Abstract
The regulation of higher-order chromosome structure is central to cell division and sexual reproduction. Heterochromatin assembly at the centromeres facilitates both kinetochore formation and sister chromatid cohesion, and the formation of specialized chromatin structures at telomeres serves to maintain the length of telomeric repeats, to suppress recombination, and to aid in formation of a bouquet-like structure that facilitates homologous chromosome pairing during meiosis. In fission yeast, genes encoding the Argonaute, Dicer, and RNA-dependent RNA polymerase factors involved in RNA interference (RNAi) are required for heterochromatin formation at the centromeres and mating type region. In this study, we examine the effects of deletions of the fission yeast RNAi machinery on chromosome dynamics during mitosis and meiosis. We find that the RNAi machinery is required for the accurate segregation of chromosomes. Defects in mitotic chromosome segregation are correlated with loss of cohesin at centromeres. Although the telomeres of RNAi mutants maintain silencing, length, and localization of the heterochromatin protein Swi6, we discovered defects in the proper clustering of telomeres in interphase mitotic cells. Furthermore, a small proportion of RNAi mutant cells display aberrant telomere clustering during meiotic prophase. This study demonstrates that the fission yeast RNAi machinery is required for the proper regulation of chromosome architecture during mitosis and meiosis.
The dynamic regulation of chromosome structure governs diverse cellular processes. To accurately pass on genetic information to daughter cells, newly replicated sister chromatids must become linked by cohesion and attach their respective kinetochores bilaterally to the microtubules that emanate from the spindle pole bodies. The presence of specialized heterochromatic structure at centromeres is required for this process. In a variety of systems, mutants that affect the formation of heterochromatin within centromeres adversely affect chromosome segregation (1–3). In the fission yeast Schizosaccharomyces pombe, silencing factors such as the heterochromatin protein 1 homolog Swi6, the histone H3 Lys-9 methyltransferase Clr4, and histone deacetylases are required for centromere function and chromosome segregation (1, 4, 5).
The proper function of telomeres likewise depends on higher-order chromatin assembly. In addition to telomere maintenance, the formation of specialized structures at telomeres plays a crucial role in telomeric clustering and chromosome dynamics in fission yeast (6–9). For example, in meiotic prophase, chromosomal ends become clustered and attach to the spindle pole body through a mechanism that requires silent chromatin assembly. This telomere bouquet configuration is conserved in most eukaryotic organisms, and is thought to facilitate meiotic pairing and subsequent homologous recombination by aligning chromosomes (10–12).
RNA interference (RNAi) is the process by which double stranded RNA triggers the destruction of cognate mRNAs (13, 14). Studies in diverse species have implicated proteins involved in the RNAi pathway in viral resistance (15), posttranscriptional gene regulation (16, 17), transcriptional gene silencing (18–20), transposon suppression (21, 22), and programmed genome rearrangement (23). In plants and animals, mutants display defects in somatic and germ-line development (24, 25). Furthermore, phenotypic analysis has implicated the RNAi machinery in stem cell maintenance (26), cell fate determination (27), and nonrandom chromosome segregation (28). Because some of the phenotypes displayed by RNAi mutants are genetically separable (29), it is likely that components of the RNAi machinery in higher eukaryotes operate in multiple overlapping pathways.
The fission yeast genome contains a single gene corresponding to each of the Argonaute (ago1), Dicer (dcr1), and RNA-dependent RNA polymerase (rdp1) factors required for RNAi in other systems. Recently, we have shown that the fission yeast RNAi machinery is required for epigenetic gene silencing at centromeres (30), and for the initiation of heterochromatin formation at the silent mating type region (31). In both cases, defects in silencing in the RNAi deletion strains correlate with loss of histone H3 Lys-9 methylation and chromatin-bound Swi6 protein from these loci. In this study, we characterize the effects of deletion of the fission yeast RNAi components on chromosome segregation, centromere cohesion, Swi6 localization, and telomere function during mitosis and meiosis.
Materials and Methods
Strains and Culture Conditions.
Standard conditions were used for growth, sporulation, tetrad analysis, and construction of diploids from haploid strains (32).
Chromatin Immunoprecipitation (ChIP).
ChIPs were performed as described (33). DNA recovered from immunoprecipitated chromatin fractions or whole cell crude extracts was subjected to multiplex PCR analysis using the dh383 primer pair (34) recognizing the dh centromeric repeat and the control ade6DN/N primer pair fragment from ade6+ gene. Fold enrichment was calculated by taking the ratio of intensities of the dh band and the ade6+ band from the ChIP fraction, and dividing that by the ratio of their intensities in the whole cell crude extracts.
Immunofluorescence Analysis (IF).
IF was carried out as previously described (33). For meiotic IF, mating-type switching-competent (h90) mid-log phase cells were concentrated by centrifugation, spotted on solid pombe minimal medium (PMA+), and allowed 15–20 h at 26°C for sporulation.
Fluorescent in Situ Hybridization (FISH).
FISH was performed as described (35). Briefly, log-phase cells were fixed in 3% paraformaldehyde for 2 min at room temperature followed by the addition of 25% glutaraldehyde to a final concentration of 0.2%, and incubated at 26°C for 1 h. After fixation, cells were washed sequentially, treated with 10 μg/ml RNase A for 2 h, and hybridized overnight with the Cy3-labeled 15-kb insert spanning the dg and dh centromeric repeats, contained on the pRS140 plasmid (36).
Microscopic Analysis.
Samples were analyzed with a Zeiss Axioplan2 fluorescent microscope. For deconvolution, images were collected at 0.2-μm intervals along the z axis and subjected to volume deconvolution using the nearest three neighbors method. openlab software (Improvision) was used for all analyses.
Southern Blot Analysis.
Genomic DNA was digested with EcoRI, separated by agarose gel electrophoresis, and transferred to a nitrocellulose membrane by standard procedures. The blot was probed with a α-32P-labeled 300-bp fragment derived from the terminal telomeric repeat contained on the pAMP002 plasmid (8).
Results and Discussion
RNAi Machinery Is Required for the Fidelity of Chromosome Segregation.
Deletion of ago1, dcr1, or rdp1 results in loss of epigenetic silencing at centromeres, and concomitant loss of H3 Lys-9 methylation and Swi6 protein (30). Because heterochromatin formation has been linked to centromere function in fission yeast (1), and in other systems (2, 3, 37), and because we noticed a variety of abnormal segregation patterns in crosses involving the RNAi mutant strains, it was of interest to investigate whether chromosome segregation was disrupted. Our analysis revealed that mutations in the RNAi machinery render the cells hypersensitive to microtubule destabilizing drug thiabendazole, as compared with wild-type cells (Fig. 1A), indicating that the process of chromosome segregation is not robust in the mutant strains.
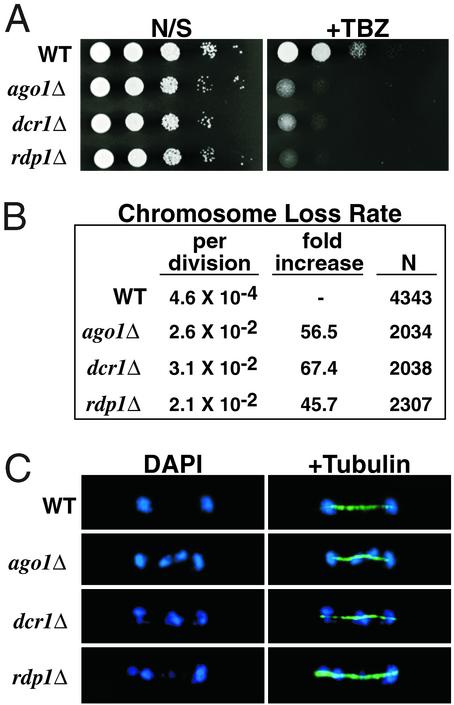
Figure 1.
Mitotic chromosome segregation is impaired in the RNAi mutants. (A) Serial dilutions of indicated cultures were spotted onto yeast extract adenine (YEA) medium or YEA medium supplemented with 10 μg/ml thiabendazole (TBZ). (B) Rates of chromosome loss for wild-type and mutant strains as measured by the breakdown of homozygous diploids. Rates of chromosome loss per cell division and relative fold increases compared with wild-type are shown. N refers to the total number of white plus half-sectored colonies included in the analysis. (C) Segregation of chromosomes during late anaphase as visualized by DAPI staining and immunofluorescence with the anti-tubulin TAT1 antibody.
We next investigated whether RNAi mutant strains show increased rates of chromosome loss due to missegregation events. Diploids containing the ade6-M210 and ade6-M216 alleles form white colonies because of intragenic complementation, but give rise to red and pink colonies, respectively, when present alone in a haploid, allowing the rate of chromosome loss to be estimated (1). The results presented in Fig. 1B demonstrate that diploids homozygous for ago1Δ, dcr1Δ, or rdp1Δ have significantly higher rates of nondisjunction (45- to 67-fold) than their wild-type counterparts, indicating that RNAi mutant strains have defects in centromere function.
To directly observe the process of mitotic chromosome segregation in wild-type and mutant cells, we performed immunoflourescence with anti-tubulin antibodies to visualize microtubules and observed chromosomes by staining with 4′,6-diamidino-2-phenylindole (DAPI). We frequently observed lagging chromosomes in late anaphase mutant cells with a fully elongated spindle (Fig. 1C), and noticed additional phenotypes including highly elongated cells containing multiple and/or fragmented nuclei, and late mitotic cells in which the majority of DNA was segregated to one of the two daughter nuclei. These results demonstrate that the ago1, dcr1, and rdp1 gene products play an important role in the proper segregation of chromosomes through mitosis.
Interestingly, we also noticed novel chromosome segregation phenotypes in the RNAi mutant strains that differ from those observed in other silencing-defective strains, such as swi6 and clr4. For example, we consistently observed asymmetric segregation of chromosomes in diploid strains, such that otherwise heterozygous diploid colonies became homozygous for single chromosomes in a clonal fashion (data not shown). The exact cause of these segregation defects in the RNAi mutants is under investigation, but appears to be due to either inappropriate reductional division during mitosis or random segregation of chromatids. It can be imagined that the RNAi machinery affects multiple aspects of chromosome segregation.
Centromeric Cohesion Is Defective in the RNAi Mutant Strains.
In fission yeast, the preferential recruitment of the cohesin complex at centromeres depends on the presence of heterochromatin, as Swi6 directly recruits cohesin to the outer repeats (38, 39). To test whether the observed segregation defects in RNAi mutants were due in part to defects in centromeric cohesion, we used a haploid strain that expresses a LacI-GFP fusion protein, and contains the LacO DNA repeat inserted at the lys1 locus linked to centromere 1 (cen1-GFP) (40). Because fission yeast cells spend most of the cell cycle in the G2 phase, the single GFP spot commonly observed in wild-type cells corresponds to the sister chromatids of chromosome I joined at the centromere. In mutant strains, we observed two GFP foci in a large proportion of cells, indicating defects in centromere cohesion (Fig. 2A). These two cen1-GFP foci were often in the general vicinity of each other, likely indicating that sister chromatid cohesion is not disrupted along the entire length of the chromosome. This finding is consistent with independent mechanisms regulating the recruitment of cohesin to chromosome arms and centromeres (38, 39).
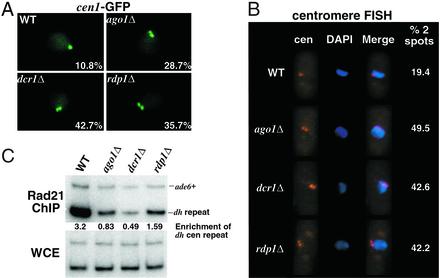
Figure 2.
Sister chromatid cohesion at centromeres is disrupted in the RNAi mutant strains. (A) Localization of cen1 in live cells as visualized by accumulation of LacI-GFP at the LacO array inserted at the lys1 locus linked to cen1. GFP spots were counted on a computer screen after capturing serial images of fields of cells at 0.4-μm intervals along the z axis. Spots were deemed distinct when their midpoints were separated by a distance greater than or equal to their respective radii. The percentage with which each genotype displayed two GFP spots is noted in the lower right corner of each image. More than 100 cells were counted for each strain. (B) FISH analysis of wild-type and mutant cells using a 15-kb probe that hybridizes to the outer repeats of all three centromeres. The number of spots were counted by microscopic inspection of >100 cells for each strain. (C Upper) ChIP analysis of Rad21-HA in wild-type and mutant strains using the 12CA5 antibody. Relative fold enrichments of dh centromeric repeats are indicated beneath each lane. (Lower) DNA prepared from whole cell extracts (WCE).
The three pairs of sister centromeres present in a G2 cell normally colocalize in a cluster visible as a single spot by FISH (41). To further examine the localization of centromeres in the RNAi mutant strains, we performed FISH using a probe that recognizes the outer region of all three centromeres. Consistent with our analysis using cen1-GFP, RNAi mutant cells often displayed two spots in contrast to the one spot observed in the majority of wild-type cells (Fig. 2B). The presence of greater than two spots was exceedingly rare, indicating that, to the level of detection provided by FISH, the clustering of nonhomologous centromeres was not severely affected.
To directly examine the concentration of cohesin at centromeres, we performed ChIP analysis with strains carrying an epitope-tagged version of the cohesin subunit Rad21. Although Rad21 was preferentially enriched at the centromeres of wild-type cells, we observed a considerable reduction in its localization at the centromeres of ago1Δ, dcr1Δ, or rdp1Δ cells (Fig. 2C). This defect in centromere cohesion is most likely a product of the observed defects in Swi6 localization to centromeres in the RNAi mutant strains (30). This finding demonstrates that one possible cause for aberrant segregation is defects in the recruitment of cohesin to centromeres.
RNAi Mutants Are Defective in Telomere Clustering.
Recent studies have implicated RNA in the formation of higher-order chromosomal structures (42). In fission yeast, heterochromatin is found at the centromeres, telomeres, and mating type region, and can be visualized by immunostaining for Swi6. Previous studies have shown two to five discrete Swi6 foci in interphase cells, of which approximately one is a cluster of all three centromeres. Telomeres form two to four foci of Swi6, with two being most common, and a faint spot corresponds to the mating type region (43). In some mutants defective in centromeric silencing, such as clr4 and rik1, Swi6 becomes entirely delocalized from the chromosomes and is present in a diffuse pattern throughout the nucleus and nucleolus (43).
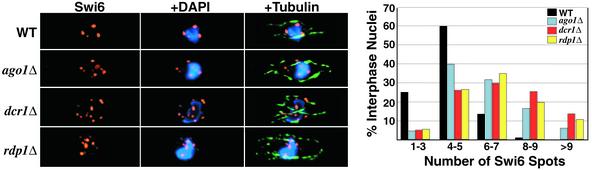
Because the heterochromatic regions of fission yeast cluster together into higher order structures reminiscent of the pericentric heterochromatin of higher eukaryotes, we analyzed the effects of deletion of the RNAi machinery on Swi6 localization by using immunofluorescence. Surprisingly, most interphase mutant cells had a greater number of Swi6 foci than wild-type cells (Fig. 3), though these foci were generally smaller in size and less intense. Considering that the RNAi mutants are defective in Swi6 localization at centromeres (30), we investigated whether the additional foci were a result of defective telomere clustering. Taz1 is a protein that binds exclusively to telomere repeats, and is commonly used as a marker for their localization (44). We performed coimmunofluorescence experiments with Swi6 and Taz1, and found that most of the Swi6 foci that we observed in interphase mutant cells colocalized with Taz1 (Fig. 4A), indicating that the mitotic clustering of telomeres is defective in the mutant strains. However, the localization of telomeres to the nuclear periphery did not appear to be affected.
Figure 3.
Mutant cells exhibit a greater number of Swi6 foci. (Left) Deconvolved images of wild-type and mutant cells subjected to immunofluorescence for Swi6 (red) and tubulin (green), and stained withDAPI (blue). (Right) A graph of the frequency of Swi6 foci number. More than 150 cells were counted for each strain.
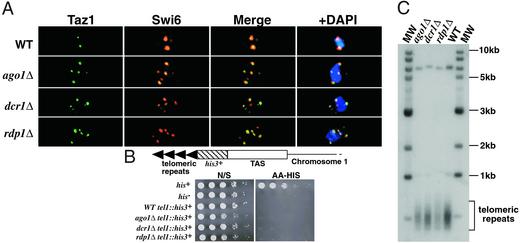
Figure 4.
Mitotic telomeric clustering is disrupted in RNAi mutant strains, but telomeric silencing and telomere length are unaffected. (A) Deconvolved images of interphase cells subjected to immunofluorescence for Swi6 (red) and Taz1-HA (green). (B) Serial dilution analysis of wild-type and mutant strains containing the his3+ reporter gene inserted at the telomere of the left arm of chromosome 1. Cells were grown on nonselective medium (N/S) and medium lacking histidine (AA-HIS). (C) Telomere length is not affected in mutant strains. Genomic DNA from wild-type and mutant strains was digested with EcoRI and probed with α-32P-labeled telomere repeat DNA (8). “MW” signifies the 1-kb molecular mass marker.
How the RNAi machinery affects mitotic telomere clustering is not known. We speculate that RNA intermediates produced by RNAi promote the chromosomal association of telomeres, perhaps by acting as a “glue” to hold distinct heterochromatic regions from dispersed genomic locations into a common structure. In this regard, it should be noted that the formation of higher-order heterochromatic structures in eukaryotes with more complex genomes requires that loci from multiple chromosomal regions cluster together. In fission yeast, similar processes may operate in the clustering of telomeres.
It is interesting that, in contrast to telomeres, we did not observe any defects in the clustering of nonhomologous centromeres (Fig. 2B). This is consistent with previous studies showing that the fission yeast centromeres are divided into two distinct domains, and that the factors that interact with these domains might contribute to redundant mechanisms responsible for centromeric clustering (45). Although the RNAi machinery is required for localization of Swi6 and at the highly repetitive outer centromeric region, it is dispensable for silencing at the central core, which is the site of kinetochore formation and occupied by entirely different factors such as Mis6 and Cnp1 (46, 47).
RNAi Machinery Is Dispensable for Telomere Maintenance.
To further examine the role of factors involved in RNAi on telomere regulation, we studied the effects of ago1Δ, dcr1Δ, and rdp1Δ on silencing of a his3+ reporter gene inserted within a telomere of chromosome 1 (tel1∷his3+) (6). Serial dilution analyses of the strains containing the tel1∷his3+ reporter revealed that maintenance of telomeric silencing is not affected in the RNAi mutant background (Fig. 4B). We also examined telomeric DNA by Southern blot and found no difference in the length of telomeric repeats in wild-type and mutant cultures (Fig. 4C), indicating that the pathways that maintain the length of telomeres are intact. The lack of silencing defects in the RNAi mutant strains probably reflects the ability of telomeric repeat DNA to directly recruit telomere-specific silencing factors.
In light of the above results showing that telomere maintenance does not appear to be affected in RNAi mutants, the precise role of mitotic telomeric clustering remains an open question. It is possible that clustering facilitates the establishment of heterochromatin by concentrating telomere ends in specialized nuclear compartments that are enriched in silencing factors. Another possibility is that mitotic telomere clustering facilitates the transition to meiosis, where the clustering of telomeres at the spindle pole body during prophase helps align homologous chromosomes (11, 12).
Meiotic Chromosome Segregation Is Severely Perturbed in the RNAi Mutants.
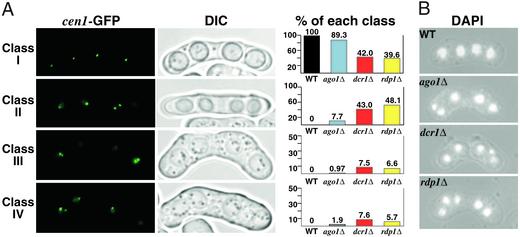
Chromosome segregation during meiosis and mitosis are distinct processes. At the first reductional meiotic division, sister chromatids remain associated at their centromeres and move together to the same pole. During the second meiotic division, sister chromatids separate from each other and segregate to opposite poles. This process requires tight regulation of kinetochore orientation and meiotic centromere cohesion (48). To examine meiotic chromosome segregation, we sporulated strains carrying the cen1-GFP marker described above to follow the segregation of the cen1 locus in live tetrads. A normal meiosis results in an ascus in which each of the four spores contain a single cen1-GFP spot. Missegregation of cen1 will result in a tetrad in which one spore contains two cen1-GFP spots, and another spore lacks a GFP spot. Because fission yeast undergoes an ordered meiosis (the two adjacent spores at each end of the tetrad are products of the same second meiotic division), it can be deduced whether a given missegregation event occurred during the first or second meiotic division. Our analysis revealed that all three RNAi mutants missegregated chromosomes during the second meiotic division, though the effect was more pronounced in dcr1Δ and rdp1Δ strains (Fig. 5A). We also observed a small but consistent proportion of mutant cells that missegregated single chromatids during the first meiotic division.
Figure 5.
Meiotic segregation is defective in the RNAi mutants. (A) Segregation of cen1-GFP through meiosis. Strains were sporulated and subjected to fluorescent microscopy for cen1-GFP and differential interference contrast microscopy. Class I represents normal meiotic segregation, where each spore receives one copy of chromosome I. Class II represents a missegregation event during one of the two second meiotic divisions. Class III is caused by missegregation during both of the second meiotic divisions. Class IV is caused by missegregation of a single cen1-GFP chromatid during the first meiotic division. (Right) The frequency with which the respective phenotypic classes were observed. Only tetrads with four visible GFP spots were scored. More than 100 tetrads were counted for each genotype. Note different scales along the y axis. (B) Aberrant meiotic segregation as visualized by DAPI staining.
Consistent with analysis above, examination of tetrads after DAPI staining revealed frequent aberrations in the distribution of DNA to daughter spores (Fig. 5B). The most common phenotype observed was the presence of DNA outside of mature spores, indicating that one or more chromosomes failed to reach the meiotic pole and were not incorporated into the developing spore. The formation of these aberrant tetrads is consistent with the frequent spore inviability in RNAi mutant backgrounds (data not shown).
One possible explanation for the observed meiotic segregation defects is that, as during mitosis, loss of heterochromatin compromises centromere cohesion. Consistent with this idea, preliminary analysis suggests that the meiotic cohesin subunit Rec8 is less abundant at the centromeres of RNAi mutants than in wild-type strains. Furthermore, we frequently observed precocious separation of cen1-GFP during the early stage of meiosis II in mutant strains. These results implicate the RNAi machinery in the fidelity of meiotic chromosome segregation, an extremely important process that ensures the genomic integrity of future generations, and lies at the heart of many heritable human disorders.
RNAi Mutants Show Defects in Meiotic Telomere Clustering.
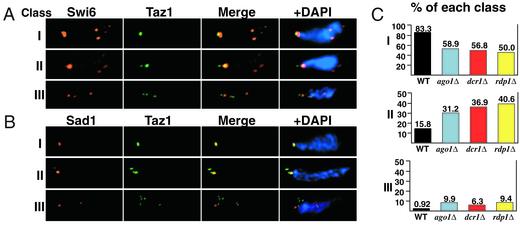
During the meiotic prophase of fission yeast, all of the telomeres cluster together near the spindle pole body and the nucleus becomes elongated and oscillates between the cells poles, led by the spindle pole body. This is referred to as the “horsetail” stage, and corresponds to the meiotic telomere bouquet observed in a wide range of eukaryotic systems (10). Mutants affecting telomeric silencing have been shown, to varying degrees, to impair the meiotic clustering of telomeres near the spindle pole body, and generally to alter the progression of the horsetail stage (6–8, 49). Because our mutants showed a defect in telomere clustering in mitotic cells, we performed immunofluorescence with antibodies for Swi6 and Taz1 on meiotic cells in the horsetail stage, as identified by DAPI staining (Fig. 6A). Although the majority of mutant cells exhibited wild-type morphology, we observed aberrant phenotypes that we summarize into three classes. Wild-type cells predominantly displayed morphology corresponding to class I, in which Taz1 and Swi6 colocalize to a single spot in the horsetail nucleus. In the mutant strains, we observed an ≈2-fold elevation in the frequency of class II morphology, in which two distinct Taz1 spots were visible either as a doublet or as completely separate foci (Fig. 6C). Furthermore, we observed a small but significant proportion of mutant cells in which greater than two telomere spots were visible (class III) indicating a loss of meiotic telomere organization.
Figure 6.
Meiotic telomere clustering and the attachment of telomeres to the spindle body protein Sad1 are perturbed in the RNAi mutant strains. (A) Deconvolved images of immunofluourescence with Swi6 (red) and Taz1 (green) in meiotic cells in the horsetail stage. Class I refers to cells exhibiting the wild-type morphology with all telomeres clustered together into one spot. Class II refers to meiotic cells in which two telomere spots were clearly visible, or in which a doublet occurred such that the midpoints of the respective spots were separated by a distance greater than or equal to the longer of the two radii. Class III refers to all cells in which greater than two telomere spots were observed. (B) Deconvolved images of immunofluorescence analysis with the spindle pole body component Sad1 (red) and Taz1 (green). Representative Sad1 staining for each class is shown. (C) Frequency of classes as observed by immunofluorescence for Taz1 and Swi6 during meiosis. More than 100 meiotic cells in the horsetail stage were counted for each strain. Note different scales along the y axis.
Mutants affecting meiotic telomere clustering frequently show aberrations in the localization of telomeres near the spindle pole body (SPB), the fungal equivalent to the centrosome (50). To characterize the attachment of telomeres to the SPB in the RNAi mutant strains, we performed immunofluorescence with Taz1 and Sad1, an essential component of the SPB (51). Representative staining for each phenotypic class is shown (Fig. 6B). As expected, the single telomere cluster observed in wild-type cells and in mutant cells with class I morphology was always adjacent to the SPB. In cells containing two telomere clusters, each was associated with a distinct Sad1 spot, though frequently one of the two clusters was smaller than the other. In cells containing more than two telomere spots, no more than two of those telomere spots were associated with a visible Sad1 spot. These results indicate that mutations in the RNAi machinery lead to a mild but consistent disruption of meiotic telomere clustering and SPB integrity.
Chromosomal Defects May Provide a Link to Other Systems.
This study connects RNAi to the regulation of chromosome dynamics and genomic integrity. We show that the fission yeast RNAi machinery is required for the fidelity of chromosome segregation during mitosis and meiosis, and demonstrate that one possible cause for chromosome missegregation in the mutant strains is the loss of centromeric cohesion. This analysis extends our previous observation of centromeric silencing defects in the mutant strains to show that centromere function is compromised when RNAi components are deleted. Moreover, the disruption of telomeric clustering indicates that, as in mammals, an RNA component may be required for the higher-order organization of heterochromatic regions.
The observed defects in segregation and chromosome organization in RNAi mutants are most likely a product of changes in chromatin structure at centromeres and telomeres. This is consistent with a role for the RNAi machinery in the targeting of histone modifying activities (30, 31). However, given the diverse functions of RNAi-related pathways in other systems, it is possible that some of the phenotypes reported in this study might be linked to the involvement of RNAi machinery in gene regulation and/or microRNA processing. In this regard, it should be noted that multiple meiosis-specific noncoding RNAs have been cloned in fission yeast, and they are predicted to contain extensive secondary structure including stable hairpins (52). The extent to which RNAi regulates endogenous genes in fission yeast is a cause for future study.
We speculate that aberrant segregation patterns caused by loss of centromere function may underlie some of the developmental phenotypes observed in the RNAi mutants of other systems. For example, reported defects in stem cell maintenance and proliferative capacity (27, 53, 54) could be due in part to frequent missegregation events that deplete the stem cell population. Furthermore, it has been proposed that specific segregation events are required for proper development (55). In this respect, it may be noteworthy that one Argonaute homolog in Drosophila specifically causes defects in the asymmetric division of germ line stem cells (26), and that another causes meiotic drive (28).
This study provides evidence that the RNAi machinery participates in the regulation of diverse chromosomal processes during mitosis and meiosis in fission yeast. We expect that future mechanistic analysis in fission yeast will yield important new information about the role of RNAi machinery in diverse chromosomal functions.
Acknowledgments
We thank Mitsuhiro Yanagida, Yoshinori Watanabe, Fuyuki Ishikawa, Julia Cooper, and Robin Allshire for providing strains and reagents. We also thank Guoping Xiao for technical assistance, and Joshua Chang Mell for helpful discussion and critical reading of the manuscript. I.M.H is an Arnold and Mabel Beckman Fellow, and K.N. is supported by Leukemia and Lymphoma Society. This work was supported by National Institutes of Health Research Grant GM59772 (to S.I.S.G.).
Abbreviations
- RNAi
RNA interference
- FISH
fluorescence in situ hybridization
- ChIP
chromatin immunoprecipitation
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Cranston G. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 2.Murphy T D, Karpen G H. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellum R, Alberts B M. J Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- 4.Grewal S I, Bonaduce M J, Klar A J. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grewal S I S, Elgin S C. Curr Opin Genet Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 6.Nimmo E R, Pidoux A L, Perry P E, Allshire R C. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 7.Cooper J P, Watanabe Y, Nurse P. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- 8.Kanoh J, Ishikawa F. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto A, Hiraoka Y. BioEssays. 2001;23:526–533. doi: 10.1002/bies.1072. [DOI] [PubMed] [Google Scholar]
- 10.Zickler D, Kleckner N. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 11.Niwa O, Shimanuki M, Miki F. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherthan H, Bahler J, Kohli J. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Hannon G J. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 15.Angell S M, Baulcombe D C. EMBO J. 1997;16:3675–3684. doi: 10.1093/emboj/16.12.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grishok A, Pasquinelli A E, Conte D, Li N, Parrish S, Ha I, Baillie D L, Fire A, Ruvkun G, Mello C C. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 17.Lau N C, Lim L P, Weinstein E G, Bartel D P. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 18.Pal-Bhadra M, Bhadra U, Birchler J A. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen R A, Cluster P D, English J, Que Q, Napoli C A. Plant Mol Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 20.Matzke M, Matzke A J, Kooter J M. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 21.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 22.Ketting R F, Plasterk R H. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki K, Fine N, Fujisawa T, Gorovsky M. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 24.Ketting R F, Fischer S E, Bernstein E, Sijen T, Hannon G J, Plasterk R H. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight S W, Bass B L. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox D N, Chao A, Baker J, Chang L, Qiao D, Lin H. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt A, Palumbo G, Bozzetti M P, Tritto P, Pimpinelli S, Schafer U. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morel J B, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volpe T A, Kidner C, Hall I M, Teng G, Grewal S I, Martienssen R A. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 31.Hall I, Shankaranarayana G, Noma K, Ayoub N, Cohen A, Grewal S I S. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 32.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama J, Allshire R C, Klar A J, Grewal S I. EMBO J. 2001;20:2857–2866. doi: 10.1093/emboj/20.11.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa H, Lee J, Hurwitz J, Allshire R C, Nakayama J, Grewal S I, Tanaka K, Murakami Y. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuura A, Naito T, Ishikawa F. Genetics. 1999;152:1501–1512. doi: 10.1093/genetics/152.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 37.Peters A H, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 38.Bernard P, Maure J F, Partridge J F, Genier S, Javerzat J P, Allshire R C. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 39.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal S I, Watanabe Y. Nat Cell Biol. 2001;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 40.Nabeshima K, Nakagawa T, Straight A F, Murray A, Chikashige Y, Yamashita Y M, Hiraoka Y, Yanagida M. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Funabiki H, Hagan I, Uzawa S, Yanagida M. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maison C, Bailly D, Peters A H, Quivy J P, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 43.Ekwall K, Nimmo E R, Javerzat J P, Borgstrom B, Egel R, Cranston G, Allshire R. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 44.Cooper J P, Nimmo E R, Allshire R C, Cech T R. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 45.Partridge J F, Borgstrom B, Allshire R C. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 46.Saitoh S, Takahashi K, Yanagida M. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi K, Chen E S, Yanagida M. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe Y, Nurse P. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 49.Chikashige Y, Hiraoka Y. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, Uzawa S, Cande W Z. Genetics. 2002;160:861–876. doi: 10.1093/genetics/160.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagan I, Yanagida M. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe T, Miyashita K, Saito T T, Yoneki T, Kakihara Y, Nabeshima K, Kishi Y A, Shimoda C, Nojima H. Nucleic Acids Res. 2001;29:2327–2237. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin H, Spradling A C. Development (Cambridge, UK) 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 54.Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klar A J. Schizophr Res. 1999;39:207–218. doi: 10.1016/s0920-9964(99)00075-4. [DOI] [PubMed] [Google Scholar]