Abstract
Rotavirus is a nonenveloped virus with a three-layered capsid. The inner layer, made of VP2, encloses the genomic RNA and two minor proteins, VP1 and VP3, with which it forms the viral core. Core assembly is coupled with RNA viral replication and takes place in definite cellular structures termed viroplasms. Replication and encapsidation mechanisms are still not fully understood, and little information is available about the intermolecular interactions that may exist among the viroplasmic proteins. NSP2 and NSP5 are two nonstructural viroplasmic proteins that have been shown to interact with each other. They have also been found to be associated with precore replication intermediates that are precursors of the viral core. In this study, we show that NSP5 interacts with VP2 in infected cells. This interaction was demonstrated with recombinant proteins expressed from baculovirus recombinants or in bacterial systems. NSP5-VP2 interaction also affects the stability of VP6 bound to VP2 assemblies. The data presented showed evidence, for the first time, of an interaction between VP2 and a nonstructural rotavirus protein. Published data and the interaction demonstrated here suggest a possible role for NSP5 as an adapter between NSP2 and the replication complex VP2-VP1-VP3 in core assembly and RNA encapsidation, modulating the role of NSP2 as a molecular motor involved in the packaging of viral mRNA.
Rotaviruses, members of the Reoviridae family, are the major cause of severe gastroenteritis in infants and young children (18). The rotavirus genome consists of 11 segments of double-stranded RNA (dsRNA) and is surrounded by three concentric layers of protein (28). The outer layer is made up of 60 spikes formed by dimers of VP4 and of 260 trimers of the glycoprotein VP7. The middle layer consists of 260 trimers of VP6. The inner layer has a T = 1 symmetry and is made of 60 dimers of the capsid protein VP2, which shows nonspecific single-stranded RNA and dsRNA binding activities (21). The amino terminus of VP2 is essential for the incorporation of the RNA-dependent RNA polymerase VP1 and guanylyltransferase methylase VP3 into the core of the virion (23). The RNA-dependent RNA polymerase (VP1) has both transcriptase and replicase activities, which catalyze the synthesis of viral mRNA and dsRNA genome, respectively. Synthesis of dsRNA occurs in association with subviral particles, since free dsRNA cannot be detected in infected cells. Furthermore, the packaging and replication of the viral genome must be a highly coordinated process, given that the 11 dsRNA segments are present in equimolar concentrations in virions and that the ratio of number of virus particles to infectious units is low (16, 25).
Although several reports have described the characterization of rotavirus replication intermediates (RI), molecular details of the replication mechanisms remain unclear (12). Structural proteins VP1 and VP2 are essential components of the in vitro replicase activity (33). Two nonstructural proteins, NSP2 and NSP5, are associated with the RI in vivo, suggesting that they could participate in the early events of RNA replication (3, 26). In infected cells, these structural and nonstructural proteins have been shown to accumulate in large definite structures referred to as viroplasms. NSP2 has helix-destabilizing and nucleoside triphosphatase activity, suggesting a possible role in unwinding and packaging of the viral RNA (17, 29, 30). NSP5 is an O-glycosylated phosphoprotein that self-assembles into dimers and has nonspecific RNA-binding protein activity (15, 31). The protein also has an autokinase activity (5). NSP5 is present in infected cells and in the form of several phosphorylated isomers with apparent molecular masses ranging from 28 to 34 kDa. (2, 27). NSP5 can be chemically cross-linked in living cells with a complex made up of VP1 and NSP2 (1). In previous work NSP2 has been found associated with VP1 (19). The interaction of NSP5 with NSP2 was demonstrated in yeast by two-hybrid assays and was confirmed in virus-infected cells (27). Coexpression of NSP2 and NSP5 in uninfected cells generates viroplasm-like structures and up-regulates hyperphosphorylation of NSP5 (1, 11).
To better understand RNA packaging and replication, we have investigated the role of NSP5 in this process and its interaction with rotavirus structural proteins. We have shown that NSP5 interacts with VP2 in rotavirus-infected MA104 epithelial cells. This result was confirmed in reconstituted systems based on recombinant proteins expressed in baculovirus and bacterial systems. This interaction may be relevant to the function of NSP5 in replication complexes. It also suggests that, by binding to cores in the infected cell, NSP5 may block or delay outer capsid assembly and make it possible for these particles to continue replicase activity.
MATERIALS AND METHODS
Cells and virus.
The RF strain of rotavirus was propagated in MA104 cells in Eagle's minimum essential medium in the presence of trypsin (0.5 μg/ml). Infections were carried out at a multiplicity of infection of 10 PFU/cell. Sf9 cells infected with each recombinant baculovirus at a multiplicity of infection of 5 PFU/cell were incubated in Hink’s medium supplemented with 1% fetal calf serum. Recombinant baculovirus encoding the following rotavirus proteins were used: VP1 (pVL941/RF-1) (9), VP2 (BacRF2A) (22), VP2 with the first 92 amino acids deleted (BacΔ92VP2) (32), green fluorescent protein (GFP) fused to VP2 (BacGFP-Δ92VP2) (7), VP3 (BacRF3) (A. Charpilienne et al., unpublished data), VP6 (BacRF6) (22), and NSP5 (BacNSP5RF) (27). NSP5 fused to the carboxyl terminus of glutathione S-transferase (GST) was expressed in Escherichia coli BL21/DE3 transformed with recombinant plasmid pET41a+RF11.
Purification of rotavirus double-layered VLP.
Double-layered virus-like particles (VLP) composed of VP1, VP2, VP3 and VP6 (VLP1/2/3/6); of VP2 and VP6 (VLP2/6); of Δ92VP2 and VP6 (VLP2Δ92/6); and of VP2 fused to GFP and VP6 (VLP2GFP/6) were produced and purified as described previously (22). Briefly, Sf9 cells were collected at 7 days postinfection and treated with Freon 113. Cell extract was then mixed with CsCl, adjusted to obtain a refraction index of 1.360, and centrifuged for 16 h at 35,000 rpm in a Beckman SW55 rotor.
Expression of recombinant NSP5.
NSP5 was expressed in Sf9 cells infected with BacNSP5RF and collected at 72 h postinfection. Cells (4 × 106) were swollen in 1 ml of hypotonic buffer (HB) (3 mM HEPES [pH 8], 3 mM NaCl, 0.5 mM MgCl2, 0.5 μg of leupeptin per ml, 2 μg of aprotinin per ml) and lysed with a Dounce homogenizer. Extracts were clarified by centrifugation at 5,000 × g for 15 min.
BL21pET41RF11 bacteria were grown at 37°C in Luria-Bertani medium to an optical density at 600 nm of 0.2 and then induced by addition of 5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated overnight at 28°C. Cells were collected, treated with 1 mg of lysozyme per ml, and lysed by two rounds of sonication in phosphate-buffered saline containing 10 mM dithiothreitol, 1% Triton X-100, and 1% Tween 20. The lysate was clarified at 14,000 × g for 15 min, and the supernatant was incubated with glutathione-agarose beads overnight at 4°C. The beads were washed with phosphate-buffered saline, and proteins bound to glutathione-agarose were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining.
Coimmunoprecipitation and immunostaining.
Rotavirus-infected MA104 cells (3 × 106) were collected at 5 h postinfection and lysed in 0.5 ml of 50 mM Tris-HCl (pH 8)-125 mM NaCl-30 mM KCl-0.1% Triton X-100-0.05% SDS-20 mM EDTA-2 μg of aprotinin/ml (IP buffer). Cell debris was pelleted by centrifugation at 100,000 × g for 15 min. Monoclonal antibody (MAb) 158G37 directed against NSP5 (27) (1 μl of ascitic fluid) was added to 300 μl of cell lysate supernatant and incubated overnight at 4°C. Alternatively, Sf9 cell lysate supernatant (150 μl) prepared as described above from cells infected with various recombinant baculoviruses were incubated with MAb 158G37. Complexes were precipitated with protein A bound to Sepharose 4B and washed with IP buffer or HB depending on whether they were obtained from MA104 or Sf9 cells, respectively.
Immunoprecipitated proteins were immunodetected after SDS-PAGE analysis and blotting on polyvinyl difluoride membranes by transverse electrophoresis in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer, pH 11. MAbs 164E22 (22) and RV138 (20) were used for immunodetection of VP2 and VP6, respectively.
EM.
Specimens for electron microscopy (EM) were prepared from CsCl gradient fractions containing VLP after desalting by passage through Sephadex G25 spun columns. Samples were applied to air-glow discharged carbon coated grids, blotted immediately with filter paper, and negatively stained with 2% uranyl acetate solution. Thin sections were prepared as described previously (8) from Sf9 cells infected with recombinant baculoviruses allowing the expression of one or several rotaviral proteins and fixed at 72 h postinfection. Grids were examined in a Philips CM12 electron microscope operated at 80 kV.
Viral particles binding to recombinant NSP5.
Double-layered particles (DLP) and viral cores were purified as previously described (4). Glutathione-agarose beads loaded with recombinant GST-NSP5 were incubated overnight with 80 μg of CsCl gradient-purified VLP, DLP, or cores in a volume of 500 μl of 50 mM KCl-5 mM MgCl2-20 mM HEPES (pH 7)-0.5% Triton X-100 at 4°C. The beads were washed with the same buffer, and protein complexes were analyzed by SDS-PAGE, Coomassie blue stained, and further identified by immunostaining as described above.
Alternatively, 100 μl of NSP5-containing lysates prepared from BacNSP5RF-infected Sf9 cells as described above were mixed with 80 μg of CsCl-purified rotavirus particles (VLP2/6, VLP2GFP/6, VLP2Δ92/6, VLP1/2/3/6, and DLP), resuspended in HB, and incubated for 1 h on ice. Samples were layered onto a 4.5-ml CsCl density gradient and spun in a Beckman ultracentrifuge for 16 h at 35,000 rpm at 4°C in a SW55 rotor. As control, lysate prepared from Sf9 cells infected with a recombinant baculovirus allowing the expression of NSP2 was used under the same conditions.
RESULTS
NSP5 and VP2 coimmunoprecipitate.
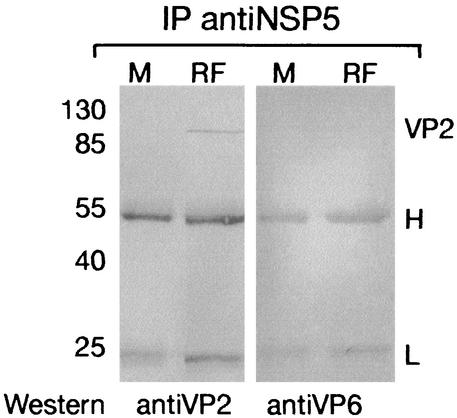
To look for interactions between NSP5 and VP2, lysates of MA104 cells infected with rotavirus for 5 h were incubated with anti-NSP5 MAb. Immunoprecipitated protein complexes were analyzed by Western blotting with MAb directed against either VP2 or VP6. As shown in Fig. 1, VP2 was detected in these complexes (lanes RF). By contrast, VP6 was not detected in the immunoprecipitated complexes containing NSP5. Mock-infected cells were used as controls (lanes M). Reverse experiments, i.e., immunoprecipitation with anti-VP2 MAb and Western blotting with anti-NSP5, gave lower signals (not shown). This result could be explained by the possibility that only a subfraction of NSP5 is complexed with viral cores.
FIG. 1.
Coimmunoprecipitation of VP2 with NSP5 in infected cells. MA104 cells infected with the RF strain of rotavirus (lanes RF) or mock infected (lanes M) were immunoprecipitated (IP) with an anti-NSP5 MAb, 158G37. VP2 and VP6 were immunodetected by Western blotting with anti-VP2 MAb 164E22 (left panel) or anti-VP6 MAb 138 (right panel), respectively. Sizes of molecular weight markers are indicated on the left in thousands. H and L, heavy and light chains of immunoglobulins, respectively.
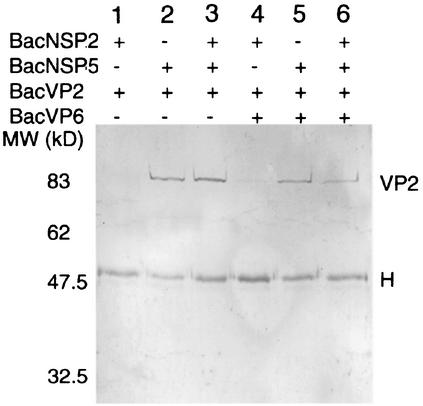
Interaction between NSP5 and VP2 was confirmed by coexpressing both proteins in insect cells (Fig. 2). Using the same assay as described above, we confirmed the presence of VP2 in the complexes immunoprecipitated with a MAb directed against NSP5 (Fig. 2, lane 2). The same results were obtained when the deletion mutant Δ92VP2 was substituted for wild-type VP2, indicating that the interaction between the two proteins is not mediated by the amino end of VP2 (not shown).
FIG. 2.
Coimmunoprecipitation of VP2 with NSP5 in baculovirus-infected cells. Sf9 cells infected with various recombinant baculoviruses were immunoprecipitated with an anti-NSP5 MAb (158G37), and VP2 present in complexes was detected with an anti-VP2 MAb (164E22) by Western blotting.
Rotavirus proteins are all highly expressed in the baculovirus system, and it is possible to express more than two proteins in insect cells. We used this versatile expression system to study the influence of other rotaviral viroplasmic proteins on VP2-NSP5 interactions. As shown in Fig. 2, interaction between VP2 and NSP5 is not prevented when NSP2 (lane 3), VP6 (lane 5), or both (lane 6) are coexpressed with VP2 and NSP5 in insect cells. In the absence of infection with the NSP5-expressing baculovirus, VP2 was not detected (lanes 1 and 4).
NSP5 modifies the subcellular distribution of VP2.
In rotavirus-infected cells, NSP5 and VP2 are colocalized in viroplasms (14). To explore the possible role of the interaction shown above in the colocalization of VP2 and NSP5 in the viroplasms, we used insect cells infected with recombinant baculoviruses. We used amino-terminally deleted VP2 fused to GFP (GFP-Δ92VP2) instead of VP2, since this fusion protein has the same assembly properties with itself and with VP6 as wild-type VP2 (7). As shown in Fig. 3A, expression of GFP-Δ92VP2 in Sf9 cells resulted in evenly distributed small fluorescent inclusions. In contrast, coexpression of GFP-Δ92VP2 and NSP5 resulted in fewer but much larger fluorescent VP2 inclusions (Fig. 3B). These large inclusions contained both VP2 and NSP5 (Fig. 3C1 to C3). Confocal microscopy examination of these insect cells demonstrated that small inclusions present when GFP-Δ92VP2 was expressed alone clustered together only when NSP5 was coexpressed with GFP-Δ92VP2. The pattern of fluorescence of GFP-Δ92VP2 was not modified when it was coexpressed with NSP2 (Fig. 3D). Interestingly, the distribution of GFP-Δ92VP2 was also modified when VP6, which is known to interact with VP2, was coexpressed (Fig. 3E). The large inclusions characteristic of coexpression of GFP-Δ92VP2 and NSP5 were not modified when the three proteins VP6, NSP5, and GFP-Δ92VP2, were coexpressed (Fig. 3F).
FIG. 3.
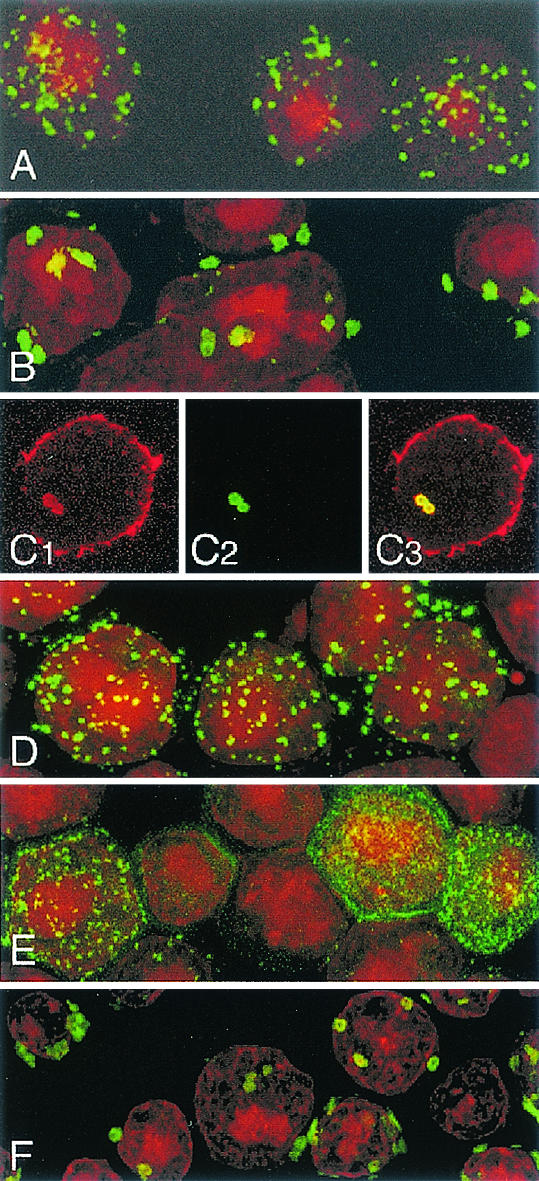
Distribution of GFP-Δ92VP2 expressed alone or in combination with other viral proteins. Sf9 cells infected with BacGFP-Δ92VP2 only (A) or coinfected with BacGFP-Δ92VP2 and either BacNSP5 (B), BacNSP2 (D), BacVP6 (E), or BacNSP5 and BacVP6 (F) were fixed with paraformaldehyde at 72 h postinfection and observed by confocal fluorescence microscopy. Colocalization of VP2 and NSP5 was verified by immunostaining of insect cells coexpressing GFP-VP2 and NSP5 with the anti-NSP5 MAb 158G37 and a Cy3-labeled donkey anti-mouse immunoglobulin G secondary antibody. Acquisitions of images for each emission wavelength (anti-NSP5, red [C1]; GFP-VP2, green [C2]) were performed sequentially, and the two images were superimposed (C3). Propidium iodide was used for nuclear staining.
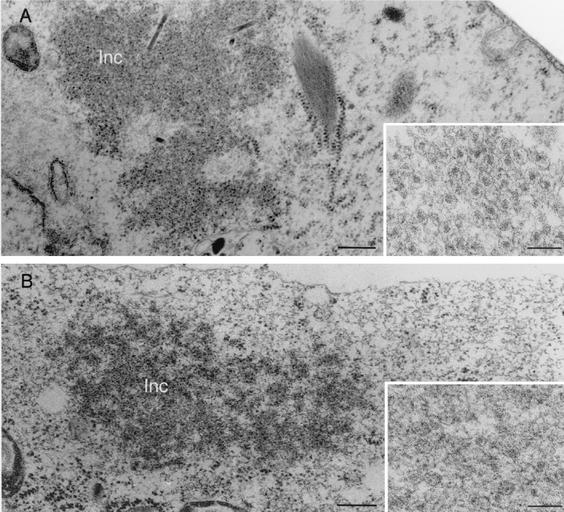
EM of thin sections of Sf9 cells infected with baculovirus expressing VP2 clearly showed inclusions containing spherical structures evoking VP2 pseudocores (Fig. 4A). In contrast, when VP2 and NSP5 were coexpressed, inclusions were larger and contained less-structured viral assemblies (Fig. 4B).
FIG. 4.
EM of inclusions present in insect cells expressing VP2 or VP2 and NSP5. Sf9 cells were infected either with BacVP2 (A) or with BacVP2 and BacNSP5 (B). Cells were fixed at 72 h postinfection and processed for thin sectioning. Inclusions (Inc) were observed in both cases, but at higher magnifications (insets) it can be observed that the fine structure is different and presents spherical motifs when VP2 is expressed alone. Bars, 300 nm (main panels) and 100 nm (insets)
Recombinant NSP5 binds cores and VLP but not DLP.
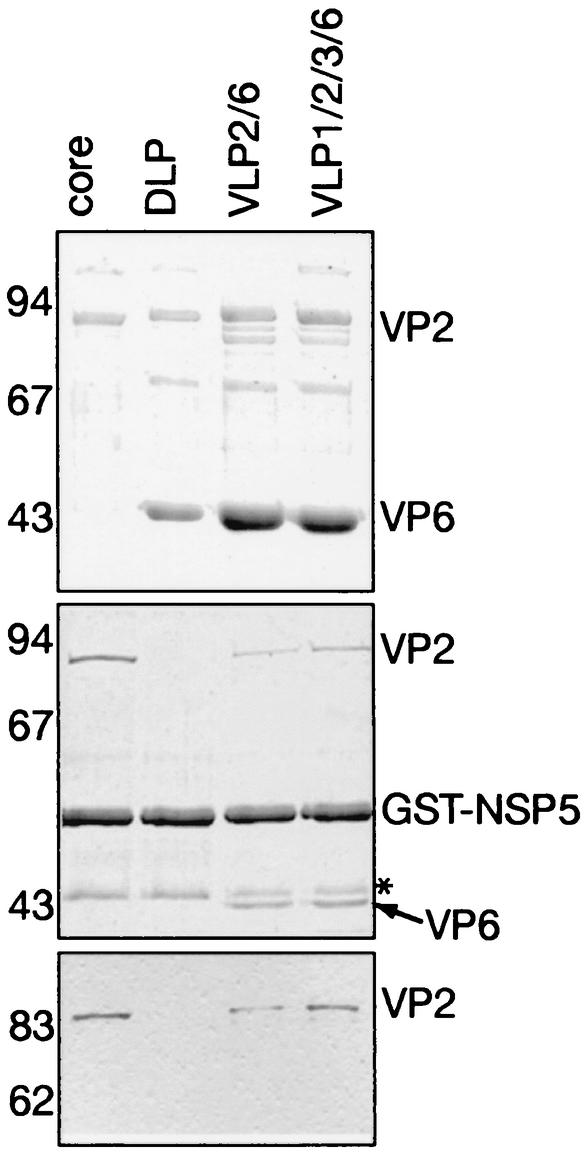
The experiments described above do not provide information about the polymeric structure of VP2 (monomeric or polymeric) that interacts with NSP5. To clarify this point, purified recombinant GST-NSP5 bound to glutathione-agarose beads was incubated with similar amounts of various types of purified viral particles, namely DLP, cores, VLP2/6, and VLP1/2/3/6. Cores and DLP were derived from purified virus and contain genomic RNA. Recombinant VLP2/6 and VLP1/2/3/6 were purified from insect cells. The input of the various types of particles used in the pull-down assay is shown in Fig. 5 (top panel). Proteins bound to GST-NSP5 glutathione-agarose beads were analyzed by SDS-PAGE and stained with Coomassie blue (Fig. 5, middle panel). Figure 5 (middle panel) clearly illustrates that DLP were not pulled down by NSP5 bound to agarose beads. In contrast, NSP5 beads pulled down cores, VLP2/6, and VLP1/2/3/6. It can be observed that the ratio of VP2 bound to VP2 present in the particle input is always lower when VP6 is present in the particles interacting with NSP5 beads (VLP2/6 and VL1/2/3/6). When VP6 is absent, i.e., in cores, the intensity of the VP2 band is maximum, suggesting that the binding of particles to NSP5 is optimal. This assay also showed that the binding of VLP2/6 and VLP1/2/3/6 to NSP5 resulted in a change of the stoichiometry of VP6 in the pulled down material (compare the intensity of VP6 bands with that of VP2 bands in the upper and middle panels of Fig. 5). Immunostaining with a MAb directed against VP2 confirmed the identity of the protein migrating as a 90-kDa band and the absence of VP2 when DLP were incubated with NSP5 beads (Fig. 5, lower panel). As a control, pull-down experiments were performed with GST-bound beads. None of the various types of viral particles showed any binding to the GST beads (data not shown).
FIG. 5.
Binding of GST-NSP5 to rotavirus particles. (Upper panel) Various types of rotavirus particles (indicated at the top) were analyzed by SDS-PAGE and Coomassie blue staining. They were incubated with a fixed amount of GST-NSP5 bound to glutathione-agarose. (Middle panel) Proteins bound to the solid phase were analyzed by SDS-PAGE and Coomassie blue staining. The protein indicated by an asterisk is a contaminant associated with GST NSP5. (Lower panel) The presence of VP2 bound to GST-NSP5 was confirmed by blotting on polyvinylidene difluoride and immunostaining with anti-VP2 MAb 164E22. Numbers on the left are molecular weights in thousands.
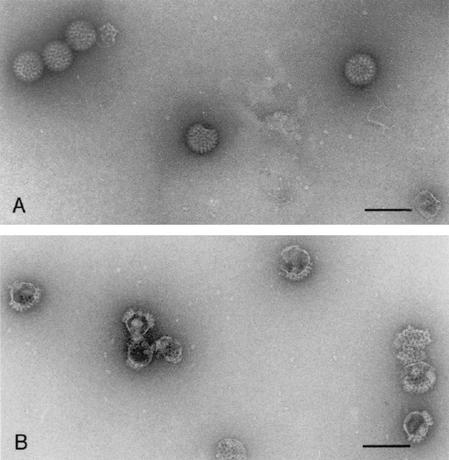
The pulling down of VLP2/6 and the absence of pulling down of DLP by GST-NSP5 beads could be due either to the absence of a limited number of VP6 trimers on VLP or to a stronger interaction between VP2 and VP6 in DLP than in VLP. Since both situations provide access to VP2, we compared the stabilities of the two types of viral particles at high pH in order to clarify this point. As shown in Fig. 6A, DLP were essentially resistant to such a treatment, but VLP2/6 were transformed into pseudocores (Fig. 6B).
FIG. 6.
EM of viral particles after negative staining. Purified DLP (A) and VLP2/6 (B) were treated for 30 min at pH 11 in 20 mM CAPS buffer and then negatively stained and observed at a magnification of ×35,000. Bars, 100 nm.
NSP5 dislodges VP6 from purified VLP2/6.
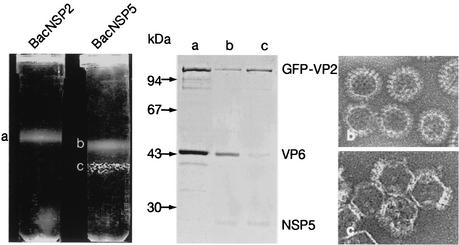
To characterize the complexes made of VP2 and NSP5, we analyzed purified VLP2/6 incubated with crude NSP5 from recombinant baculovirus-infected insect cell lysate by centrifugation in a CsCl gradient. A control experiment was performed with an Sf9 cell lysate containing NSP2. As shown in Fig. 7, incubation of purified VLP2GFP/6 with BacNSP2-infected Sf9 cell lysate did not change the density of VLP (Fig. 7, left panel, band a). In contrast, when the Sf9 cell lysate contained NSP5, an additional band was observed. The upper band (Fig. 7, left panel, band b) is similar to the band (band a) observed in the control, but the lower band contained flocculate fluorescent material (Fig. 7, left panel, band c). Both bands were fluorescent and thus contained GFP-Δ92VP2. Proteins present in these bands were analyzed by SDS-PAGE. As shown in Fig. 7 (middle panel), the lower band containing flocculate fluorescent material consisted essentially of GFP-Δ92VP2 with only trace amounts of VP6 (Fig. 7, middle panel, lane c). The upper band contained GFP-Δ92VP2 and VP6 in the same proportions as the control (Fig. 7, middle panel, lanes a and b). A small amount of NSP5 was present in both bands obtained when VLP2GFP/6 was incubated with NSP5-containing Sf9 lysate. In contrast, the protein profile of VLP2GFP/6 incubated with Sf9 lysate containing NSP2 was not altered and was identical to the profile obtained before incubation (Fig. 7, middle panel, lane a). EM examination of CsCl bands revealed that the flocculate material corresponded to aggregated VLP essentially devoid of the VP6 layer (Fig. 7, lower right panel), whereas the upper band contained normal VLP (Fig. 7, upper right panel). The same results were obtained when VLP2/6, VLP2Δ92/6, and VLP1/2/3/6 were substituted for VLP2GFP/6 (data not shown). Incubation of DLP with Sf9 lysate containing NSP5 did not change either their density in CsCl centrifugation or their protein composition (data not shown).
FIG. 7.
Analysis of purified VLP2GFP/6 in a CsCl density gradient after incubation with NSP5. Purified VLP2GFP/6 was incubated with a lysate of Sf9 cells infected with BacNSP5 or BacNSP2 and analyzed by CsCl gradient density centrifugation. (Left panel) A flocculate band can be seen when VLP2GFP/6 is incubated with a lysate from Sf9 cells infected with BacNSP5 but not when it is incubated with a lysate from Sf9 cells infected with BacNSP2. (Middle panel) Protein composition of each band of these gradients after Coomassie blue staining. (Right panels) VLP2/6 and incomplete VLP were observed by EM after staining of bands b (upper panel) and c (lower panel), respectively.
DISCUSSION
Viral factories referred to as viroplasms are observed in the cytoplasm of rotavirus-infected cells as early as 5 h postinfection. They contain several structural and nonstructural viral proteins, including NSP5 and VP2. In this study we have shown a direct interaction between VP2 and NSP5 early after infection. This interaction was confirmed in insect cells when VP2 and NSP5 were coexpressed in the absence of another viral protein and viral RNA. In this expression system, VP6 and NSP2, which are both components of viroplasm, did not inhibit VP2-NSP5 interactions. We did not observe interactions between NSP2 and VP2 in insect cells. NSP5 interaction with VP2 is not required for assembly of inner capsid proteins (VP1 and VP3 [possibly viroplasmic]) or outer capsid proteins (VP7 and VP4 [not viroplasmic]), since it has been demonstrated that all of these proteins assemble in the absence of NSP5 in insect cells (10, 32). We also demonstrated that NSP5 interacts with purified polymeric assemblies of VP2, including cores, VLP2/6, and VLP1/2/3/6.
In rotavirus-infected MA104 cells, most of the NSP5 is hyperphosphorylated, and its interaction with VP2 as demonstrated in these cells is probably due to this form. In this study, we also used nonphosphorylated NSP5 derived from E. coli and phosphorylated (but not hyperphosphorylated) NSP5 derived from insect cells. It was possible to demonstrate the interaction of VP2 with all forms of NSP5. This indicates that the phosphorylation state of NSP5 is not essential for the interaction, but it cannot be excluded that the interaction could be modulated by various degrees of phosphorylation of NSP5. The amino-terminal domain of VP2 is not necessary for the interaction, since a deletion mutant lacking the first 92 amino acids of VP2 interacted with NSP5 as well as wild-type VP2 did. The amino end of VP2 corresponds to the nucleic acid binding domain of VP2. This also suggests that the interaction between the two proteins does not involve nucleic acid.
It has previously been shown that NSP5 expressed alone is evenly distributed into the cytoplasm (1, 27). In this study we demonstrated that VP2 alone is not evenly distributed in insect cells and that coexpression of both proteins resulted in a dramatic change in the size of VP2 inclusions. The increased size of inclusions could simply result from the accumulation of NSP5 and VP2 proteins in the same area. NSP5 is a small oligomeric protein (27) that could link VP2 inclusions in large aggregates. The increased size of VP2 inclusions when NSP5 is present could also be the consequence of a conformational change of VP2 induced by NSP5. This hypothesis is supported by EM observations of an altered morphology of VP2 pseudocores when NSP5 is coexpressed with VP2 in insect cells. A similar increase in the size of viral inclusions has been observed when the core protein of hepatitis C virus is coexpressed with NSP5A in COS-7 cells (13).
In this study, we have shown that NSP5 interacts with VP2 and that VP6 hinders this interaction. The modulation of VP2-NSP5 interaction by VP6 could be due to a strong interaction between VP6 and VP2 that itself could be dependent on the presence of RNA inside viral particles. The interaction between VP2 and VP6 is stronger in DLP than in VLP2/6 as demonstrated by the respective resistances of the two types of particles to high-pH treatment. It can be hypothesized that the binding of NSP5 to core RI that do not yet have their complete set of genomic RNA segments prevents the binding of VP6 that would result in a defective particle containing a partial set of genomic segments, based on the observation that purified DLP did not interact with NSP5. The possibility that some VP6 trimers are absent and allow access to VP2 in VLP2/6 cannot be totally excluded either. The binding of NSP5 to rotavirus cores is reminiscent of the binding of μNS to reovirus cores (6). In reovirus-infected cells, μNS reduces the formation of recoated cores that are transcriptionally active, by the binding of outer capsid proteins μ1 and sigma 3 (which could be considered VP6 and VP7 counterparts, respectively), whereas NSP5, which dislodges VP6 from VLP and binds to transcriptionally inactive cores, could prevent an early start of transcriptase activity of newly formed DLP. It is therefore possible that μNS and NSP5 could both play a role in the delicate switch between transcriptase and replicase activities.
It has already been demonstrated that NSP5 interacts with NSP2 (1, 27). The data presented here are the first evidence of an interaction between NSP5 and a structural protein and the first evidence of an interaction between VP2 and a nonstructural protein. This interaction could be significant in the context of RNA replication and capsid morphogenesis. In a recently proposed model (17, 24), interaction between NSP5 and NSP2 in a complex consisting of VP1, VP3, VP2, NSP2, NSP5, and RNA was considered. Direct interaction between VP2 and NSP5, as demonstrated here, would modify this model and place NSP5 as an intermediate between VP2 and NSP2. Moreover, our results suggested that the competition between VP6 and NSP5 for VP2 could release the NSP2-NSP5 complex from the core RI (26). This could most likely occur at the edge of the viroplasm, where VP6 seems to accumulate (14).
Acknowledgments
We are grateful to J. Lepault for stimulating discussions and to N. Castagné and A. Charpilienne for their technical assistance.
This work was supported by a Programme de Recherche Fondamentale en Microbiologie sur les Maladies Infectieuses et Parasitaires grant (Réseau de Recherche sur les Gastro-entérites à rotavirus) from the Ministère de l'Enseignement de la Recherche et de la Technologie and by a 5th Programme Cadre de Recherche et Développement grant from the Union Européenne (QLRT 1999-00634).
REFERENCES
- 1.Afrikanova, I., E. Fabbretti, M. C. Miozzo, and O. R. Burrone. 1998. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 79:2679-2686. [DOI] [PubMed] [Google Scholar]
- 2.Afrikanova, I., M. C. Miozzo, S. Giambiagi, and O. Burrone. 1996. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 77:2059-2065. [DOI] [PubMed] [Google Scholar]
- 3.Aponte, C., D. Poncet, and J. Cohen. 1996. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2. J. Virol. 70:985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bican, P., J. Cohen, A. Charpilienne, and R. Scherrer. 1982. Purification and characterization of bovine rotavirus cores. J. Virol. 43:1113-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackhall, J., A. Fuentes, K. Hansen, and G. Magnusson. 1997. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J. Virol. 71:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein muNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charpilienne, A., M. Nejmeddine, M. Berois, N. Parez, E. Neumann, E. Hewat, G. Trugnan, and J. Cohen. 2001. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J. Biol. Chem. 276:29361-29367. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, J., A. Charpilienne, S. Chilmonczyk, and M. K. Estes. 1989. Nucleotide sequence of bovine rotavirus gene 1 and expression of the gene product in baculovirus. Virology 171:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, S. E., M. Labbe, J. Cohen, M. H. Burroughs, Y. J. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 12.Gallegos, C. O., and J. T. Patton. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172:616-627. [DOI] [PubMed] [Google Scholar]
- 13.Goh, P. Y., Y. J. Tan, S. P. Lim, S. G. Lim, Y. H. Tan, and W. J. Hong. 2001. The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290:224-236. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, R. A., R. Espinosa, P. Romero, S. Lopez, and C. F. Arias. 2000. Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch. Virol. 145:1963-1973. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, S. A., and O. R. Burrone. 1991. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology 182:8-16. [DOI] [PubMed] [Google Scholar]
- 16.Hundley, F., B. Biryahwaho, M. Gow, and U. Desselberger. 1985. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology 143:88-103. [DOI] [PubMed] [Google Scholar]
- 17.Jayaram, H., Z. Taraporewala, J. T. Patton, and B. V. Prasad. 2002. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature 417:311-315. [DOI] [PubMed] [Google Scholar]
- 18.Kapikian, A. Z. 1996. Overview of viral gastroenteritis. Arch. Virol. Suppl. 12:7-19. [DOI] [PubMed] [Google Scholar]
- 19.Kattoura, M. D., X. Chen, and J. T. Patton. 1994. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology 202:803-813. [DOI] [PubMed] [Google Scholar]
- 20.Kohli, E., L. Maurice, C. Bourgeois, J. B. Bour, and P. Pothier. 1993. Epitope mapping of the major inner capsid protein of group A rotavirus using peptide synthesis. Virology 194:110-116. [DOI] [PubMed] [Google Scholar]
- 21.Labbe, M., P. Baudoux, A. Charpilienne, D. Poncet, and J. Cohen. 1994. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. J. Gen. Virol. 75:3423-3430. [DOI] [PubMed] [Google Scholar]
- 22.Labbe, M., A. Charpilienne, S. E. Crawford, M. K. Estes, and J. Cohen. 1991. Expression of rotavirus VP2 produces empty corelike particles. J. Virol. 65:2946-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawton, J. A., C. Q. Zeng, S. K. Mukherjee, J. Cohen, M. K. Estes, and B. V. Prasad. 1997. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J. Virol. 71:7353-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patton, J. T. 2001. Rotavirus RNA replication and gene expression. Novartis Found. Symp. 238:64-81. [DOI] [PubMed] [Google Scholar]
- 25.Patton, J. T., and C. O. Gallegos. 1990. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J. Gen. Virol. 71:1087-1094. [DOI] [PubMed] [Google Scholar]
- 26.Patton, J. T., and C. O. Gallegos. 1988. Structure and protein composition of the rotavirus replicase particle. Virology 166:358-365. [DOI] [PubMed] [Google Scholar]
- 27.Poncet, D., P. Lindenbaum, R. L'Haridon, and J. Cohen. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 71:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad, B. V., and W. Chiu. 1994. Structure of rotavirus. Curr. Top. Microbiol. Immunol. 185:9-29. [DOI] [PubMed] [Google Scholar]
- 29.Taraporewala, Z., D. Chen, and J. T. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taraporewala, Z. F., and J. T. Patton. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vende, P., Z. F. Taraporewala, and J. T. Patton. 2002. RNA-binding activity of the rotavirus phosphoprotein NSP5 includes affinity for double-stranded RNA. J. Virol. 76:5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng, C. Q., M. K. Estes, A. Charpilienne, and J. Cohen. 1998. The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. J. Virol. 72:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng, C. Q., M. J. Wentz, J. Cohen, M. K. Estes, and R. F. Ramig. 1996. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J. Virol. 70:2736-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]