Abstract
Despite worldwide eradication of naturally occurring variola virus, smallpox remains a potential threat to both civilian and military populations. New, safe smallpox vaccines are being developed, and there is an urgent need for methods to evaluate vaccine efficacy after immunization. Here we report the identification of an immunodominant HLA-A*0201-restricted epitope that is recognized by cytotoxic CD8+ T cells and conserved among Orthopoxvirus species including variola virus. This finding has permitted analysis and monitoring of epitope-specific T cell responses after immunization and demonstration of the identified T cell specificity in an A*0201-positive human donor. Vaccination of transgenic mice allowed us to compare the immunogenicity of several vaccinia viruses including highly attenuated, replication-deficient modified vaccinia virus Ankara (MVA). MVA vaccines elicited levels of CD8+ T cell responses that were comparable to those induced by the replication-competent vaccinia virus strains. Finally, we demonstrate that MVA vaccination is fully protective against a lethal respiratory challenge with virulent vaccinia virus strain Western Reserve. Our data provide a basis to rationally estimate immunogenicity of safe, second-generation poxvirus vaccines and suggest that MVA may be a suitable candidate.
The anthrax attacks in the United States changed a theoretical scenario of bioterrorism into a realistic threat. Variola virus, the cause of human smallpox, emerged as one of the infectious agents being listed with highest priority for research on prevention and therapy (1). Its malevolent use was predicted to result in a rapid epidemic in human populations because little immunity exists since vaccination with the Orthopoxvirus relative vaccinia virus has ceased (2). Efficacy of replication-competent vaccinia virus as live viral vaccine was demonstrated impressively by the worldwide eradication of naturally occurring variola virus (3). Because of adverse events associated with vaccinia virus vaccination, safer second-generation vaccines are an urgent need (4). However, this need is accompanied by the challenge to determine vaccine efficacy in the absence of naturally occurring smallpox disease and the question of vaccine safety in individuals with immunodeficiencies caused by HIV infection or iatrogenic immunosuppression (5). Because smallpox vaccination ended more than 20 years ago, we have only limited knowledge about immunologic correlates of protective vaccination. Although there are means to monitor induced antibodies after vaccination, cell-mediated immunity has scarcely been assessed (for review see ref. 6). Yet, experience with individual human vaccinees and work in animal models suggest an important role of cell-mediated immune responses in protection (7, 8). Long-lasting memory of cellular immunity is demonstrated by virus-specific cytotoxic T cells (CTLs), being detectable even decades after primary vaccination (9). However, substantial information about relevant Orthopoxvirus antigens and epitope specificities of virus-specific CTL responses is still lacking.
In the present study, we identified an immunodominant HLA-A*0201-restricted vaccinia virus-specific peptide epitope, VP35#1, which is encoded by the vaccinia virus gene H3L and can be recognized by murine and human CD8+ T cells. We demonstrated the usefulness of this epitope to directly compare specific CD8+ T cell responses induced by different vaccinia virus strains in HLA-A*0201 transgenic mice. Although vaccination of HLA-A*0201 transgenic mice with VP35#1 peptide alone resulted in high levels of VP35#1-specific CD8+ T cells, immunized animals were not protected against a lethal poxvirus challenge. In contrast, we demonstrated the protective capacity of vaccination with highly attenuated, replication-deficient vaccinia virus [modified vaccinia virus Ankara (MVA)] and compared this protection against lethal respiratory infection to the one mediated by a conventional replication-competent vaccine strain. Here we are able to show that immunizations that were sufficient for protection also allowed ex vivo detection of VP35#1-specific CD8+ T cells. Taken together, our data demonstrate that identification of an HLA-A*0201-restricted vaccinia virus-derived epitope permits direct peptide-specific immunologic comparison of different vaccine strains and evaluation of Orthopoxvirus-specific immune memory.
Materials and Methods
Viruses.
Vaccinia virus strains New York City Board of Health (Wyeth), Western Reserve (WR) (both provided by Bernard Moss, National Institutes of Health, Bethesda), chorioallantois vaccinia virus Ankara (CVA) at second passage on chicken embryo fibroblasts, or MVA (cloned isolate F6) at 582nd passage on chicken embryo fibroblasts were used for this study. All viruses were propagated and titered following standard methodology (10).
Screening for HLA-A*0201-Restricted Peptides.
HLA-A*0201-restricted peptide epitopes derived from vaccinia virus proteins were selected by epitope prediction using the SYFPEITHI database (11). Converted internal tyrosinase 369–377 peptide YMDGTMSQV (hTyr369), influenza virus A/PR/8/34 matrix protein M1 58–66 peptide GILGFVFTL, or HIV-1 reverse transcriptase 476–484 peptide ILKEPVHGV, all of which were known to bind to HLA-A*0201 (12), were used as negative controls. Purity of peptides was ascertained by mass spectrometry and HPLC to be >90%.
Animal Models.
Transgenic HHD+/+ β2m−/− Db−/− mice (HHD, 6–8 weeks old) (13) were used for immunizations inoculating 0.5- or 0.1-ml volumes of virus vaccine by the i.p. or i.m. route and monitoring for HLA-A*0201-restricted T cell responses 10 days after vaccination. For protection assays, groups of BALB/c or HHD mice were vaccinated once i.m. with 0.1 ml of virus vaccine. Alternatively, HHD mice were immunized s.c. with 0.3 mg of VP35#1 peptide in 10 nM CpG oligodeoxynucleotides (ODN) 1668 (14). Three weeks after immunization with virus or 2 weeks after peptide immunization, animals were infected intranasally with vaccinia virus WR diluted in 30 μl of PBS and monitored for 3 weeks with daily measurement of individual body weights and scoring of signs of illness as described (15). Animals suffering from severe systemic infection and having lost >30% of body weight were killed. The mean change in body weight was calculated as percentage of the mean weight for each group on the day of challenge.
Intracellular Cytokine Staining.
Splenocytes from vaccinia virus- or peptide-immunized or naive HHD mice were prepared and incubated for 5 h with A*0201-binding peptides at 10−6 M. After 2 h, brefeldin A was added at a final concentration of 1 μg/ml (GolgiPlug, PharMingen/Becton Dickinson, Heidelberg). Cells were live/dead-stained in PBS/1% BSA/1 μg/ml ethidium monoazide bromide and blocked with 5 μg/ml anti-CD16/CD32 (Fc Block, PharMingen/Becton Dickinson) for 20 min at 4°C. Cell-surface staining was performed with phycoerythrin-anti-CD8 (53-6.7), peridinin chlorophyll protein-anti-CD4 (RM4-5), and allophycocyanin-anti-CD62L (Mel-14) for 30 min at 4°C. Intracellular cytokine staining was performed for 30 min at 4°C by applying FITC-anti-IFN-γ (XMG1.2), FITC-anti-tumor necrosis factor α (MP6-XT22), or FITC-labeled IgG1 isotype control (R3-34) using Cytofix/Cytoperm (PharMingen/Becton Dickinson). Splenocytes were analyzed by four-color flow cytometry (FACSCalibur) using CELLQUEST software (Becton Dickinson).
Chromium-Release Assays.
Ten days after vaccination, spleen cells were cocultured with irradiated (3,000 rad) and lipopolysaccharide-activated syngeneic spleen cells pulsed with VP35#1 or tyrosinase 369–377 peptide at 5 μg/ml and human β2-microglobulin (Sigma) at 10 μg/ml. On day 6 cytolytic activity was determined at various effector/target (E/T) ratios in a 4-h standard 51Cr-release assay as described (12). The A2.1-restricted murine CTL lines specific for human tyrosinase peptide 369–377 (CTL A2Kb hTyr) or influenza M1 peptide 58–66 (CTL CD8xA2Kb FluM1) served as negative controls and were established and maintained as described (12). T2 target cells were pulsed with 10−6 M VP35#1 peptide. Tyrosinase 369–377 peptide or influenza M1 58–66 peptide served as controls. Human cell lines A375/H3L or A375/LV were generated by transfection of tyrosinase-negative HLA-A*0201-positive melanoma cell line A375 (ATCC CRL 1619) with plasmid pZeoSV2+ (Invitrogen Life Technologies, Karlsruhe, Germany) containing full-length vaccinia virus H3L (ORF 093L) prepared by PCR from MVA genomic DNA (A375/H3L) or with the empty control vector (A375/LV).
Generation of CD8+ VP35#1-Specific Human T Cells.
Human HLA-A*0201-restricted VP35#1-specific T cells were generated by repetitive stimulation with VP35#1 peptide-pulsed mature monocyte-derived dendritic cells as described (16). The stimulator-to-responder cell ratio was 1:20 for priming and 1:40 for restimulation. After two stimulations, VP35#1-specific T cells were stained by using A2/VP35#1 multimers as described (17). For sorting of VP35#1-specific T cells, cells were resuspended in buffer without propidium iodide and filtrated through nylon, applied to a MoFlo cell sorter (Cytomation, Fort Collins, CO), and sorted with 25,000 events per s at a maximum of 1 psi (1 psi = 6.89 kPa). Sorted T cells were expanded in the presence of anti-CD3 (Okt-3, Janssen–Cilag), rIL-2 (Chiron Behring), and allogeneic irradiated lymphoblastoid cell lines and peripheral blood mononuclear cells (PBMCs) as described (16, 18). After 2 weeks, expanded T cells were stained with A2/VP35#1 multimers. The lytic activity of expanded T cells was determined at various E/T ratios in 4- to 5-h standard 51Cr-release assays as described (12). The E/T ratio was adapted to the actual amount of VP35#1-specific T cells as determined by multimer staining.
Results
Identification of an Epitope for HLA-A*0201-Restricted Orthopoxvirus-Specific CD8+ T Cell Responses.
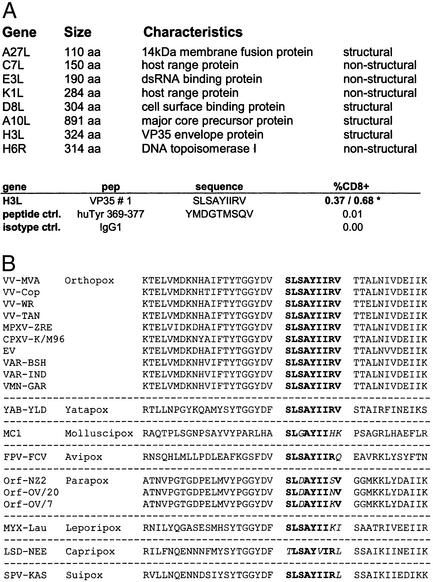
The genome of vaccinia virus encodes for >200 proteins that may serve as possible antigens (19). In our attempt to identify immune correlates of successful poxvirus-specific immunization, we concentrated on vaccinia virus gene products that (i) share high amino acid homologies to variola virus antigens (20), (ii) were known to be immunodominant with regard to induction of virus-specific antibodies (21, 22), or (iii) are nonstructural virus proteins being produced during early viral gene expression and possibly more suitable for MHC class I-associated immune presentation (23). Because of dominant representation of HLA-A*02 in human populations, we sought to identify poxvirus polypeptides allowing for HLA-A*0201-specific restriction of peptide epitopes. Using epitope prediction offered by the SYFPEITHI database of MHC ligands and peptides (11), we preselected eight vaccinia virus polypeptide sequences (Fig. 1A) and defined peptides with highest probability for association with HLA-A*0201. These peptides were tested for their ability to specifically induce cytokine production in stimulated splenocytes from vaccinia virus-infected HLA-A*0201-transgenic mice (HHD) (13). Monitoring T cells by fluorescence-activated cell sorter analysis after IFN-γ-specific intracellular staining, we could identify multiple peptides that induced IFN-γ release above background levels (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Peptide VP35#1 had particular stimulating capacity for up to 0.7% of all CD8+ T cells (Fig. 1A), whereas for weaker responses detected with other peptides additional work is required to demonstrate significance. VP35#1 is part of a 35-kDa vaccinia virus envelope protein encoded by ORF H3L and described as an immunodominant antigen (22, 24). Comparison of VP35#1 sequence with known H3L gene homologues derived from various poxviruses revealed complete conservation of the peptide among Orthopoxvirus species including variola major and variola minor. Interestingly, VP35#1 is also conserved in more heterologous protein sequence from Yaba-like disease virus, a member of the Yatapoxvirus genus, which can cause infections in humans (25), whereas amino acid substitutions occur in sequences of poxviruses from other genera (Fig. 1B).
Figure 1.
Identification of HLA-A*0201-restricted peptide epitope VP35#1. (A) Vaccinia virus polypeptides selected for analysis of peptides with highest probability for HLA-A*0201 binding are shown. Incubation of splenocytes from vaccinated HHD mice with peptide VP35#1 results in specific secretion of IFN-γ. T cell response is depicted as percentage of cytokine-secreting CD8+ cells (%CD8+). An irrelevant HLA-A*0201-binding peptide (huTyr 369) was used as a negative control. The numbers marked by an asterisk refer to results from two independent experiments. (B) Alignment of VP35#1-peptide sequences derived from poxvirus gene products homologous to vaccinia virus H3L protein: vaccinia virus strains MVA (VV-MVA), Copenhagen (VV-Cop), WR (VV-WR), and Tian Tan (VV-Tan); monkeypox virus Zaire-96-I-16 (MPXV-ZRE); camelpox virus M-96 (CPXV-K/M96); ectromelia virus Naval (EV); variola major viruses Bangladesh-1975 (VAR-BSH) and India-1967 (VAR-IND); variola minor virus Garcia-1966 (VMN-GAR); yaba-like disease virus (YAB-YLD); molluscum contagiosum virus 1 (MC1); fowlpox virus (FPV-FCV); orf viruses NZ2 (Orf-NZ2), OV/20 (Orf-OV/20), and OV/7 (Orf-OV/7); myxoma virus Lausanne (MYX-LAU); lumpy skin disease virus Neethling 2490 (LSD-NEE); and swinepox virus 17077-99 (SPV-KAS).
Quantitation and Characterization of Epitope-Specific CD8+ T Cells Induced by Immunization with Different Vaccinia Viruses.
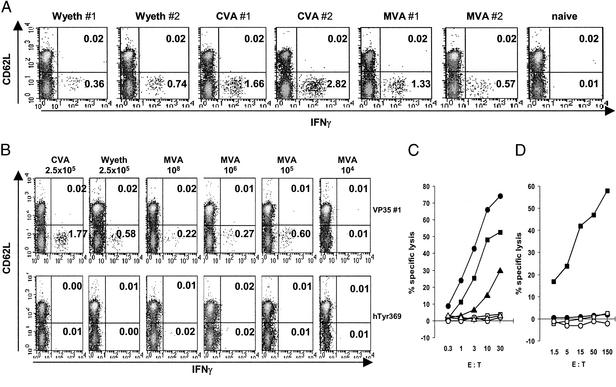
To comparatively characterize VP35#1-specific T cell responses induced after immunization, we vaccinated HHD mice with the different vaccinia virus strains Wyeth, CVA, or MVA, all of them having been used previously as smallpox vaccines. Wyeth and CVA are replication-competent viruses that were grown on live calves or primary bovine tissue culture for production of smallpox vaccine and used for multipuncture intradermal vaccination in humans (5, 26). MVA was developed originally from CVA by serial passage in chicken embryo fibroblasts and tested in Germany as a safe, tissue culture-prepared vaccine (27). MVA is severely host range-restricted and serves as vector virus for recombinant gene expression and as candidate vaccine for delivery of heterologous antigens (28, 29).
Relying on previous experience using MVA vaccines in HLA-A*0201-transgenic mice, we immunized animals i.p. with 108 infectious units (IU) of MVA (12). In contrast, vaccines based on replication-competent viruses CVA and Wyeth were given i.p. at 2.5 × 105 plaque-forming units (PFU), corresponding to an infectious dose of CVA that when previously used in mice resulted occasionally in generalizing infections but not in mortality (30). A single inoculation of all vaccinia viruses was able to induce VP35#1-specific activated CD8+ T cells as revealed by fluorescence-activated cell sorter analysis of freshly prepared splenocytes after intracellular staining for IFN-γ (Fig. 2A) and tumor necrosis factor α (data not shown). The highest levels of VP35#1-reactive T cells secreting IFN-γ as well as tumor necrosis factor α were detected after vaccination with CVA, whereas immunizations with Wyeth or MVA elicited comparable responses e.g., ranging from 0.36% to 1.33% IFN-γ-releasing CD8+ T cells. We confirmed the suitability of our initially chosen vaccine dosage after repetitive in vivo titration of Wyeth and MVA vaccines for their ability to induce VP35#1-specific activated CD8+ T cells after i.p. immunization. For replication-competent Wyeth we found optimal levels of IFN-γ-releasing CD8+ T cells (0.71–1.46%) after vaccination with doses in the range of 105 to 106 PFU, whereas MVA immunizations inoculating 107 to 108 IU yielded the highest responses (0.69–1.27% CD8+ T cells) (data not shown).
Figure 2.
HLA-A*0201-restricted epitope-specific cellular immunogenicity of various smallpox vaccines. (A and B) Peptide-specific intracellular cytokine release of splenocytes from vaccinated HHD mice. Mice were immunized i.p. with 2.5 × 105 PFU of CVA or Wyeth or 108 IU of MVA (A) or i.m. with 2.5 × 105 PFU of CVA or Wyeth or 108, 106, 105, and 104 IU of MVA (B). #1 and #2 refer to individual mice analyzed. Splenocyte preparations were analyzed for the presence of VP35#1 peptide-specific, activated (CD62Llow) CD8+ T cells. The magnitude of the induced T cell response is depicted as percentages of cytokine-secreting CD8+ T cells within the live CD8+ cell population. (C) Specific lysis of peptide-pulsed human target cells by VP35#1-specific CTLs from vaccinated HHD mice. Effector CTLs were generated by single i.p. immunization of HHD mice with MVA (■ and □), CVA (● and ○), or Wyeth (▴ and ▵). CTLs were assayed for cytotoxicity at the indicated E/T ratio against T2 targets pulsed with either VP35#1 (filled symbols) or influenza M1 peptide (open symbols). (D) Processing and presentation of peptide epitope VP35#1 after heterologous expression of the vaccinia virus H3L gene. Human A375/H3L (■ and □) or control A375 (● and ○) cells were tested against A*0201-restricted murine CTLs specific for VP35#1 (filled symbols) or human tyrosinase 369–377 epitopes (open symbols) as effector cells at the indicated E/T ratio.
Interestingly, we were able to repetitively detect comparable VP35#1-specific T cell responses >6 months after a single i.p. inoculation with either 108 IU of MVA or 2.5 × 105 PFU of CVA. We found 0.21–0.78% IFN-γ-releasing splenic CD8+ memory T cells in MVA-immunized animals and 0.30–0.61% VP35#1-reactive T cells secreting IFN-γ in CVA-vaccinated mice. In all experiments, background levels obtained with irrelevant stimulator peptides were <0.03% (Fig. 6, which is published as supporting information on the PNAS web site). This result suggests potential usefulness of the VP35#1 epitope for immune monitoring of vaccinia virus-specific CD8+ T cell memory responses.
To assess vaccination in a more realistic scenario, we analyzed immune responses in HHD mice inoculated once by the i.m. route. Additionally, we wished to monitor for possible effects of MVA replication deficiency and included animals that had received decreasing doses of MVA vaccine. Despite the less “systemic” nature of this vaccination route we found easily detectable levels of VP35#1-specific T cell responses (Fig. 2B). Vaccination with MVA by using 105 to 108 IU elicited comparable immune responses, whereas we failed to detect VP35#1-specific T cells after immunization with less infectivity (Fig. 2B, 104 IU). To test the peptide-reactive T cells for their cytotoxic capacities, we assayed splenocytes from HHD mice vaccinated with MVA, CVA, or Wyeth against VP35#1-specific human target cells (Fig. 2C). Efficient specific lysis was found with effector cells from all vaccinees. There was no lysis of target cells pulsed with the A*0201-binding but irrelevant influenza M1 58–66 peptide. Again, the highest levels of cytotoxicity were obtained after vaccination with CVA, closely followed by CTL responses induced with MVA and Wyeth. In addition, various concentrations (10−6 to 10−9 M) of peptide VP35#1 sensitized target cells to VP35#1-specific CTLs, whereas these targets were not lysed by A*0201-restricted murine CTLs specific for the influenza M1 58–66 peptide (data not shown).
Finally, we wished to confirm that VP35#1 is truly derived and properly processed from vaccinia virus H3L protein. We cloned the H3L gene into plasmid pZeoSV2+ and used this heterologous eukaryotic expression vector to generate stably H3L-transfected human A*0201-positive A375 cells. When tested as target cells in chromium-release assays, A375/H3L cells were specifically lysed by VP35#1-specific A*0201-restricted murine CTLs (Fig. 2D). Only background cytotoxicity was detected in assays using either untransfected A375 cells or A*0201-restricted murine CTLs specific for an irrelevant antigen.
Isolation, Expansion, and Characterization of Human VP35#1-Specific CD8+ T Cells.
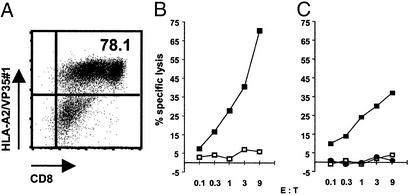
Having exclusively monitored for A*0201-restricted but murine CTLs, we wished to demonstrate that A*0201-positive humans possess the identified T cell specificity. We analyzed PBMCs from a A*0201-positive healthy donor who had been vaccinated against smallpox during childhood. Repetitive stimulation of PBMCs with VP35#1-pulsed dendritic cells followed by multimer-guided sorting allowed us to expand VP35#1-specific T cells, resulting in a T cell population containing 78.1% of A*0201/VP35#1-binding CD8+ T cells (Fig. 3A). To test the multimer-binding T cells for their functional activity, we assessed cytotoxic capacities using VP35#1 peptide-pulsed human T2 cells (Fig. 3B) or stably H3L-transfected human A375/H3L cells (Fig. 3C) as target cells in chromium-release assays. We found efficient specific lysis with the expanded effector T cells against VP35#1 peptide-pulsed target cells (Fig. 3B) or target cells endogenously processing and presenting the VP35#1 epitope (Fig. 3C). Only background lysis was detectable in assays using untransfected or control vector-transfected human A375 cells or T2 cells pulsed with A*0201-binding but irrelevant peptide.
Figure 3.
Isolation and expansion of human CD8+ VP35#1-specific T cells from peripheral blood and lysis of VP35#1-presenting human target cells. Human HLA-A*0201+ PBMCs were stimulated with autologous dendritic cells pulsed with VP35#1. T cells were stained with HLA-A2/VP35#1 multimers after two in vitro stimulations and multimer-guided sorting (A). Numbers in the upper right quadrant represent the percentage of multimer-binding CD8+ T cells gated on propidium iodide-negative PBMCs. T2 targets pulsed with either the VP35#1 peptide (■) or HIV-1 RT 476–484 peptide (□) at 1 μM (B) or human A375/H3L (■), A375/LV (□), or A375 cells (●) (C) were tested against the human A*0201-restricted, VP35#1-specific T cells as effector cells at the indicated E/T ratio.
Induction of VP35#1 Epitope-Specific T Cells by Peptide Immunization and Protection Against Lethal Virus Challenge by MVA Vaccination.
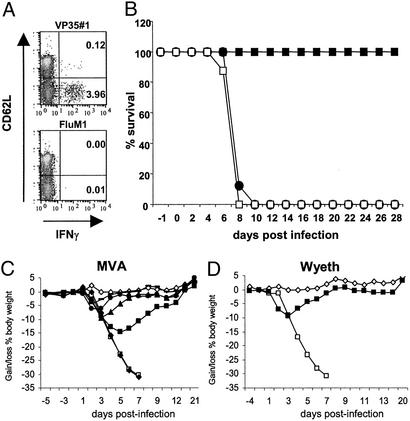
To investigate a potential protective capacity of single epitope-specific T cell responses, we established a heterologous immunization protocol allowing for induction of potent VP35#1-specific T cells. We vaccinated HHD mice s.c. with a single dose of VP35#1 peptide plus CpG-ODN as adjuvant. This immunization elicited notable peptide-specific CD8+ T cell responses as demonstrated by detection of almost 4% of all activated CD8+ T cells producing IFN-γ and tumor necrosis factor α (data not shown) after stimulation with VP35#1 (Fig. 4A). To assess how our data regarding vaccine immunogenicities in HLA-A*0201-transgenic mice would compare with vaccine protection, we used the respiratory infection with highly virulent vaccinia virus WR as a model (15, 31). We immunized HHD mice s.c. with the VP35#1/CpG-ODN vaccine, and groups of animals being vaccinated i.m. with 107 IU of MVA or PBS served as controls. We chose this dose of MVA vaccine because it had consistently induced VP35#1-specific CD8+ T cells in our previous experiments. After vaccination, we challenged animals by intranasal inoculation of 2.5 × 106 PFU of WR corresponding to at least 50 LD50 of this vaccinia virus strain in BALB/c mice (31). The infection resulted in mock-vaccinated HHD mice in the onset of respiratory illness, substantial loss of body weight, and death within 8 days postchallenge, whereas all animals inoculated with 107 IU of MVA were protected (Fig. 4B). In contrast, mice immunized with VP35#1/CpG-ODN complex did not survive day 9 postinfection, an outcome that, also with regard to signs of illness and weight loss, was practically indistinguishable from the one observed in the mock-vaccinated group. This result suggested that the chosen VP35#1-peptide vaccination alone, despite its capacity to potently induce VP35#1 epitope-specific CD8+ T cells, was not sufficient to achieve protection against a vigorous challenge infection with WR.
Figure 4.
Analysis of VP35#1-peptide vaccine-induced immunity and protection in mouse challenge models. (A) Induction of HLA-A*0201-restricted VP35#1 peptide-specific CD8+ T cell responses after peptide immunization. HHD mice were immunized once with VP35#1 peptide-CpG-ODN. Splenocytes were stimulated with VP35#1 or influenza M1 peptide and stained for intracellular cytokines. Cells were analyzed by flow cytometry for the presence of VP35#1 peptide-specific, activated (CD62Llow) CD8+ T cells. The magnitude of the induced T cell response is depicted as percentages of cytokine-secreting CD8+ T cells within the live CD8+ cell population. (B) Protective efficacy of VP35#1/CpG-ODN vaccination in HLA-A*0201-transgenic mice. HHD mice (n = 8) were immunized i.m. with 107 (■) IU of MVA or s.c. with 0.3 mg of VP35#1 peptide plus 10 nM CpG-ODN 1668 (●). Mock-vaccinated (□) mice served as control. Two (peptide) or 3 (MVA) weeks after vaccination animals were challenged with 2.5 × 106 PFU of WR and monitored daily for a 4-week period. The survival (%) of the animals was plotted. (C and D) Protective efficacy of vaccination in BALB/c mice. Groups of mice (n = 10) were immunized i.m. with 104 (♦), 105 (■), 106 (▴), 107 (●), or 108 IU of MVA (−) (C) or 105 PFU of Wyeth (■) (D). Three weeks after vaccination animals were challenged intranasally with 1 × 106 PFU of WR. Individual animal weights were monitored daily and are expressed as means for each group. Mock-vaccinated (□) and mock-challenged (⋄) mice served as control.
To investigate the protective capacity of MVA vaccination in more detail, we chose to test different vaccines also in the well established intranasal challenge model using BALB/c mice (15). Matching our immunization experiments to determine CTL responses in HHD mice, we vaccinated BALB/c mice by single i.m. injections of 102 to 108 IU of MVA. Three weeks after immunization, we submitted animals to a respiratory infection with 106 PFU of WR corresponding to at least 20 LD50 (31). All animals having received 105 IU or higher doses of MVA vaccine survived the challenge, whereas no protection from severe disease resulting in death of all animals was seen after inoculation with 104 or less IU of MVA. The monitoring of body weight (Fig. 4C) and typical signs of disease (Fig. 7A, which is published as supporting information on the PNAS web site) suggested that the levels of protection correlated well with increased dosage of the vaccine. Mice in the group vaccinated with 105 IU of MVA demonstrated a >10% reduction of body weight and mild signs of illness, whereas no signs of illness and only a minor transient reduction in body weight was seen in mice immunized with 106 IU of MVA. This latter level of protection closely resembled the efficacy of vaccinations with 105 PFU of Wyeth, which we used as control vaccine representing a replication-competent vaccinia virus with well established protective capacity (Figs. 4D and Fig. 7B).
Discussion
Here we identified a vaccinia virus-derived peptide, VP35#1, as an epitope for HLA-A*0201-restricted CD8+ T cell responses. This epitope may allow for more detailed experimental monitoring of cellular immune responses induced by poxvirus-specific immunization including vaccination of HLA-A*0201-positive humans. Importantly, the usefulness of VP35#1-specific immune monitoring is supported strongly by our isolation of functionally active VP35#1-specific T cells from PBMCs of an HLA-A*0201-positive human donor. The highly immunogenic and well conserved peptide VP35#1 is processed from vaccinia virus protein H3L (Fig. 2D) that has been described as a target antigen of immunodominant antibody responses (22). Our use of the VP35#1 epitope for direct comparison of T cell responses induced by live vaccinia viruses in a single antigen-specific manner suggests that different vaccinia virus strains may vary to some degree in their ability to elicit epitope-specific T cells. We detected peak VP35#1-specific T cell responses after immunization with CVA, which was described to prompt a higher incidence of multiple lesions developing at the site of inoculation when tested as smallpox vaccine in Germany (26). After vaccination with less virulent but replication-competent strain Wyeth or highly attenuated replication-deficient MVA, we found comparable amounts of VP35#1-specific T cells. Although we had inoculated a notably higher dose of MVA compared with vaccination with replicating vaccinia virus Wyeth, this result was very reminiscent of the comparable immunogenicities determined for Wyeth or MVA vector vaccines after vaccination of rhesus macaques against simian immunodeficiency virus (32). Similarly, when we analyzed MVA- or CVA-vaccinated HHD mice >6 months after immunization, we could still detect almost equal sustained levels of VP35#1-specific CD8+ T cells. Moreover, after monitoring for total vaccinia antigen-specific T cell responses after vaccination with replication-competent vaccinia virus CVA or MVA, we detected very similar levels of virus-specific CD8+ and CD4+ T cells (Fig. 8, which is published as supporting information on the PNAS web site).
Data from experimental infections of mice with ectromelia virus strongly suggest that both CD4+ and CD8+ T lymphocytes are important for in vivo control of Orthopoxvirus infections (8). In an attempt to assess the role of single epitope-specific CD8+ T cells in vaccine protection, we were able to effectively induce VP35#1-specific CD8+ T cells in HHD mice using a VP35#1 peptide-based vaccine. Similar vaccinations were known to trigger antiviral CTL responses successfully in mouse models for infection with lymphocytic choriomeningitis virus or herpes simplex virus (14, 33). However, VP35#1 peptide-immunized HHD mice were not protected against a lethal challenge with vaccinia virus WR. This outcome was in contrast to MVA-immunized HHD mice being protected after one inoculation with 107 IU of MVA, an immunization which in our hands consistently elicited lower frequencies of VP35#1-specific CD8+ T cells as compared with vaccination with peptide-CpG-ODN. Our data could indicate that a single epitope-specific T cell response may not be sufficient to control a lethal dose of poxvirus infection even when specific T cells are present at high frequency. Nevertheless, we believe that these results do not automatically preclude a possible contribution of VP35#1-specific CD8+ T cells in vaccine protection. Because we used as read-out a high-dose, acutely lethal vaccinia virus infection, it will be interesting to perform lower-dose challenge experiments, e.g., to assess viral loads in organs, an approach that has been used to demonstrate antiviral activity of epitope-specific CD8+ T cells in the mouse lymphocytic choriomeningitis virus model (14, 33). Importantly, successful vaccination of HHD mice with MVA demonstrated that protection against a harsh challenge infection is possible. Further, this result allows the assumption that immune responses directed against multiple specificities, likely including both humoral and cell-mediated responses, are required for solid immunity against poxvirus infections. Finally, we could confirm protective capacity of MVA vaccination in BALB/c mice. Respiratory infection of these nontransgenic animals with vaccinia virus can be considered as a particularly well established challenge model in which the impact of infection is reflected most accurately by changes in animal body weights followed by the onset of signs of illness (15, 31). Interestingly, our in vivo titrations of MVA demonstrate that inoculation of vaccine doses that induced detectable VP35#1-specific CD8+ T cells also protected against lethal respiratory infection with virulent vaccinia virus WR. These data, together with the fact that we found quite prominent VP35#1 epitope-specific T cell responses after immunization with 108, 106, or 105 IU of MVA but failed to detect any specific T cells giving lower doses of MVA vaccine, may point toward the requirement for threshold levels of antigen to induce specific CD8+ T cells. Yet in contrast to parameters measured to demonstrate in vivo protection (e.g., body weight and signs of illness), this T cell response seemed not to increase proportionally with vaccine dosage. Most likely, additional contribution of other vaccine-induced humoral and cellular immune responses to protection may account for this observation. Importantly, we show that highly attenuated MVA, despite its likely incapability to replicate in vivo, can provide levels of vaccine protection equal to those induced by the conventional vaccine virus Wyeth yet with the requirement for at least 10-fold higher dosage. The latter may be interpreted as compensation for the in vivo replication of Wyeth naturally providing higher levels of antigen after immunization.
Taken together, the data described here define VP35#1 as an Orthopoxvirus-derived epitope recognized by HLA-A*0201-restricted CD8+ T cells, which allows vaccinia virus-specific immune monitoring. Its future use will likely include study of T cell kinetics after vaccination, comparison to epitope-specific responses elicited against recombinant antigens, and evaluation of Orthopoxvirus-specific immune responses in humans. Further, our results presented here suggest that MVA can serve as an excellent candidate of second-generation vaccine against smallpox that is safer yet likely as effective than the current vaccine.
Supplementary Material
Acknowledgments
We thank Christopher C. Broder for helpful advice and Marianne Löwel and Ronny Ljapoci for excellent technical assistance. The work was supported by Deutsche Forschungsgemeinschaft Grants SFB455-A10 and SFB456-B7 (to G.S.). G.S., V.E., D.H.B., and H.B. are members of the GSF Clinical Cooperation Group “Vaccinology.”
Abbreviations
- CTL
cytotoxic T cell
- MVA
modified vaccinia virus Ankara
- WR
Western Reserve
- CVA
chorioallantois vaccinia virus Ankara
- ODN
oligodeoxynucleotides
- E/T
effector/target
- PBMC
peripheral blood mononuclear cell
- IU
infectious unit(s)
- PFU
plaque-forming unit(s)
References
- 1.Lane H C, Montagne J L, Fauci A S. Nat Med. 2001;7:1271–1273. doi: 10.1038/nm1201-1271. [DOI] [PubMed] [Google Scholar]
- 2.Gani R, Leach S. Nature. 2001;414:748–751. doi: 10.1038/414748a. [DOI] [PubMed] [Google Scholar]
- 3.Fenner F, Henderson D A, Arita I, Jezek Z, Ladnyi I. Smallpox and Its Eradication. Geneva: W.H.O.; 1988. [Google Scholar]
- 4.Birmingham K, Kenyon G. Nat Med. 2001;7:1167. doi: 10.1038/nm1101-1167b. [DOI] [PubMed] [Google Scholar]
- 5.Rosenthal S R, Merchlinsky M, Kleppinger C, Goldenthal K L. Emerg Infect Dis. 2001;7:920–926. doi: 10.3201/eid0706.010602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson D A, Moss B. In: Vaccines. Plotkin S A, Orenstein W A, editors. Philadelphia: Saunders; 1999. pp. 74–97. [Google Scholar]
- 7.Freed E R, Duma R J, Escobar M R. Am J Med. 1972;52:411–420. doi: 10.1016/0002-9343(72)90031-9. [DOI] [PubMed] [Google Scholar]
- 8.Karupiah G, Buller R M L, Vanrooijen N, Duarte C J, Chen J H. J Virol. 1996;70:8301–8309. doi: 10.1128/jvi.70.12.8301-8309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demkowicz W E, Jr, Littaua R A, Wang J, Ennis F A. J Virol. 1996;70:2627–2631. doi: 10.1128/jvi.70.4.2627-2631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss B, Earl P L. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. New York: Wiley; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 11.Rammensee H, Bachmann J, Emmerich N P, Bachor O A, Stevanovic S. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 12.Drexler I, Antunes E, Schmitz M, Wolfel T, Huber C, Erfle V, Rieber P, Theobald M, Sutter G. Cancer Res. 1999;59:4955–4963. [PubMed] [Google Scholar]
- 13.Pascolo S, Bervas N, Ure J M, Smith A G, Lemonnier F A, Perarnau B. J Exp Med. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vabulas R M, Pircher H, Lipford G B, Hacker H, Wagner H. J Immunol. 2000;164:2372–2378. doi: 10.4049/jimmunol.164.5.2372. [DOI] [PubMed] [Google Scholar]
- 15.Alcami A, Smith G L. Proc Natl Acad Sci USA. 1996;93:11029–11034. doi: 10.1073/pnas.93.20.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer zum Büschenfelde C, Metzger J, Hermann C, Nicklisch N, Peschel C, Bernhard H. J Immunol. 2001;167:1712–1219. doi: 10.4049/jimmunol.167.3.1712. [DOI] [PubMed] [Google Scholar]
- 17.Knabel M, Franz T J, Schiemann M, Wulf A, Villmow B, Schmidt B, Bernhard H, Wagner H, Busch D H. Nat Med. 2002;8:631–637. doi: 10.1038/nm0602-631. [DOI] [PubMed] [Google Scholar]
- 18.Riddell S R, Watanabe K S, Goodrich J M, Li C R, Agha M E, Greenberg P D. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 19.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 20.Massung R F, Liu L-I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez J F, Paez E, Esteban M. J Virol. 1987;61:395–404. doi: 10.1128/jvi.61.2.395-404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinoviev V V, Tchikaev N A, Chertov O Y, Malygin E G. Gene. 1994;147:209–214. doi: 10.1016/0378-1119(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 23.Bronte V, Carroll M W, Goletz T J, Wang M, Overwijk W W, Marincola F, Rosenberg S A, Moss B, Restifo N P. Proc Natl Acad Sci USA. 1997;94:3183–3188. doi: 10.1073/pnas.94.7.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Fonseca F G, Wolffe E J, Weisberg A, Moss B. J Virol. 2000;74:7508–7517. doi: 10.1128/jvi.74.16.7508-7517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H J, Essani K, Smith G L. Virology. 2001;281:170–192. doi: 10.1006/viro.2000.0761. [DOI] [PubMed] [Google Scholar]
- 26.Herrlich A, Mayr A. Arch Gesamte Virusforsch. 1957;7:284–296. [PubMed] [Google Scholar]
- 27.Mayr A, Hochstein-Mintzel V, Stickl H. Infection. 1975;3:6–14. [Google Scholar]
- 28.Sutter G, Moss B. Proc Natl Acad Sci USA. 1992;89:10847–10851. doi: 10.1073/pnas.89.22.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutter G, Wyatt L S, Foley P L, Bennink J R, Moss B. Vaccine. 1994;12:1032–1040. doi: 10.1016/0264-410x(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 30.Meyer H, Sutter G, Mayr A. J Gen Virol. 1991;72:1031–1038. doi: 10.1099/0022-1317-72-5-1031. [DOI] [PubMed] [Google Scholar]
- 31.Lee M, Roos J, McGuigan L, Smith K, Cormier N, Cohen L, Roberts B, Payne L. J Virol. 1992;66:2617–2630. doi: 10.1128/jvi.66.5.2617-2630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsch V, Fuerst T, Sutter G, Carroll M, Yang L, Goldstein S, Piatak M, Elkins W, Alvord G, Montefiori D, Moss B, Lifson J. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gierynska M, Kumaraguru U, Eo S K, Lee S, Krieg A, Rouse B T. J Virol. 2002;76:6568–6576. doi: 10.1128/JVI.76.13.6568-6576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.