Abstract
Adeno-associated virus (AAV) is a classification given to a group of nonpathogenic, single-stranded DNA viruses known to reside latently in primates. During latency in humans, AAV type 2 (AAV2) preferentially integrates at a site on chromosome 19q13.3ter by targeting a sequence composed of an AAV Rep binding element (RBE), a spacer, and a nicking site. Here, we report the DNA sequence of an African green monkey AAV integration site isolated from CV-1 cells. Overall, it has 98% homology to the analogous human site, including identical spacer and nicking sequences. However, the simian RBE is expanded, having five perfect directly repeated GAGC tetramers. We carried out a number of in vitro and in vivo assays to determine the effect of this expanded RBE sequence on the Rep-RBE interaction and AAV targeted integration. Using electromobility shift assays it was demonstrated that AAV4 Rep68 bound the expanded RBE with a sixfold-greater affinity than the human RBE. To determine the basis for the affinity increase, DNase I protection and methylation interference (MI) assays were performed. Comparison of footprints on both the human and simian RBEs revealed nearly identical protection; however, MI analysis suggested greater interaction with the guanine nucleotides of the expanded RBE, thus providing a biochemical basis for the increased binding activity. In vivo, integration targeted to the simian RBE was demonstrated by PCR analysis of latently infected Cos-7 cells. Interestingly, the frequency of site-specific integration was twofold greater in Cos-7 cells than in HeLa cells. Overall, these experiments establish that the simian RBE, identified in CV-1 cells, functions analogously to the human RBE and provide further evidence for a developing model that proposes individual roles for the RBE and the spacer and nicking site elements.
To date, eight different but closely related adeno-associated virus (AAV) serotypes (types 1 to 8) which infect primates have been identified (reviewed in references 6 and 16). These viruses are members of the family Parvoviridae, and they have a unique biology that makes them particularly well suited as gene therapy vectors (1, 6, 16, 35, 42-44). Additionally, AAV (specifically type 2 [AAV2]) has been shown to be the only known mammalian virus having the potential to site-specifically integrate into the human genome, preferentially targeting a site on human chromosome 19q13.3qter (ch19) (6, 9, 20, 21, 25, 33, 34, 41)
The AAV genome is composed of linear single-stranded DNA ∼4.7 kb long (42, 49), and while AAV2 is the most thoroughly characterized AAV, studies of other serotypes are under way (10, 12, 38). These include monkey-specific AAV4, which was recently cloned and found to have a high degree of homology to AAV2 (12). The AAV4 genome has two open reading frames, like that of AAV2, and codes for similar Rep proteins (Rep78 [78 kDa], -68, -52, and -40). For example, the AAV4 Rep68 protein has 90% amino acid identity to AAV2 Rep68, with 5% of the divergent amino acids being similar in hydrophobicity or structure (12). There is more divergence in capsid proteins (Vp1, -2, and -3), with only 60% homology at the amino acid level. In the inverted terminal repeats (ITRs), similarity between the putative hairpin structures in the two viruses is retained, but the AAV4 ITR sequences diverge at two specific regions important in Rep binding. The AAV4 ITR Rep binding element (RBE) is expanded, with four direct GAGY tetramer repeats (GAGTGAGTGAGCGAGC), not three perfect GAGC tetramers as seen in type 2 (12, 30). Additionally, type 4 virus diverges at another possible Rep contact site, the flip hairpin secondary structure, having a 5′-CTCTG-3′ at the apex (12, 40). The exact importance of these changes and their impact on the virus life cycle are still unknown. However, the interaction of the AAV2 Rep proteins with the RBE, spacer, and terminal resolution site (trs) of both AAV2 and human ch19 has been previously characterized. DNase I footprinting of type 2 Rep68 binding to linear AAV2 demonstrated protection of 32 nucleotides (nt), which included the RBE, the 13-nt spacer, and the trs (11), and evidence of Rep-specific interaction with nucleotides of these sites was confirmed by chemical interference (40). Similar to the linear ITR, DNase I footprinting of the ch19 site demonstrated 33 nt (non-trs strand) protected by type 2 Rep68 (24). Additionally, other studies suggest that stable binding is required for Rep to be functionally active (8, 31). In vivo studies have determined the biological importance of these sequences in targeting AAV2 to ch19 (13, 14, 27, 28, 59). Therefore, it is not clear how changes in these important sequences will affect targeted integration. Also, sequence divergence is not restricted to type 4 virus. The type 5 ITR has four direct GAGY repeats, one being imperfect (GAGCCAGCGAGCGAGC). The trs of this serotype is also distinct, as is the spacer between the RBE and the trs, which has an additional 5 nt. The impact of these sequence changes on AAV replication has been measured by the inability of either type 2 or 5 Rep to cleave the other's ITR substrate (10). No studies have determined the impact of these divergent ITR elements on site-specific integration.
Central to AAV2 site-specific integration is the presence of these elements (RBE, spacer, and trs) on human ch19 (41, 50). Not only are they present, but each component is required for type 2 targeted integration (13, 14, 27, 28, 59). Similar to type 2 Rep cleavage activity, evidence for nicking and covalent linkage on ch19 during targeted integration has been reported (22, 23). Almost all experiments characterizing AAV latency have been performed using the AAV2 serotype and human cell lines (6, 13, 14, 20, 26, 28, 39, 48). Demonstration of AAV2 site-specific integration in nonhuman targets has generally employed “knock-in” animal models carrying the human ch19 sequences (39, 58). The biology of the monkey-specific AAV4, with its expanded RBE, has not been thoroughly studied, nor has the role of the simian chromosomal site-specific integration locus. Only Southern analysis, which demonstrated the presence of sequences with homology to the ch19 targeting locus in the monkey genome, has been carried out (45). To date, no information about the suitability of this sequence for AAV site-specific integration has been obtained. With the known divergence of the type 4 ITR, it was of specific interest to determine the exact sequence present in the monkey genome and, more importantly, to test the abilities of these elements to carry out site-specific integration as seen with type 2 in human cells.
In this report, we have identified an AAV integration locus in CV-1 cells from the African green monkey and characterized its interaction with the AAV4 Rep68 protein. The AAV4 integration site has perfect homology to the human trs and spacer; however, the simian RBE diverges, adding two additional GAGC tetramer repeats, creating five perfect direct repeats. We constructed an AAV4 Rep68-histidine fusion protein (AAV4 Rep68H) and demonstrate that this protein will specifically bind the simian RBE with a sixfold increase in affinity compared to the human RBE. Analysis of binding by methylation interference (MI) suggested that the increase is due in part to specific interaction of Rep68H with the extra guanine (G) nucleotides of the expanded RBE, as well as an increased interaction overall. Electron microscopy (EM) analysis demonstrated sequence-specific protein-DNA complexes indistinguishable from those generated using type 2 Rep and human DNA substrates. These in vitro studies were followed up by in vivo analysis, which demonstrated AAV site-specific integration into the simian Cos-7 cells, establishing that the simian RBE, spacer, and trs function analogously to the human ch19 integration locus. It is interesting that we consistently observed integration frequencies in Cos-7 cells that were twofold greater than those in HeLa cells. Although further studies are required to rule out the potential roles of additional genomic elements, variability in type 2 and type 4 Rep proteins, or host proteins that may influence targeted integration, the expanded simian RBE was the only obvious difference. Overall, these results establish AAV site-specific integration in the African green monkey and are consistent with previous studies that determined that the RBE, spacer, and trs DNA elements individually influence site-specific integration.
MATERIALS AND METHODS
Plasmids.
pRE2 contains a 2.7-kb fragment from human ch19 (45). pStump68 contains the gene for AAV2 Rep68 protein (45). pHIV68 contains the AAV2 Rep68 gene (56). pDD-neo, pDD-puro, and pDD-GFP (56) contain the neomycin resistance gene, the puromycin resistance gene, and the green fluorescent protein (GFP) gene, respectively, and all contain the AAV2 ITR sequences.
Cloning the African green monkey AAV integration site.
pMkRBE contains the RBE, spacer, and trs from the African green monkey genome. The plasmid was constructed by TA cloning a PCR-amplified fragment (302 bp) from CV-1 cells into pCR2.1 (Invitrogen). CV-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum and penicillin-streptomycin. PCR was performed as described by the manufacturer with TaKaRa Ex Taq (PanVera Corp.) and 100 ng of CV-1 genomic DNA in a standard reaction mixture supplemented with 30 mM MgCl2 and deaza-dCTP. Temperature cycling consisted of 2 cycles of 97°C for 30 s, 58°C for 30 s, and 72°C for 1.2 min, 35 cycles of 94°C for 1 min, 58°C for 30 s, and 72°C for 1 min, and 1 cycle of 94°C for 1 min, 58°C for 30 s, and 72°C for 7 min. PCR was performed initially with the primers 5′-GACATCGCACCGCCCGCCCG-3′ and 5′-TACTTACTTACTTACCC-3′. Then a second round of PCR was performed with the primers 5′-GACATCGCACCGCCCGCCCG-3′ and 5′-TGACTTCAGGCGTCGA-3′. The clones were sequenced at the University of North Carolina (UNC) Sequencing Facility.
Viral propagation and purification.
AAV4 was purchased from the American Type Culture Collection and amplified in Cos-7 cells as previously described (12). The Cos-7 cells were maintained in DMEM supplemented with 10% fetal calf serum and penicillin-streptomycin. The cells were grown at 37oC in a 5% CO2 humidified environment. The virus was purified using a cesium chloride (CsCl2) gradient, and titers were determined by dot blotting (3).
AAV4 Rep68H protein production.
AAV4 pRep68H contains the gene for the AAV4 Rep68 protein inserted into pQE-70 (Qiagen). This vector places a six-histidine tag on the C terminus of the protein. To clone the AAV4 Rep68 gene, PCR was performed using Takara Ex Taq polymerase in a standard reaction mixture containing AAV4 genomic DNA (100 ng) with the oligonucleotides 5′-TCTATCTAGACTTGGCCACTCCCTCTCTGCGCGC-3′ and 5′-AGGCCTTAAGAGCAGTCGTCCACCACCTTGTTCC-3′. The unspliced gene was then digested with SphI and BglII (all restriction enzymes were purchased from New England BioLabs) and ligated into pQE70 according to the manufacturer's instructions. To construct the intron minus AAV4 Rep68H, the clone was digested with AatII and BglII and ligated to the correctly spliced C-terminal synthetic DNA. The oligonucleotides used to construct the C-terminal synthetic DNA were 5′-GATCTGAGAGTTGTCCTCTAGCCAACCTGTCCGCGTAGTCCACCGGAGCTTCCGCGTCTGACGT-3′ and 5′-CAGACGCGGAAGCTCCGGTGGACTACGCGGACAGGTTGGCTAGAGGACAACCTCTCA-3′.The AAV4 Rep68H gene was sequenced at the UNC Sequencing Facility. The amino acid sequence was wild type except for the six C-terminal histidines and a proline-to-alanine change at amino acid 536. The AAV4 and AAV2 Rep68H proteins were expressed and purified from SG13009 (Qiagen) as previously described (58).
EMSAs.
Competition electromobility shift assays (EMSAs) were performed as previously described (31). Briefly, DNA substrates (200 bp) were made by digesting pMkRBE with the restriction enzymes HindIII-AatII and digesting pRE2 with PvuII-AatII; then, the fragments were gel purified following standard procedures (3). For experiments using synthetic DNA as the substrate, oligonucleotides (41 bp) were annealed. Rep68H (1 pmol) was placed in a 20-μl binding reaction mixture containing cold competitor DNA, 10 mM HEPES-KOH [pH 7.5], 8 mM MgCl2, 40 mM KCl, 0.2 mM dithiothreitol, 8% glycerol, and 0.1 μg of poly(dI-dC)/μl. Next, a [γ-32P]ATP end-labeled DNA substrate (6,000 cpm) was added, and the reaction mixture was incubated for 20 min. Separation was done at 90 V for 1 to 2 h using nondenaturing 4% polyacrylamide gel electrophoresis (PAGE). The free and bound fractions were quantified using a PhosphorImager (Molecular Dynamics).
DNase I protection assay.
Protection assays were performed as previously described (3, 31). Briefly, DNA substrates were made by restriction enzyme digestion of pMkRBE and pRE2 with BspE1-DraIII and BspE1-EcoRI, respectively. Rep68H (15 pmol) was incubated with the [γ-32P]ATP end-labeled DNA substrate (50,000 cpm) in the EMSA binding buffer for 20 min at room temperature. Then, DNase I (diluted to 0.5 U/μl) was added and allowed to hydrolyze the DNA for 30 to 60 s at room temperature. The sample was separated using 10% denaturing PAGE and run at 1,600 V for approximately 1 h, and then the gel was exposed to film (Kodak).
MI.
For the MI assay, 300-bp substrates were produced by restriction enzyme digestion of pMkRBE and pRE2 with BspE1-AatII and BspE1-AccI, respectively. MI assays were performed as previously described (3, 40). Briefly, after being [γ-32P]ATP end labeled, the substrate was treated with dimethyl sulfate (Sigma). The Rep protein was incubated with the methylated substrate (100,000 cpm) and electrophoresed as in the EMSA protocol. The free and bound fractions were gel purified (3); then, Maxam-Gilbert chemical sequencing was performed (3). The bands were quantified using a PhosphorImager, and the relative band intensity ratio for each G residue was plotted as previously described (40).
EM.
EM analysis was performed at the UNC EM Core Facility as described previously (58). Purified AAV4 Rep68H was incubated with a 2.2- or 2.3-kb DNA fragment containing the African green monkey or human ch19 RBE, respectively. The binding reaction used a 1:1 mass ratio of protein-DNA, and the conditions for binding were the same as for the EMSA.
Detection of latent AAV.
Cos-7 and HeLa cells were transfected with either pDD-neo, pDD-puro, or pDD-GFP alone or with AAV4 or AAV2 pHIVRep68 using Superfect reagent (Qiagen). All cells were maintained in DMEM supplemented with 10% fetal calf serum and penicillin-streptomycin. Colonies transfected with either pDD-neo or pDD-puro were selected with G418 and puromycin, respectively (3, 56). After selection, genomic DNA was extracted following standard protocols (3). For GFP-expressing cells, a modular flow cytometer (MoFlo; Cytomation Inc.) at the UNC Flow Cytometer Facility was used to detect fluorescence and to place GFP-expressing cells into individual wells. To demonstrate the presence of latent AAV, 100 ng of the DNA was analyzed by PCR as previously described (52). Briefly, Deep Vent Exo(−) polymerase (New England Biolabs) and the primers 5′-CGGGGAGGATCCGCTCAGAGGACA-3′ and 5′-CGGCCTCAGTGAGCGAGCGAGC-3′ were used along with genomic DNA in standard PCR. The temperature cycle was 99°C for 1 min, followed by 35 cycles of 99°C for 10 s and 72°C for 4 min. A fraction of the PCR product was blotted and probed with a ch19 probe. As a negative control, each primer was run separately in a reaction with genomic DNA to quantify the background signal. Colonies were positive for integrated AAV if a signal greater than that of background was obtained for the PCR (53).
RESULTS
Identification of an AAV integration site in CV-1 cells from African green monkeys.
To isolate the simian AAV integration site, primers were designed using the human ch19 sequence and PCRs were carried out with genomic DNA from CV-1 cells (an African green monkey kidney cell line). The PCR consistently amplified a 302-bp fragment. Sequencing and alignment revealed significant homology to the AAV integration locus of human ch19 (Fig. 1). Seven independent clones were sequenced and aligned, all of which were identical, suggesting accurate amplification of the simian DNA. Overall, the cloned sequence demonstrated 98% identity between the human and African green monkey sequences, and excluding the RBE, the divergent nucleotides occurred randomly throughout the cloned fragment. At the Rep binding site, the simian DNA has perfect homology to the human ch19 trs and spacer. However, the simian RBE diverges in length from the analogous human sequence by having two additional GAGC tetramers, creating five perfect direct repeats. The identification of nearly identical genomic sequences carrying the RBE, spacer, and trs in CV-1 cells made it reasonable to expect that AAV4 recombination would be directed to this site. Since previous studies had determined that the repeating GAGY motif of the RBE was important for Rep binding, we carried out a series of experiments to determine the role of the expanded simian motif.
FIG. 1.
Sequence alignment of the AAV integration site from human ch19 and the 302-bp DNA fragment identified in the African green monkey genome (Monkey) as cloned from CV-1 cells (Vector NTI software; InforMax). The three elements of the AAV integration locus, the RBE, spacer, and trs, are underlined. The human ch19 RBE has three direct GAGC tetramers (dark shading), while the simian RBE is expanded, having two additional GAGC tetramers (light shading). The simian trs is also darkly shaded, and an 8-bp spacer is located between this element and the simian RBE, just as in the human ch19. Sequence homology is indicated by dots, dashes represent gaps in the sequence, and nonconsensus bases for the monkey sequence are given (bottom line).
Characterization of AAV4 Rep68H binding to the simian target sequence.
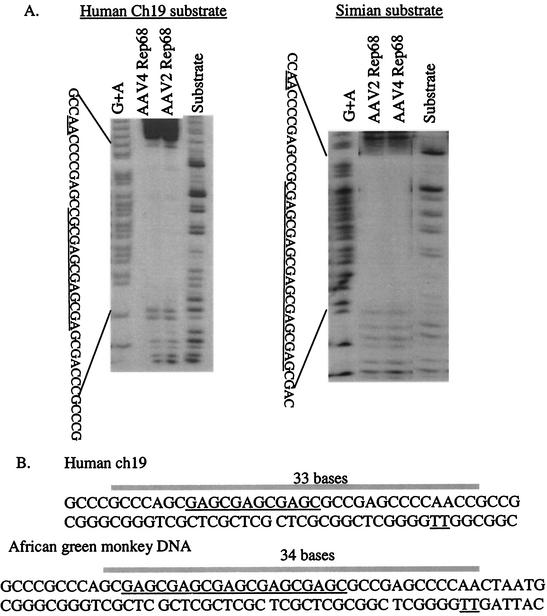
To investigate the interaction of AAV4 Rep68H with the simian RBE, DNase I protection assays were performed. In reciprocal experiments, AAV4 or AAV2 Rep68H protein was incubated with either the simian or the human ch19 AAV integration sequence (non-trs strand). The results (Fig. 2A) demonstrated that the proteins produced nearly identical protection patterns on either substrate. Both the AAV4 and AAV2 Rep68H proteins protected approximately 33 to 34 nt of the expanded sequence, beginning 2 nt downstream of the first GAGC repeat and extending to 1 bp upstream of the nicking site complement. Evaluation of binding to the human RBE produced similar results, with protection extending 33 nt and including the RBE, spacer, and trs sequences (summarized in Fig. 2B). For the AAV2 protein, bound to the human RBE, the footprinting results are consistent with previously reported data (24).
FIG. 2.
(A) Reciprocal DNase I protection analysis of AAV2 and AAV4 Rep68H proteins on the human ch19 and African green monkey DNA substrates containing the RBE (underlined), spacer, and trs complement (underlined). The [γ-32P]ATP end-labeled substrate (non-trs strand) was incubated with 15 pmol of Rep before DNase I digestion and denaturing PAGE analysis. For lanes G+A, substrate DNA was chemically sequenced using formic acid. For lanes Substrate, substrate DNA was treated with DNase I without any addition of Rep. (B) DNase I results are summarized for human (top) and simian (bottom) sequences. The bar indicates the bases fully protected by both AAV2 and AAV4 Rep68H proteins. The RBE and nicking site are both underlined.
To further characterize the interactions between the Rep proteins and their respective binding sites, we used EM to visualize AAV4 Rep68H binding to both the simian RBE and the human ch19 RBE. The AAV4 Rep68H protein was added to a binding buffer containing either a 2.2-kb DNA fragment with the simian RBE located approximately 400 bases from one end or a 2.3-kb DNA fragment containing the human ch19 RBE located approximately 800 bases from one end (Fig. 3A and B, respectively). The AAV4 Rep68H protein bound both the simian and human fragments (Fig. 3) in a location consistent with an RBE-specific interaction and formed Rep-DNA complexes seemingly identical in size and conformation to each other and to those previously described for type 2 Rep binding human ch19 RBE (58). At a 1:10 molar ratio (DNA-protein), ∼70% (16 of 23 molecules) of the fragments had single AAV4 Rep68H bound to the simian RBE. All of the remaining 30% (7 of 23 molecules analyzed) had multiple Rep proteins, generally two, with one Rep complex binding the RBE and another bound nonspecifically to the DNA and not in contact with the RBE-bound Rep. Similar results were observed for the AAV4 Rep68H protein bound to the human ch19 RBE (data not shown).
FIG. 3.
EM analysis of AAV4 Rep68H binding to the AAV site-specific integration loci from African green monkeys (A) and human ch19 (B). The simian fragment was 2.2 kb long and contained the 302-bp simian site from CV-1 cells, with the expanded simian RBE located 0.4 kb from one terminus. The human ch19 DNA fragment was 2.3 kb in length, with the RBE located 0.8 kb from one terminus. For both substrates, the AAV4 Rep68H protein covered an average of 60 bp as previously reported (58).
The results of the footprinting experiments and EM analysis demonstrate that the AAV4 Rep68H protein binds specifically and protects the simian RBE, spacer, and trs, suggesting that these sequences carry all of the cis-acting elements necessary to direct targeted integration of AAV4 in cells derived from the African green monkey.
Rep68H relative affinity to human and simian chromosomal DNAs.
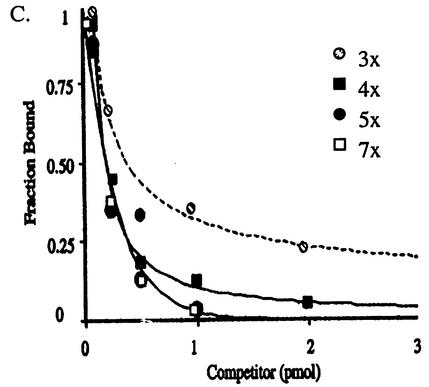
Next, we determined the relative affinities of the proteins to the human and simian RBEs by performing competitive EMSAs. The 302-bp fragment containing the simian RBE was [γ-32P]ATP end labeled, and the binding of either AAV2 or AAV4 Rep68H to this substrate was competed away by cold (either the simian or human RBE) competitor DNA (Fig. 4A). In these reciprocal assays, the concentrations of Rep68H proteins and the lengths of the DNA fragments, for the substrate and competitor, were held constant. In addition, to accurately compare affinities, the competition curve was performed over a broad range of cold-competitor concentrations. Using this range, we could perturb binding to the labeled DNA only slightly or completely abolish it, providing an accurate determination of the relative equilibrium dissociation constants (15, 46). The AAV4 and AAV2 Rep68H proteins demonstrated a 6- and 10-fold increase in relative binding affinity, respectively, for the simian RBE compared to the human RBE (Fig. 4B). To further explore the possibility that the increased affinity was due to a greater number of tetramer repeats, EMSA experiments were conducted using synthetic DNA substrates. For these, the AAV2 Rep68 protein was used to bind substrates containing either three (3×), four (4×), five (5×), or seven (7×) direct repeats of the GAGC tetramer. The results are represented graphically in Fig. 4C and suggest that relative affinity increases as the number of repeats increases, up to five tetramers. At that point, the relative affinities of Rep for the 5× and 7× substrates were essentially equivalent (Table 1). The fivefold increase in relative affinity of the AAV2 Rep protein binding the 5× versus the 3× substrate was less than the 10-fold increase demonstrated above. This may be due to the short fragment length (41 bp) of the synthetic DNA substrates destabilizing binding. For both experiments, however, the basis for the increase in affinity for substrates having a greater number of tetarmers was not apparent, considering that the simian and human substrates produced nearly identical footprints. Next, we compared the base-specific interactions of the Rep proteins on the human ch19 and the simian DNA counterpart.
FIG. 4.
(A) Reciprocal competition EMSAs were performed to determine the relative affinities of AAV4 Rep68H to the simian (left) and human (right) DNA fragments. The African green monkey substrate (200 bp) was [γ-32P]ATP end labeled and titrated with cold competitor DNA (200 bp), either the simian fragment or the human fragment, before the addition of the AAV4 Rep68H protein (1 pmol). For lanes Rep−, no Rep protein was added. Separate experiments using the AAV2 Rep68H protein were conducted in an identical manner. (B) After the bound and free fractions were quantified (using a PhosphorImager), the bound fraction was graphed as a function of the cold-competitor concentration, simian fragment (open squares) and human fragment (solid squares). (C) Additionally, AAV2 Rep68 was used in competition EMSAs with synthetic DNA as the substrate, and the results were graphed. The synthetic substrates (41 bp) contained either three (3×), four (4×), five (5×), or seven (7×) direct repeats of the GAGC tetramer. The substrates and their relative affinities to cold competitor 3× are given in Table 1. For all experiments, the competitor concentrations required to achieve a 50% reduction in the fraction bound were compared for each protein, and the relative binding affinity is the ratio of these competitor concentrations.
TABLE 1.
Synthetic RBE substrates
| Name | Sequence | Relative affinity |
|---|---|---|
| 3× | CCCCGAGCCGCGCGAGCGAGCGAGCGACCCGCCCGCCCGCC | 1 |
| 4× | CCCCGAGCCGCGAGCGAGCGAGCGAGCCGACCCGCCCGCCC | 2 |
| 5× | CCCCCGAGCCGCGAGCGAGCGAGCGAGCGAGCGACCCGCCC | 5 |
| 7× | CCCCGAGCCGCGAGCGAGCGAGCGAGCGAGCGAGCGAGCCG | 5 |
Methylation interference analysis of AAV4 Rep68H binding.
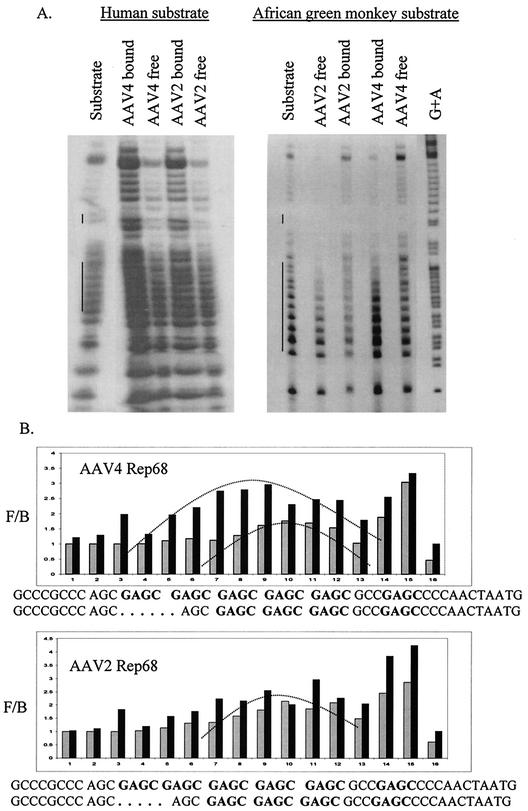
To determine a possible chemical basis for the increase in affinity toward the expanded simian RBE, MI assays were performed. AAV2 Rep68 is known to interact strongly with G nucleotides of the human RBE (32, 40), and we suspected that the expanded GAGC motif of the simian RBE was contributing to the increased binding affinity. The MI assays were done as previously described, using dimethyl sulfate to methylate the non-trs strand of the linear Rep binding site (32, 40). The intensity of each band was quantified by PhosphorImager analysis, and the free:bound ratio was plotted for each G residue of the sequence. A free/bound ratio of 1 indicates that the interaction of Rep with the particular G nucleotide contributes little to binding, while an interaction of >1 indicates that the G nucleotide contributes to binding (40). In reciprocal assays, we evaluated the interactions of AAV4 and AAV2 Rep68H proteins with either the methylated human or simian DNA (Fig. 5A). The results indicated that the interactions of both proteins with the expanded RBE were quantitatively stronger than with the human RBE. These results were consistent with the assays described above and demonstrate that both Rep proteins interacted with G residues within the DNase I protected region (Fig. 5B). At the repeating GAGC motif, both proteins produced symmetrical interaction patterns centered approximately at the middle tetramer, as previously described for AAV2 Rep68 binding (40). However, this pattern was slightly distorted for AAV2 Rep68H binding to the simian RBE. Interestingly, for any single G nucleotide, the greatest interaction for both proteins was at the single GAGC tetramer within the spacer region. Overall, the pattern produced by the interaction of both the AAV2 and AAV4 Rep68H proteins with the methylated substrates suggests that the additional G residues of the monkey RBE contribute to the greater affinity of both proteins for the simian RBE.
FIG. 5.
(A) Reciprocal MI analysis of the AAV4 and AAV2 Rep68H proteins binding the human ch19 (left) and simian (right) sequences. The substrates (non-trs strand) were [γ-32P] end labeled and methylated before being incubated with either of the Rep proteins (2 pmol). Then, the bound and free fractions were purified and cleaved before separation by denaturing PAGE. For lanes Substrate, methylated substrate DNA was run without any addition of the Rep protein. For lane G+A, substrate DNA was chemically sequenced using formic acid. The long and short vertical lines next to lanes Substrate indicate the GAGC tetramers and the trs location, respectively. Because of variable amounts of substrate in each lane, the concentration of substrate was normalized by quantification of bands outside the Rep binding site before graphical analysis. (B) MI graphical representation of AAV4 Rep68H and AAV2 Rep68H binding either the methylated simian substrate (solid bars) or the methylated human substrate (shaded bars). The curved lines on the graph indicate symmetrical patterns produced by the interaction of the Rep proteins with the substrate. The sequences for the simian and human RBE, spacer, and trs are given beneath the graphs, and the GAGC tetramers are in boldface. F/B, free/bound ratio.
Site-specific integration in Cos-7 cells.
The above-mentioned in vitro studies were followed up by in vivo experiments that demonstrated that the AAV4 Rep68 protein could mediate site-specific integration in the simian genome. Cos-7 cells were first cotransfected with AAV4 pHIV68 and pDD-neo (an AAV substrate previously characterized for AAV site-specific integration) (56, 59) and then selected for resistance to G418. Colonies were examined by PCR for AAV site-specific recombination at the simian RBE locus (53). As a negative control, pDD-neo was transfected into Cos-7 cells without a source of Rep protein. Each colony was tested in duplicate, and the results demonstrated that 7 of 11 colonies (64%) were positive for site-specific integration at the simian RBE locus (Fig. 6). These results indicate that the RBE, spacer, and trs identified in African green monkey cells are sufficient to direct site-specific recombination.
FIG. 6.
AAV4 Rep68 mediates the site-specific integration of an AAV2 ITR construct targeting the African green monkey RBE, spacer, and trs in Cos-7 cells. The cells were cotransfected with AAV4 pHIV-Rep68 and pDD-neo and selected for G418 resistance before PCR analysis. Primers ch19/AAV represent a PCR using one primer (ch19) that anneals to the simian genome and a second primer (AAV) that anneals to the AAV2 ITR sequence. After PCR, the reaction product was blotted and hybridized to a ch19 probe. The AAV and ch19 primers were used alone as negative controls in separate PCRs.
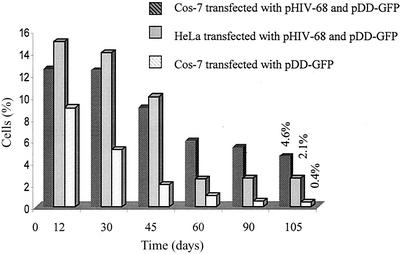
Next, we compared the integration rates in Cos-7 and HeLa cells after contransfection of the AAV2 Rep68 and AAV2 ITR constructs pDD-puro and pDD-GFP. For the pDD-puro construct, targeting was demonstrated after selection by PCR analysis in 75 (12 of 16) and 42% (5 of 12) of the Cos-7 and HeLa cells, respectively. For the GFP-expressing colonies, 3 to 5 days posttransfection the cells were enriched for GFP expression by sorting individual cells into wells using a MoFlo cytometer. This created clonal colonies and corrected for any difference in transfection efficiencies and growth rates between the two cell lines. The cells were cultured continuously and monitored for >4 months without selection. Both the Cos-7 and HeLa cells had approximately the same growth rate and number of cells at confluence (data not shown). Initially, for both cells lines, the percentage of cells expressing GFP decreased rapidly. By day 12 after sorting, the Cos-7 and HeLa cells that received AAV2 Rep68 had a slightly higher percentage of cells positive for GFP expression. At day 105, the expression of GFP had been stable for >60 days, and the flow cytometer was used again to determine the fraction of cells expressing GFP. In the absence of AAV2 Rep68, only 0.4% of Cos-7 and 0.6% of HeLa cells expressed stable GFP past 105 days. In contrast, 2.1% of the HeLa cells and 4.6% of the Cos-7 cells transfected with both AAV2 pHIV-Rep68 and pDD-GFP were stable for GFP expression. The relationship between the percentage of positive cells and time postsorting was graphed (Fig. 7). These experiments suggested that the presence of AAV2 Rep68 increased long-term stable expression of GFP in both human and African green monkey cells and that latency in Cos-7 cells was at least twofold greater than in HeLa cells. PCR analysis for site-specific integration was performed on the stable GFP-expressing cells (52). Our PCR results indicated that in 80% of the Cos-7 clones, the GFP gene was being expressed from the RBE locus, while none of the clones transfected with only pDD-GFP targeted this site. For the HeLa cells, 36% of the colonies expressed GFP from the simian RBE locus. These studies support the observation that the chromosomal RBE, spacer, and trs DNA sequences direct AAV recombination to the site in both the human and simian genomes and in addition suggest that the affinity and specificity of Rep for the chromosomal RBE are directly related to the specific integration rate.
FIG. 7.
Cos-7 and HeLa cells were cotransfected with pHIV-Rep68 and pDD-GFP and monitored for GFP expression for 105 days. Three to 5 days posttransfection, GFP-expressing cells were sorted by fluorescence-activated cell sorter into individual wells. Cells transfected with pDD-GFP alone were also monitored. The data are graphed as the fraction of green fluorescent cell colonies to the total number of colonies as determined on different days.
DISCUSSION
In this study, we report the DNA sequence of an AAV integration locus in the African green monkey genome. This is the first time that a native RBE, spacer, and trs sequence other than human has been identified and shown to support AAV targeted recombination. Previously, a knock-in mouse containing the human AAV integration locus on the mouse X chromosome was developed independently by two different laboratories (39, 58). The rodent models have a common overlapping human ch19 sequence of 1.6 kb that contains the RBE, spacer, and trs sequences, which were shown to direct targeting to this locus on the X chromosome. These studies clearly demonstrated that all necessary information required to support site-specific recombination was contained in the 1.6-kb ch19 cis element.
Architectures of human and simian integration sites.
In the African green monkey, the spacer and trs sequences are identical to the human ch19 sequence. However, the simian RBE has five rather than three direct repeats of the GAGC tetramer. This expansion of the RBE sequences has also been noted in the AAV4 ITR, where there are four rather than three GAGY repeats, as seen in AAV2. The expansion of the RBE sequence in both the simian chromosome and the AAV4 ITR suggests coevolution between virus and host. Although the simian targeting locus and the type 4 viral ITR have diverged from their human counterparts (ch19 and the AAV2 ITR, respectively) with respect to the RBE, the spacer and trs are conserved. The AAV2 and -4 ITRs, compared to the corresponding targeting loci (monkey and human ch19), contain different spacer sequences (13 versus 8 nt, respectively). It is not clear why both the human and monkey chromosomal loci have 13-nt spacers while AAV2 and -4 have evolved to carry only 8 nt. Surprisingly, AAV5, which was isolated from humans, has an 18-nt spacer which is distinct from those of all other AAV ITRs and from those of the human and simian chromosomal counterparts. Of the AAV serotypes, type 5 virus is the only example where the spacing between the RBE and trs is not conserved. In addition, Rep protein derived from serotypes 1 to 4 will utilize all other AAV ITRs except type 5 ITR for replication substrates, and type 5 Rep is unable to utilize type 1 through type 4 ITRs for replication (unpublished observations and reference 10). These observations beg the question of how the AAV origin evolved (human to virus or virus to human) (8). In fact, using information concerning the number of RBEs present in a serotype-specific ITR, one can strongly suggest the species of origin. When we compared all of the ITR sequences available (serotypes 1 to 7) against type 2 with the hope of identifying the origin of monkey versus human isolates, type 5 appeared similar to type 4 in that the ITR also carries an expanded repeat (10). If this is correct, type 5 maybe the result of a nonhuman primate virus that jumped into humans. More importantly, the newly described types 6 and 7 have sequence identity to type 1 ITRs. This may represent human isolates that have infected monkey colonies (16). Recently, it was demonstrated that a viable nicking site on the viral substrate is not necessary for site-specific recombination (59). It will be interesting to determine if type 5 can carry out targeted integration, as well as to determine what cis-acting elements may be required. In addition, to these sequence differences, we demonstrated that both AAV4 and AAV2 Rep68H proteins have greater affinity for the expanded RBE that is at least in part due to the greater interaction with the guanine bases of the added tetramers. We also documented that the simian locus could serve as a suitable target for site-specific integration in the monkey genome. Based on our in vitro and in vivo data, the expansion of the simian chromosomal RBE did not hamper the type 2 targeting frequency and may have influenced integration in a positive way (a twofold increase in targeting in Cos-7 versus HeLa cells). Although not specifically addressed in this study, it would be interesting to determine if an expansion of the RBE in the AAV2 ITR, such as occurs in AAV4, will also affect targeting. Elucidation of these factors will contribute to clarifying the integration mechanism and the complex relationship between the AAV ITR and chromosomal sequences.
Rep68-DNA stoichiometry.
The AAV2 and AAV4 Rep68H proteins demonstrated greater affinity for the expanded simian RBE, and initially we presumed that a greater number of Rep molecules may be binding the five directly repeating GAGC tetramers. Recent studies, using a Rep protein mutated in the multimerization domain, suggested that the ability of Rep to form multimers is critical to its function (8, 22, 32, 53, 55). Rep forms a multimeric protein complex when bound to the RBE, spacer, and trs, and evidence for Rep78 forming dimeric to hexameric structures on binding the AAV2 ITR has been reported (36, 47). Our DNase I protection and EM data demonstrated that the footprints Rep produced on binding either simian or human sequences were almost identical in size, suggesting that the numbers of Rep molecules forming the protein complex are equivalent. This is in agreement with our laboratories' EM results, which suggested that equivalent numbers of molecules form a Rep complex whether binding the AAV2 ITR, the human RBE, the simian RBE, or nonspecific DNA (2, 57, 58). Previously, Ryan et al. examined Rep interaction with mutated RBEs and suggested that the RBE, spacer, and trs contain a single Rep recognition site (40). Together, the data suggest that Rep does not regulate its action by forming different multimeric protein structures at its various DNA binding sites. Despite this, most investigators performing EMSAs on Rep-RBE interactions have noted the appearance of multiple (generally three to six) bands or laddering in the bound fraction. McCarty et al. suggested these bands corresponded to either multiple DNA molecules bound to a single Rep complex, multiple Rep complexes bound to a single DNA molecule, or different numbers of Rep molecules forming the Rep complex. Based on DNase I protection and EM data (2, 57, 58), a likely cause of this laddering, which occurs readily at high concentrations of Rep, may be multiple Rep complexes (with identical stoichiometries) binding a single DNA molecule. Additionally, DNA conformation transitions, such as bending or looping, may be occurring (15). Under certain conditions, the presence of Rep-induced DNA bending can be observed by EM analysis. It is unknown whether these DNA conformation changes are artifacts or have some biological significance (2, 57, 58). Protein-induced DNA bending is known to be associated with many types of protein-DNA interactions, including recombination, and additionally is known to increase a protein's binding affinity for DNA (15, 51). In the case of Rep, an increase in binding affinity for the simian RBE may positively relate to integration frequency in African green monkey cells. It is also possible that the looping previously observed by EM analysis (57, 58) affects AAV targeting by altering Rep-ch19 interaction in some manner. We suspect that these DNA conformational changes are Rep induced and as such warrant further investigation.
Rep68-RBE interaction.
In agreement with previous studies, we demonstrated that Rep has greater affinity for substrates with increased numbers of the GAGY tetramer (55). Our competition EMSAs demonstrated that AAV4 and AAV2 Rep68H proteins have 6- and 10-fold-greater affinity, respectively, for the expanded simian RBE than for the human ch19 RBE. Interestingly, we performed EMSAs with synthetic DNA containing as many as seven copies of the GAGC tetramer and determined that with more than five directly repeating tetramer copies, the relative affinity does not significantly increase (Fig. 4C). We believe that this result is due to the inability of Rep to contact more than five GAGY tetramers in series, as demonstrated by DNase I protection analysis, and its inability to form protein complexes with variable stoichiometries. This observation may indicate the importance of three, four, and five direct repeats located in the type 2 and 4 viruses and the simian targeting locus, respectively. It is of interest to note that the AAV2 ITR and human ch19 retain similar numbers of GAGY tetramers (three each), whereas the AAV4 ITR and the simian targeting locus are different (four versus five, respectively). Compared to humans, monkeys as a group are evolutionarily more divergent from each other, and additional sequence analysis of targeting loci from related and unrelated primates, as well as ITR analysis of new monkey serotypes, should facilitate the investigation of the importance of these RBE expansions.
cis-acting sequences and emerging integration models.
The basis for the greater affinity of Rep for the simian RBE was demonstrated by MI, where both AAV2 and AAV4 Rep68H proteins specifically contacted all five GAGC tetramers, producing a quantitatively stronger interaction than with the human RBE. The interaction was symmetrical about the RBE as previously described (40) and demonstrated significant sequence contacts within the spacer element. This increased interaction at the chromosome spacer may indicate the importance of Rep contact within the spacer region, as recently indicated by the in vitro integration assay (34). In that study, an RBE, trs, and spacer of human ch19 were present in an Epstein-Barr virus shuttle vector, and it was demonstrated that mutating or removing the spacer significantly decreased or abolished integration. A developing model for Rep-ch19 interaction proposes that each element—the RBE, spacer, and trs—has a particular function in AAV site-specific integration. In summary, the number of GAGC tetramers contained in the RBE affects the affinity of Rep and its ability to locate the target site, while the spacer may be important in orienting the Rep complex toward the trs.
AAV integrates site specifically, targeting the simian RBE.
Our study clearly demonstrated that the simian sequence could serve as a locus for site-specific integration when transfection assays were carried out with AAV plasmid integration substrates. Site-specific recombination in the simian genome (using Cos-7 cells) was demonstrated using AAV2 ITR neo constructs with AAV4 Rep68 and AAV2 ITR puro or GFP constructs with AAV2 Rep68. Using these substrates, we demonstrated an approximately twofold increase in targeting in Cos-7 versus HeLa cells (Table 2). Comparatively, targeting efficiency in Cos-7 cells was consistently greater than in HeLa cells. Additionally, the 42 and 36% targeting efficiencies in HeLa cells are consistent with previous reports (reviewed in reference 43). Although in this study we compared the efficiency and frequency of targeting for a relatively small number of cells (Fig. 6 and Table 2), all of the cells (HeLa and Cos-7) were treated in an identical manner to provide the most accurate comparison, and in the case of ITR-GFP constructs, there was no drug selection; instead, cell sorting was employed to normalize for transfection differences.
TABLE 2.
Summary of AAV targeting frequencies in Cos-7 and HeLa cells as determined by PCR analysis
| Cells | Frequency (%)
|
||
|---|---|---|---|
| AAV4 Rep68a | AAV2 Rep68a
|
||
| G418 | Puro | GFP | |
| Cos-7 | 64 (7 of 11) | 75 (12 of 16) | 80 (17 of 20) |
| HeLa | 42 (5 of 12) | 36 (4 of 11) | |
Mediating protein.
The GFP constructs were used to monitor long-term expression from the chromosomal integration locus and compared overall (specific and nonspecific) integration frequencies mediated by AAV2 Rep68 without drug selection. Again, Cos-7 cells demonstrated at least a twofold-greater site-specific latency than HeLa cells (4.6 versus 2.1%). This result is consistent with the twofold-greater targeted (specific) latency determined by PCR analysis. Additionally, our data were reported without compensating for the 2.7 copies of the ch19 locus in HeLa cells. Previously, Hoggan et al. reported that 20% of primary kidney cell lots from African green monkeys and 1 to 2% of human embryonic kidney cell lots examined were infected with latent AAV (19). These observations suggest that the frequency of wild-type AAV integration in the simian genome may be greater than in the human genome. The integration frequency of 20% is greater than that generally reported for wild-type AAV in culture, unless infections are performed at very high multiplicities of infection (7, 26, 43). An additional factor cited in producing low frequencies of targeted integration during in vitro experiments is that latent cells may be diluted out of a culture before it is tested for the presence of proviral AAV (43). This may affect the frequency of integration, especially at low multiplicities of infection, where the initial number of cells infected is relatively small. In our experiments, transfection efficiencies were ∼90%, and additionally, we enriched for positive GFP-expressing cells at day 5 postinfection by sorting. Therefore, diluting out cells carrying AAV proviral DNA appeared unlikely. We also noted a slow decline over months in cells expressing GFP, which is inconsistent with the dilution of episomal templates. Cells lost fluorescence intensity, suggesting a low turnover rate for the GFP protein within the cell (known half-life, 3 to 4 weeks). Regardless, our experiments were designed to compare relative integration frequencies in the Cos-7 and HeLa cells, and the results from these studies suggest that frequencies of integration in both cell lines are relatively low. This is in agreement with the results of other laboratories, which have also reported low integration frequencies in human cell lines (≤3%) with recombinant AAV2 constructs (reviewed in reference 43). Previously in our laboratory, low integration frequencies were also observed when long-term expression from the LacZ reporter gene was monitored in HeLa cells for >18 months (AAV2 Rep68-mediated targeting). In that study, 1% of the HeLa cells stably expressed the LacZ gene product 70 days after transfection, and this percentage continued unchanged for the entire 18 months of the experiment (56). In the present study, we did not have access to the AAV4 ITRs, and the integration analysis was derived from type 2 and 4 Rep68 proteins using AAV2 ITRs. This combination may have affected the true targeting frequency in simian cells (type 4 in Cos cells). unique
The experimental results were consistent for the three constructs (pDD-neo, pDD-puro, and pDD-GFP), and integration in the simian genome was consistently twofold greater than in the human genome. A similar relationship has been recognized in transposon integration, where higher affinity of the transposase for its DNA binding site correlates to greater targeting frequency (29). Comparing recombinant AAV integration in human and simian cells, other than an increased Rep binding affinity for the expanded simian RBE, we are unaware at this point of any mechanisms that may be contributing to the increase in targeting frequency, although many other factors (e.g., host proteins) could be at play. However, the RBE expansion in the African green monkey may make the site inherently more distinctive in the genome, providing more information for Rep and, we suspect, giving Rep68 a greater functional specificity (4, 5, 54). It will be interesting to investigate the AAV integration sites in other nonhuman primates to determine if the RBE expansion is universal in this diverse group. In addition, testing of artificially generated targeting loci whose RBEs, as well as other critical cis-acting elements, vary will eventually define the importance of each of these sequences in facilitating AAV site-specific recombination and the future development of AAV-targeting vectors.
Acknowledgments
This work was supported by NIH grants NHLBI 5-51687 and 5-33215 and NIDDK 5-33117.
REFERENCES
- 1.Amiss, T., and R. J. Samulski. 2001. Methods for adeno-associated virus-mediated gene transfer into muscle, p. 455-470. In M. Starkey (ed.), Methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 2.Amiss, T. J. 2001. Ph.D. thesis. University of North Carolina, Chapel Hill.
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 4.Berg, O. G., and P. H. von Hippel. 1987. Selection of DNA binding sites by regulatory proteins. J. Mol. Biol. 193:723-750. [DOI] [PubMed] [Google Scholar]
- 5.Berg, O. G., and P. H. von Hippel. 1988. Selection of DNA binding sites by regulatory proteins. J. Mol. Biol. 200:709-723. [DOI] [PubMed] [Google Scholar]
- 6.Berns, K. I. 1996. Parvoviridae: the viruses and their replication, p. 2173-2197. In B. N. Fields et al. (ed.), Fields virology, 3rd ed., vol. 2. Raven Press, Philadephia, Pa.
- 7.Berns, K. I., T. C. Pinkerton, F. F. Thomas, and M. D. Hoggan. 1975. Detection of adeno-associated virus (AAV) specific nucleotide sequences in DNA isolated from latently infected Detroit 6 cells. Virology 68:556-560. [DOI] [PubMed] [Google Scholar]
- 8.Brister, J. R., and N. Muzyczka. 2000. Mechanism of Rep-mediated adeno-associated virus origin nicking. J. Virol. 74:7762-7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. K., M. D. Hoggan, W. W. Hauswirth, and K. I. Berns. 1980. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 33:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiorini, J. A., S. Afione, and R. M. Kotin. 1999. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other serotypes. J. Virol. 73:4293-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiorini, J. A., S. M. Wiener, R. A. Owens, S. R. M. Kyostio, R. M. Kotin, and B. Safer. 1994. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J. Virol. 68:7448-7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyall, J., and K. I. Berns. 1998. Site-specific integration of adeno-associated virus into an episome with the target locus via a deletion-substitution mechanism. J. Virol. 72:6195-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyall, J., P. Szabo, and K. I. Berns. 1999. Adeno-associated virus (AAV) site-specific integration: formation of AAV-AAVS1 junctions in an in vitro system. Proc. Natl. Acad. Sci. USA 96:12849-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fried, M. G. 1989. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis 10:366-370. [DOI] [PubMed] [Google Scholar]
- 16.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavin, D. K., S. M. Young, W. Xiao, B. Temple, C. R. Abernathy, D. J. Perira, N. Muzyczka, and R. J. Samulski. 1999. Charge-to-alanine mutagenesis of the adeno-associated virus type 2 Rep78/68 proteins yields temperature-sensitive and magnesium-dependent variants. J. Virol. 73:9433-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman, S. D., and O. Kay. 1999. Replacement of integration host factor protein-induced DNA bending by flexible regions of DNA. J. Biol. Chem. 274:37004-37011. [DOI] [PubMed] [Google Scholar]
- 19.Hoggan, M. D., G. F. Thomas, F. G. Thomas, and F. B. Johnson. 1972. Continuous carriage of adenovirus associated virus genome in cell culture in the absence of helper adenovirus, p. 243-249. In Proceedings of the 4th Lepetite Colloquium, Cocoyac, Mexico.
- 20.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19 for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughoin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus type 2. J. Virol. 60:251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyostio, S. R., R. S. Wonderling, and R. A. Owens. 1995. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 Rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J. Virol. 69:6787-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamartina, S., E. Sporeno, and C. Toniatti. 1999. In vitro and in vivo analysis of the integration site in human chromosome 19. J. Gene Med. 1:68-71. [Google Scholar]
- 24.Lamartina, S., G. Ciliberto, and C. Toniatti. 2000. Selective cleavage of AAVS1 substrates by the adeno-associated virus type 2 Rep protein is dependent on topological and sequence constraints. J. Virol. 74:8831-8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamartina, S., E. Sporeno, E. Fattori, and C. Toniatti. 2001. Characteristics of the adeno-associated virus pre-integration site in human chromosome 19: open chromatin conformation and transcription competent environment. J. Virol. 74:7671-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laughlin, C. A., C. B. Cardellichio, and H. C. Coon. 1986. Latent infection of KB cells with adeno-associated virus type 2. J. Virol. 60:515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linden, R., M. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, F., and G. Churchward. 1995. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J. Bacteriol. 177:1938-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusby, E., K. H. Fife, and K. I. Berns. 1980. Nucleotide sequence of the inverted terminal repeat in adeno-associated virus DNA. J. Virol. 34:402-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarty, D. M., D. J. Pereira, I. Zolotukhin, X. Zhou, J. H. Ryan, and N. Muzyczka. 1994. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J. Virol. 68:4988-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarty, D. M., J. H. Ryan, S. Zolotukhin, X. Zhou, and N. Muzyczka. 1994. Interaction of the adeno-associated virus Rep protein with a sequence in the A palindrome of the viral terminal repeat. J. Virol. 68:4998-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaughlin, S. K., P. Collis, P. L. Hermonat, and N. Muzyczka. 1988. Adeno-associated virus general transduction vectors: analysis of proviral structures. J. Virol. 62:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meneses, P., K. I. Berns, and E. Winocour. 2000. DNA sequence motifs which direct adeno-associated virus site-specific integration in a model system. J. Virol. 74:6213-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 36.Ni, T. H., X. Zhou, D. M. McCarty, I. Zolotukhin, and N. Muzyczka. 1994. In vitro replication of adeno-associated virus DNA. J. Virol. 68:1128-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens, R. A., M. D. Weitzman, S. R. Kyostio, and B. J. Carter. 1993. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus. Rep protein. 67:997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizzuto, G., G. Gorgoni, M. Cappellitti, D. Lazzaro, I. Gloaguen, V. Poli, A. Sgura, D. Cimini, G. Ciliberto, R. Cortese, E. Fattori, and N. La Monica. 1999. Development of animal models for adeno-associated virus site-specific integration. J. Virol. 73:2517-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan, J. H., S. Zolotukhin, and N. Muzyczka. 1996. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 70:1542-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samulski, R. J. 1993. Adeno-associated virus: integration at a specific chromosomal locus. Curr. Opin. Genet. Dev. 3:74-80. [DOI] [PubMed] [Google Scholar]
- 42.Samulski, R. J., K. I. Berns, M. Tan, and N. Muzyczka. 1982. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA 79:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samulski, R. J., M. Sally, and N. Muzyczka. 1999. Adeno-associated viral vectors, p. 131-172. In T. Freeman (ed.), The development of human gene therapy. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Samulski, R. J., A. Srivastava, K. I. Berns, and N. Muzyczka. 1983. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 33:135-143. [DOI] [PubMed] [Google Scholar]
- 45.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer, R. T. 1991. Protein-DNA interactions. Methods Enzymol. 208:104-115. [PubMed] [Google Scholar]
- 47.Smith, R. H., A. J. Spano, and R. M. Kotin. 1997. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J. Virol. 71:4461-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder, R. O., D. S. Im, T. Ni, X. Xiao, R. J. Samulski, and N. Muzyczka. 1993. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J. Virol. 67:6096-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequences and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surosky, R. T., M. Urabe, S. G. Godwin, S. A. McQuiston, G. J. Kurtzman, K. Ozawa, and G. Natsoulis. 1997. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Travers, A. A. 1990. Why bend DNA? Cell 60:177-180. [DOI] [PubMed] [Google Scholar]
- 52.Trempe, J. P., E. Mendelson, and B. J. Carter. 1987. Characterization of adeno-associated virus rep proteins in human cells by antibodies raised against Rep expressed in E. coli. Virology 161:18-28. [DOI] [PubMed] [Google Scholar]
- 53.Urabe, M., Y. Hasumi, A. Kume, R. T. Surosky, G. J. Durtzman, K. Tobita, and K. Ozawa. 1999. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated virus type 2 Rep78 protein. J. Virol. 73:2682-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Von Hippel, P., and O. Berg. 1989. Facilitated target location in biological systems. J. Biol. Chem. 264:675-678. [PubMed] [Google Scholar]
- 55.Wonderling, R., and R. Owens. 1997. Binding sites for adeno-associated virus Rep proteins within the human genome. J. Virol. 71:2528-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, W. 1996. Ph.D. thesis. University of North Carolina, Chapel Hill.
- 57.Young, S. M. 2000. Ph.D. thesis. University of North Carolina at Chapel Hill, Chapel Hill.
- 58.Young, S. M., Jr., D. M. McCarty, N. Degtyareva, and R. J. Samulski. 2000. Roles of adeno-associated virus rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74:3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young, S. M., Jr., and R. J. Samulski. 2001. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. USA 98:13525-13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, X., I. Zolotukhin, D. S. Im, and N. Muzyczka. 1999. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J. Virol. 73:1580-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]