Paterson, R. G., and R. A. Lamb. 1987. Ability of the hydrophobic fusion-related external domain of a paramyxovirus F protein to act as a membrane anchor. Cell 48:441-452. [DOI] [PubMed] [Google Scholar]
- 26.Pavio, N., D. R. Taylor, and M. M. C. Lai. 2002. Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR. J. Virol. 76:1265-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peisajovich, S. G., O. Samuel, and Y. Shai. 2000. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 296:1353-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peisajovich, S. G., and Y. Shai. 2002. New insights into the mechanism of virus-induced membrane fusion. Trends Biochem. Sci. 27:183-190. [DOI] [PubMed] [Google Scholar]
- 29.Reitter, J., T. Sergel, and T. Morrison. 1995. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J. Virol. 69:5995-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 31.Sergel, T., and T. Morrison. 1995. Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology 210:264-272. [DOI] [PubMed] [Google Scholar]
- 32.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skach, W. R., M. C. Calayag, and V. R. Lingappa. 1993. Evidence for an alternate model of human P-glycoprotein structure and biogenesis. J. Biol. Chem. 268:6903-6908. [PubMed] [Google Scholar]
- 34.Toyoda, T., T. Sakaguchi, H. Hirota, B. Gotoh, K. Kuma, T. Miyata, and Y. Nagai. 1989. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology 169:273-282. [DOI] [PubMed] [Google Scholar]
- 35.Toyoda, T., T. Sakaguchi, K. Imai, N. M. Inocencio, G. Gotoh, M. Hamaguchi, and Y. Nagai. 1987. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology 158:242-247. [DOI] [PubMed] [Google Scholar]
- 36.Wang, C., G. Raghu, T. Morrison, and M. E. Peeples. 1992. Intracellular processing of the paramyxovirus F protein: critical role of the predicted amphipathic alpha helix adjacent to the fusion domain. J. Virol. 66:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, C., T. Connolly, T. Morrison, and R. Gilmore. 1988. Integration of membrane proteins into the endoplasmic reticulum requires GTP. J. Cell Biol. 107:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, C., R. Gilmore, and T. Morrison. 1987. Translation and membrane insertion of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. Mol. Cell. Biol. 7:1386-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, J.-T., M. Duthle, and V. Ling. 1993. Membrane topology of the N-terminal half of the hamster P-glycoprotein. J. Biol. Chem. 268:15101-15110. [PubMed] [Google Scholar]