Abstract
Human immunodeficiency viruses (HIVs) and the related bovine lentiviruses bovine immunodeficiency virus (BIV) and Jembrana disease virus (JDV) utilize the viral Tat protein to activate viral transcription. The arginine-rich RNA-binding domains of the Tat proteins bind to their cognate transactivation response element (TAR) RNA hairpins located at the 5′ ends of the viral mRNAs, resulting in enhanced processivity of RNA polymerase II. It has previously been shown that HIV type 1 (HIV-1) Tat requires the cellular cyclin T1 protein for high-affinity RNA binding whereas BIV Tat and JDV Tat bind with high affinity on their own and adopt distinct β-hairpin conformations when complexed to RNA. Here we have engineered the BIV and JDV Tat-TAR interactions into HIV-1 and show that the heterologous interactions support viral replication, correlating well with their RNA-binding affinities. Viruses engineered with a variant TAR able to bind all three Tat proteins replicate efficiently with any of the proteins. In one virus containing a noncognate Tat-TAR pair that neither interacts nor efficiently replicates (HIV-1 TAR and BIV Tat), viral revertants were isolated in which TAR had become mutated to generate a functional BIV Tat binding site. Our results support the view that incremental changes to TAR structure can provide routes for evolving new Tat-TAR complexes while maintaining active viral replication.
RNA-protein interactions play critical roles in the life cycles of viruses. The human immunodeficiency viruses (HIVs) and related lentiviruses encode an RNA-binding transcriptional activator, Tat, that is essential for virus replication. In HIV type 1 (HIV-1), Tat binds to the transactivation response element (TAR) located at the 5′ end of the viral mRNAs and enhances transcription from the HIV promoter in the long terminal repeat (LTR) (4). Tat activation occurs primarily at the level of transcriptional elongation whereby Tat converts RNA polymerases that are poorly processive into more-processive forms (17, 32, 35, 39, 42). Recent studies indicate that Tat induces hyperphosphorylation of the carboxyl-terminal domain of RNA polymerase II, primarily through recruitment of positive transcription elongation factor b (P-TEFb), which contains a cyclin-dependent kinase (Cdk9), and perhaps through recruitment of other carboxyl-terminal domain kinases (16, 22, 26, 27, 29, 38). One of the components of P-TEFb, cyclin T1, has been identified as an essential Tat cofactor that participates in TAR RNA binding in addition to activating Cdk9 (24, 56).
HIV-1 TAR forms a stable hairpin of 59 nucleotides (nt) and contains a 3-nt bulge and 6-nt loop in the upper part of the hairpin that are both essential for function (4, 18, 47). Tat binds primarily to the bulge region, utilizing one arginine within an arginine-rich RNA-binding domain (residues 49 to 57) to make a sequence-specific RNA contact and requiring surrounding charged residues to enhance binding affinity (1, 8, 37, 44, 48, 51, 55). In addition to its RNA-binding domain, Tat contains an activation domain (residues 1 to 48) that interacts with cyclin T1 and recruits the complex to TAR (5, 11, 23, 25, 30, 34, 56). In the context of cyclin T1, RNA binding specificity is extended to the loop and RNA binding affinity is enhanced, although it is not yet clear whether cyclin T1, Tat, or both proteins make loop-specific contacts (25, 45, 46, 56).
Bovine immunodeficiency virus (BIV) is a lentivirus related to HIV that causes persistent lymphocytosis in cattle (28). BIV encodes a TAR similar to that of HIV-1 and an analogous Tat protein that contains an arginine-rich RNA-binding domain (residues 68 to 81) and closely related activation domain (9, 19, 36). In contrast to the HIV-1 Tat-TAR interaction, however, the BIV RNA-binding domain recognizes BIV TAR with high affinity and specificity in a loop-independent manner and in the absence of cyclin T1 or other cellular proteins (2, 6, 14, 15). In the BIV complex, the peptide adopts a β-hairpin conformation and utilizes eight amino acids to specifically recognize the bulge region of TAR (15, 43, 57). Another bovine lentivirus, Jembrana disease virus (JDV), encodes a Tat protein closely related to that of BIV (10, 13), and interestingly, its RNA-binding domain exhibits chameleon-like behavior that allows recognition of BIV TAR in the β-hairpin binding mode or of HIV-1 TAR in the loop- and cyclin T1-dependent binding mode (49). Consistent with this observation is the finding that JDV Tat is able to activate the HIV-1 LTR in addition to its own LTR (12).
The structural comparisons among the HIV, BIV, and JDV Tat-TAR interactions and the observation that hybrid HIV/BIV TAR RNAs can be recognized in the two different binding modes (50) suggest that Tat-TAR interactions may readily evolve in actively replicating viruses where two or more binding modes are simultaneously maintained. To explore this hypothesis further and to develop a viral replication system in which new Tat-TAR interactions might be discovered, we have engineered hybrid HIV-1 proviruses with heterologous Tat-TAR interactions and have measured their replication properties. We show that the different binding modes all support robust viral replication, consistent with the view that Tat and TAR can coevolve to generate new binding specificities and different dependencies on host factors while maintaining functional intermediates.
MATERIALS AND METHODS
Proviral plasmids.
Wild-type HIV-1 proviral (R7/3) and Tat-defective (R7/3Δtat) clones used in this study have been previously described (17). The Tat mutant has a deletion of the second nucleotide (U) of the initiation codon and an insertion of an MluI linker containing two termination codons. To engineer proviruses with heterologous Tat-TAR interactions, we reconstructed the R7/3 clones to facilitate replacement of the 5′ and 3′ TAR elements and inserted another tat gene (encoding HIV-1 Tat residues 1 to 72) into the Nef coding region, which has little effect on viral replication in tissue culture (17). These plasmid reconstructions also eliminated some flanking cellular sequences present in the original R7/3 proviral clones.
To facilitate provirus construction, the >9-kb HIV-1 genome was subcloned in three segments (5′ LTR, 3′ LTR, and central genome) by PCR of the R7/3 or R7/3Δtat templates and cloning into the pGEM-11Zf(+) vector (Promega). The vectors designated 5′LTR were constructed with sense (5′-ACG CGT CGA ATT CTG GAA GGG CTA ATT C-3′) and antisense (5′-ACG CGT CTC TAG ACC CAT CGA TCT AAT TC-3′) primers and EcoRI and XbaI cloning sites (underlined). The vectors designated 3′LTR were constructed with sense (5′-ACG CGT CTC TAG ATT AGT GAA CGG ATC CT-3′) and antisense (5′-GCT CTA GAG CGG CCG CTA GAG ATT TTC CAC-3′) primers and XbaI and NotI cloning sites (underlined). The new tat gene (designated HIV tat 1-72) was amplified from pSV2tat72 (21) with sense (5′-ACT CTA GGC GCG CAT GGA ACC GGT CGA CCC GC-3′) and antisense (5′-GAG CTT ACG CGTCAC TGT TTA GAC AGA GAA ACC TGG TGG GTC TGC GAT CCC TGC-3′) primers and was cloned into the nef region of the 3′LTR vectors by digestion with BssHII and MluI (underlined) and ligation into the compatible MluI sites. The 5′LTR- and 3′LTR/tat 1-72 vectors were then combined to generate two-LTR vectors (also containing the inserted HIV tat 1-72 gene) by digestion and ligation of EcoRI-XbaI fragments. To complete the provirus clones, the central portion of the genome was cloned into the two-LTR vectors with ClaI-BamHI restriction fragments from R7/3 and R7/3Δtat.
Hybrid proviruses were constructed in which HIV-1 TAR was replaced with the BIV, JDV, and H/B (defined below) TAR elements. Fragments containing the TAR elements were amplified from LTR reporter plasmids (50) with sense (5′-GAG AGC TGC ATC CGG AGT ACT TC-3′) and antisense (BIV, 5′-GCT TTA TTG AGG CTT AAG CAG TGG GTT CCC TAG TTA GCC TCG GAG CTA ATG AGC TAC ACG AGG TCT AAC CAG AGA GAC CC-3′; JDV, 5′-GCT TTA TTG AGG CTT AAG CAG TGG GTT CCC TAG TTA GCC TCG GAG CTG TCA GCT ATC CAG AGG TCT AAC CAG AGA GAC CC-3′; H/B, 5′-GCT TTA TTG AGG CTT AAG CAG TGG GTT CCC TAG TTA GCC TCG GAG CTT CCC AGA GCT CAA CGA GGT CTA ACC AGA GAG ACC C-3′) primers, digested with AflII and BspEI (underlined), and ligated into the 5′LTR and 3′LTR/tat 1-72 vectors. Plasmids were then combined as described above to generate the corresponding two-LTR vectors and proviral clones.
Hybrid proviruses were constructed in which the HIV tat 1-72 gene (inserted into the nef region) was replaced by fusion proteins containing the arginine-rich RNA-binding domains of BIV or JDV Tat (residues 65 to 81) in place of the RNA-binding domain of HIV-1 Tat (residues 49 to 57) (Fig. 1). The tat genes were amplified from fusion protein expression vectors (49) with sense (BIV, 5′-ACC AAA GCC CTA GGT ATC TCT TAC GGC AGC GGA CCG CGG CCT AGA GGT ACC AGA GGA AAG GGA AGG AGG ATC AGG AGA C-3′; JDV, 5′-ACC AAA GCC CTA GGT ATC TCT TAC GGC GGA AGA AGG AAG AAA AGA GGA ACC AGA GGA AAG GGG AGA AAA ATC CAC TAT C-3′) and antisense (BIV, 5′-GAG CTT ACG CGT CAC TGT TTA GAC AGA GAA ACC TGG TGG GTC TGC GAT CCC TGC GGC GGT CTC CTG ATC CTC CTT CCC-3′; JDV, 5′-GAG CTT ACG CGT CAC TGT TTA GAC AGA GAA ACC TGG TGG GTC TGC GAT CCC TGC GGC GGA TAG TCG ATT TTT CTC CCC-3′) primers, and fragments were extended with T4 DNA polymerase, digested with AvrII and MluI (underlined), and ligated into the 3′LTR/tat 1-72 vector. Proviral clones were constructed with all combinations of Tat fusion proteins and TAR elements by ligation of appropriate restriction fragments (Fig. 2).
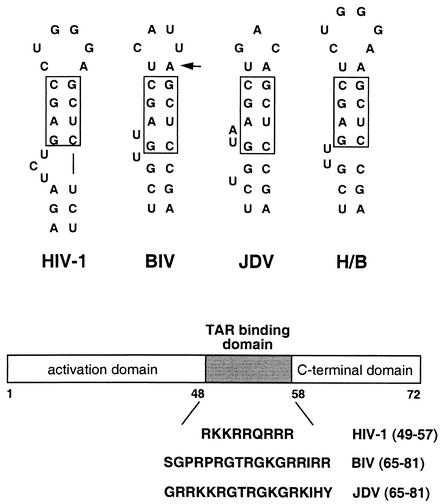
FIG. 1.
Secondary structures of the upper stems of HIV-1, BIV, JDV, and H/B TAR RNAs. The boxes indicate conserved base pairs in the upper stem required for HIV-1 or BIV Tat binding, and the additional U:A base pair above this region (arrow) is important for BIV binding (50). The bulge architectures of the various RNAs are different. H/B TAR is a hybrid consisting of the HIV-1 loop (required for cyclin T1 binding) and the optimal BIV Tat binding site that is able to bind HIV-1 Tat in a cyclin T1-dependent manner and the BIV Tat RNA-binding domain in a β-hairpin conformation (50). For the proviral constructs described in this paper, the lower portion of the HIV-1 TAR hairpin (∼20 bp not shown) was kept constant and only these upper regions were used to replace nt 20 to 42 of HIV-1 TAR. A schematic of the Tat protein structure is shown below. For the proviral constructs, the activation and C-terminal domains of HIV-1 Tat were kept constant and the RNA-binding domains shown were inserted to create the BIV and JDV fusion proteins.
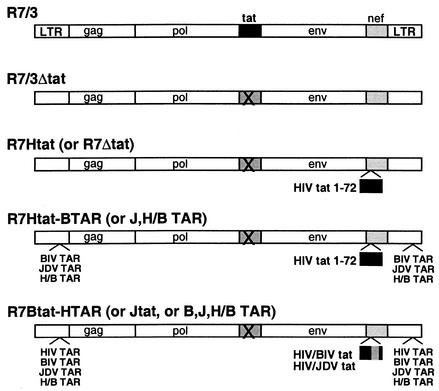
FIG. 2.
Genomic arrangements of the HIV-1 proviral clones and chimeras. R7/3 is the proviral clone previously described (17), and R7/3Δtat is defective for Tat expression. The tat gene from pSV2tat72 (21) was inserted into the nef region of the virus to restore a functional Tat in the R7Htat clone. The R7Δtat clone does not have the tat gene inserted but is in the same vector background. Chimeric proviral clones containing BIV, JDV, and H/B TAR elements, and the BIV and JDV Tat fusion proteins, were constructed as described in Materials and Methods. Parentheses in the provirus nomenclature indicate the different individual chimeras.
Cell lines and infections.
Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (FCS), 100 U of penicillin G/ml, and 100 μg of streptomycin/ml. The lymphocytic T-cell lines SupT1 and MT-4 were maintained in RPMI 1640 medium supplemented with 10% FCS, penicillin, and streptomycin. U373-MAGI cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, penicillin, streptomycin, 0.2 mg of G418/ml, and 0.1 mg of hygromycin B/ml. U373-MAGI cells express CD4 linked to neo and an HIV-1-LTR-β-galactosidase reporter linked to hyg and were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health) (from Michael Emerman and Adam Geballe).
Viral stocks were prepared by transfection of 293T cells with 10 μg of the various proviral plasmids in 100-mm-diameter culture plates by calcium phosphate precipitation. Supernatants were removed 2 days after transfection, filtered through 0.45-μm-pore-size polyethersulfone membranes, and stored in aliquots at −80°C for use as virus stocks. Levels of p24 capsid protein were determined by antigen-capture enzyme-linked immunosorbent assays (ELISAs) (Abbott Laboratories, Abbott Park, Ill.) according to the manufacturer's protocol, and equal p24 levels were used for subsequent infections.
HIV-1 infections were performed at 37°C with SupT1 (106 cells, 15 ng of p24) or MT-4 (106 cells, 4 ng of p24) cells in 2 ml of RPMI medium. After viral adsorption for 2 h, the cells were extensively washed to remove unadsorbed virus particles and cultured in 10 ml of medium. Cultures were split at a 1:10 ratio every 3 to 4 days. Virus replication was monitored by p24 ELISA and reverse transcriptase (RT) activity (31) assays with cell-free culture fluid at the indicated time points. Data are representative of three independent experiments.
Virus titer determination by immunofluorescence.
We utilized an immunofluorescence assay in addition to p24 and RT assays to measure viral levels. Various amounts of infected cell culture medium were diluted to 250 μl and added to 104 U373-MAGI cells plated in chamber slides (performed in duplicate). Virus was adsorbed for 2 h at 37°C in 5% CO2, 1 ml of fresh culture medium was added, and cells were incubated for 48 h. After being washed with phosphate-buffered saline (PBS), cells were fixed in 1 ml of cold (−20°C) 50% methanol-50% acetone for 5 min and washed three times with PBS. An 0.25-ml quantity of mouse anti-p24 antibody (1:400 dilution; obtained from the AIDS Research and Reference Reagent Program; 183-H12-5C monoclonal antibody from Bruce Chesebro and Kathy Wehrly) was added for 1 h at room temperature, cells were washed three times with PBS, 0.25 ml of fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G (1:400 dilution; RPC, Rochester, N.Y.) was added for 1 h at room temperature, and cells were washed three more times with PBS. p24-positive cells were counted by fluorescence microscopy, and values were normalized to the amount of culture supernatant used. Syncytia with multiple nuclei were counted as a single infected cell. Similar values were obtained by staining for β-galactosidase in the U373-MAGI reporter cells (54) (data not shown), but the immunofluorescence assay was more sensitive and was used for quantitation.
CAT assays.
Activities of the Tat fusion proteins expressed within the proviral clones were measured in chloramphenicol acetyltransferase (CAT) reporter assays. Fifty nanograms of an HIV-1 LTR CAT plasmid (containing HIV-1 TAR, BIV TAR, JDV TAR, or H/B TAR [49]) was cotransfected with 100 ng of a proviral plasmid into 293T cells with Lipofectamine (Gibco BRL, Gaithersburg, Md.). Total plasmid DNA was adjusted to 1 μg with pGEM-11Zf(+). Transiently transfected cells were collected by trypsinization after 48 h, extracts were prepared by three cycles of freezing-thawing (−80°C-37°C), and CAT activities were measured with an amount of extract adjusted to be within the linear range of the assay (7). Activities were quantitated with a Molecular Dynamics PhosphorImager, and levels of Tat activation were calculated relative to transfections with the reporter plasmids alone.
RESULTS
Activation of the HIV-1 LTR by heterologous lentiviral Tat RNA-binding domains.
Previous studies of the HIV-1 and BIV Tat-TAR complexes have shown that, while the primary, secondary, and tertiary structures of the two TAR elements are quite similar (Fig. 1) (50), the modes of Tat recognition are distinct. HIV-1 Tat primarily uses one arginine within its RNA-binding domain to contact the bulge region of HIV-1 TAR and requires cyclin T1 for loop recognition, whereas BIV Tat binds BIV TAR with high affinity and specificity on its own, adopting a β-hairpin conformation in the complex (8, 15, 43, 57). Despite the structural similarities of the TAR elements, BIV Tat is unable to bind HIV-1 TAR with high affinity, and conversely, HIV-1 Tat is unable to bind BIV TAR with high affinity (14, 50). Consequently, HIV-1 Tat efficiently activates the HIV-1 but not the BIV LTR, and BIV Tat efficiently activates the BIV but not the HIV-1 LTR (36). Similarly, a hybrid protein in which the RNA-binding domain of BIV Tat (residues 65 to 81) was fused to the activation domain of HIV Tat (residues 1 to 48) (Fig. 1) activated the HIV-1 LTR only when HIV-1 TAR was replaced by BIV TAR (14). The JDV Tat RNA-binding domain is a chameleon that is able to bind to both HIV-1 and BIV TAR elements with high affinity, in cyclin T1-dependent and β-hairpin modes, respectively (49), and consequently is able to activate both LTRs (12).
To examine the activities of the various Tat-TAR interactions in the context of viral replication, we constructed a set of matched HIV-1 viruses in which wild-type Tat was inactivated and hybrid Tat proteins containing the HIV-1 activation domain fused to the various RNA-binding domains were inserted into the nef region of the virus (Fig. 2). We also constructed viruses in which HIV-1 TAR was replaced by BIV TAR, JDV TAR, or H/B TAR (a hybrid RNA able to bind both HIV-1 and BIV Tat) (50) (Fig. 1 and 2). For the TAR replacements, the lower portion of the HIV-1 stem, which is not directly involved in Tat binding, was maintained. By placing the Tat-TAR interactions within the same HIV-1 background and making only small changes to Tat and TAR, we attempted to focus directly on how the mode of RNA binding influences viral replication and to minimize the possible effects of using different, albeit related, viral LTRs or Tat activation domains (12).
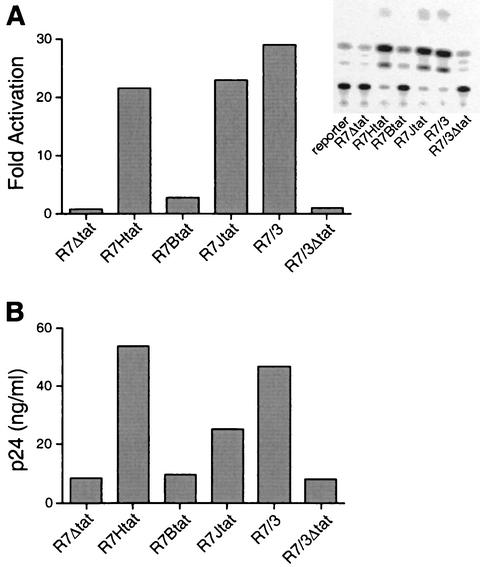
To ensure that the inserted Tat proteins were expressed in the proviral context, we first used a CAT reporter assay to monitor transcriptional activation and found that proviruses expressing HIV-1 Tat or JDV Tat RNA-binding domains were able to activate the HIV-1 promoter through HIV-1 TAR to similar, high levels, whereas the BIV Tat RNA-binding domain showed only low levels of activation (Fig. 3A), consistent with previous binding and activation studies. Tat expressed from the nef location functioned as well as that from its wild-type location (compare R7Htat to the parental R7/3 provirus [Fig. 3A]), and proviruses not expressing Tat (R7Δtat and R7/3Δtat) did not activate the reporter. Additional CAT assays with BIV, JDV, and H/B TAR reporters showed activation only with proviruses having the appropriate RNA-binding domain (data not shown). In addition, transient transfection of 293T cells (nonpermissive for infection) showed that proviruses encoding the HIV-1 and JDV Tat domains expressed significant levels of p24 whereas a provirus encoding the BIV domain expressed only background levels (Fig. 3B).
FIG. 3.
Transcriptional activation and p24 expression mediated by Tat hybrid proteins engineered into proviral plasmids. (A) Levels of activation measured in a CAT reporter assay. 293T cells were cotransfected with an HIV-1 LTR-TAR CAT reporter plasmid and variant proviral plasmids expressing HIV-1 Tat or HIV-BIV or HIV-JDV Tat hybrid proteins. CAT activities were quantitated 48 h after transfection, and fold activation was calculated by dividing the amount of CAT activity in the presence of Tat by the amount in the absence of Tat. The inset shows the results of a representative CAT assay, with some points out of the linear range of the assay. For quantitation, assays were repeated with an appropriate amount of cell extract. (B) Levels of p24 expression following transient transfection. 293T cells were transfected with 10 μg of each proviral DNA by calcium phosphate precipitation, and p24 levels in the culture supernatant were measured by ELISA as described in Materials and Methods.
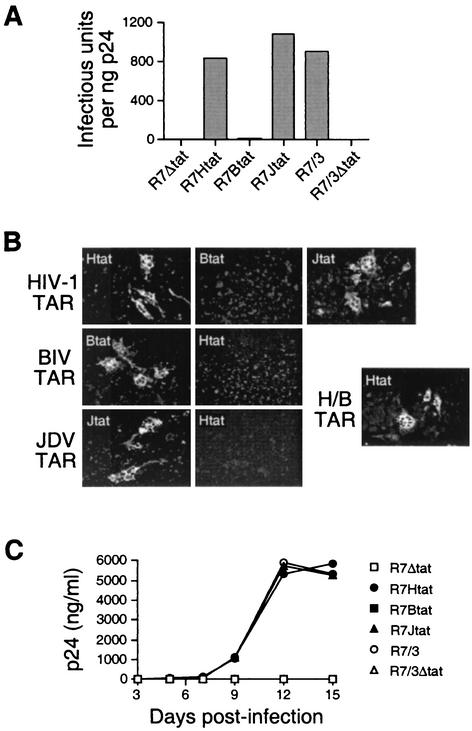
Two additional assays demonstrate that the HIV-1 and JDV Tat RNA-binding domains can support viral replication through HIV-1 TAR whereas the BIV Tat domain cannot. First, in a transfection assay, we generated virus stocks by transiently transfecting 293T cells, removing the culture supernatants, and determining viral titers by infection of U373-MAGI cells (54) and p24 immunofluorescence. Proviruses encoding the HIV-1 or JDV, but not the BIV, RNA-binding domain produced high viral titers (Fig. 4A) and induced syncytium formation (Fig. 4B, top panels). Second, in an infection assay, we monitored the kinetics of virus replication in a SupT1 T-cell line by p24 ELISA and observed robust replication for proviruses with the HIV-1 and JDV domains and no detectable replication for proviruses with the BIV domain or without Tat (Fig. 4C). Thus, viral replication correlates well with the known affinities and activities of the HIV-1, BIV, and JDV Tat RNA-binding domains (14, 49).
FIG. 4.
Replication of the chimeric viruses in U373-MAGI and SupT1 cell lines. (A) U373-MAGI-CD4+ cells were infected with a fixed amount of p24 from each viral stock, and the infectious titer was determined by counting the number of infected cells by p24 immunofluorescence (individual cells or syncytia were scored as single cells) and normalizing to the input p24 levels. (B) Syncytium formation in U373-MAGI cells by the chimeric viruses. Cells were fixed 48 h after infection and assayed by immunofluorescence (representative fields are shown at 20× magnification) or by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of these reporter cells (data not shown; see Materials and Methods). (C) Replication kinetics of the parental R7/3 virus and HIV-1 TAR-containing viruses with various Tat proteins. SupT1 cells (106) were infected with fixed amounts of each virus (15 ng of p24), and replication was monitored by determining p24 levels in cell-free supernatants at the times indicated. Data are representative of three independent experiments.
Heterologous Tat-TAR interactions can support HIV-1 replication.
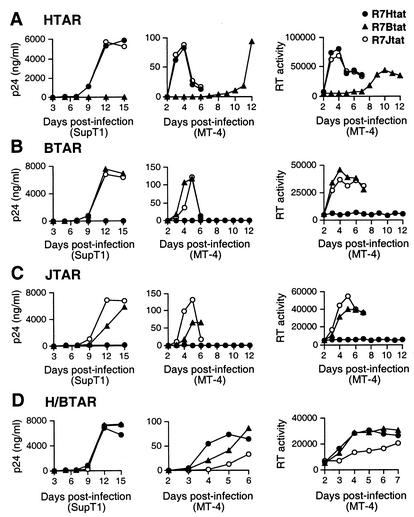
To determine whether, and how well, viruses can replicate when both Tat and TAR are replaced with heterologous interactions, including the BIV interaction, which does not depend on cyclin T1 for RNA recognition, we measured replication rates for all pairwise Tat-TAR combinations. We prepared viral stocks by transfecting 293T cells with the various chimeric proviruses (Fig. 2) and found that proviruses engineered with BIV, JDV, and H/B TAR elements produced high viral titers only when the appropriate RNA-binding domain was present (Table 1). The same amount of virus, based on p24 levels, was then used to infect SupT1 cells (15 ng of p24/106 cells) or MT-4 cells (4 ng of p24/106 cells), and rates of virus replication were monitored by p24 and RT assays (Fig. 5). In the HIV-1 TAR viruses, the HIV-1 and JDV Tat RNA-binding domains supported similar high rates of replication while the BIV domain did not (Fig. 5A), as also described above. Infections peaked at ∼12 days in SupT1 cells and at ∼4 days in MT-4 cells, with extensive cytopathic effect observed in the MT-4 cells. Virus production was eventually observed with the BIV Tat domain after 10 to 12 days of culture in MT-4 cells, and these viruses were characterized as described below. In the BIV TAR viruses, the BIV and JDV RNA-binding domains supported active replication while the HIV-1 domain did not (Fig. 5B), consistent with the high affinities of BIV and JDV Tat for BIV TAR (14, 49). Similarly in the JDV TAR viruses, the BIV and JDV, but not the HIV-1, binding domains supported replication (Fig. 5C), consistent with their known binding properties (49). In these JDV TAR viruses, the cognate interaction with JDV Tat was slightly better than that with BIV Tat, also consistent with the slightly higher affinity of JDV Tat peptides for JDV TAR (49). Finally, viruses engineered with H/B TAR, which can bind both HIV-1 and BIV Tat peptides, replicated efficiently with any of the three Tat proteins (Fig. 5D), although the virus with JDV Tat exhibited slightly slower kinetics in MT-4 cells. Thus, viral replication correlates with the affinity of the Tat-TAR interaction and can occur independently of the cyclin T1-TAR interaction in the context of HIV-1.
TABLE 1.
Infectivity of the chimeric viruses
| Provirus | Infectious units/ng of p24a |
|---|---|
| R7Δtat | 0 |
| R7Htat | 836 |
| R7Btat-HTAR | 11 |
| R7Jtat-HTAR | 1,072 |
| R7Htat-BTAR | 3 |
| R7Btat-BTAR | 969 |
| R7Jtat-BTAR | 636 |
| R7Htat-JTAR | 4 |
| R7Btat-JTAR | 96 |
| R7Jtat-JTAR | 861 |
| R7Htat-H/BTAR | 783 |
| R7Btat-H/BTAR | 992 |
| R7Jtat-H/BTAR | 412 |
293T cells were transfected with the indicated proviral plasmid, and supernatants were collected after 48 h and assayed for p24. Equal amounts of p24 were used to infect U373-MAGI cells, and viral titers were determined by counting p24-positive cells by immunofluorescence as described in Materials and Methods.
FIG. 5.
Replication kinetics of the chimeric viruses in SupT1 and MT-4 cell lines. (A) Growth kinetics of viruses containing HIV-1 TAR, determined by assaying p24 levels in SupT1 cells and p24 and RT levels in MT-4 cells. Because HIV-1 grows rapidly in MT-4 cells and induces extensive cytopathic effect, p24 levels are considerably lower than those in SupT1 cells. (B) Growth kinetics of viruses containing BIV TAR. (C) Growth kinetics of viruses containing JDV TAR. (D) Growth kinetics of viruses containing H/B TAR. Data in each panel are representative of three independent experiments.
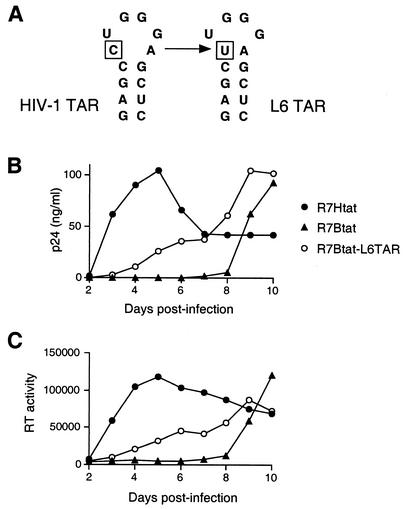
An HIV-1 TAR loop mutation allows the BIV Tat RNA-binding domain to function.
As noted above, replication-competent viruses were eventually observed in cultures infected with the hybrid virus containing HIV-1 TAR and the BIV Tat domain (Fig. 5A) even though BIV Tat binds only weakly to HIV-1 TAR (14, 49). To determine whether these viruses represented examples of forced evolution in which Tat or TAR had been mutated to generate a functional interaction, we PCR amplified several regions of the integrated proviral DNA and cloned and sequenced the fragments. Analyses of 12 clones revealed no changes to the Tat coding region or viral promoter, but all contained a C-to-U mutation at position 30 of the TAR loop (Fig. 6A). Interestingly, this mutation corresponds to a previously characterized TAR variant (L6) that is able to bind the BIV Tat domain because it contains an additional closing U:A base pair at the base of the loop (50). To confirm directly that the mutation was sufficient to allow replication with the BIV Tat domain, we constructed a provirus with L6 TAR and the BIV peptide and found that, indeed, the virus replicated with reasonable kinetics (Fig. 6B and C). Replication was slower than that with the wild-type HIV-1 Tat-TAR interaction, consistent with the observation that, while the extra base pair in the upper stem is required for BIV binding, additional changes in the lower stem are needed to fully generate a high-affinity BIV interaction (50). Nevertheless, the chimeric virus can readily evolve into a replication-competent form by a single base change to the TAR loop.
FIG. 6.
Growth and replication kinetics of a mutant virus with a change in the HIV-1 TAR loop, initially isolated as an escape mutant from MT-4 cells. (A) Secondary structure of the TAR loop and previously characterized L6 mutation (50). (B) Growth kinetics of the L6 mutant virus in MT-4 cells monitored by p24 assays. Significant replication of the R7Btat virus is observable by day 9, presumably reflecting accumulation of the L6 escape mutant, as in Fig. 5A. (C) Growth kinetics of the L6 mutant virus in MT-4 cells monitored by RT assays. Data in panels B and C are representative of three independent experiments.
DISCUSSION
We have shown that HIV-1 proviruses in which the Tat-TAR interaction has been replaced by other lentiviral Tat-TAR interactions are able to replicate as efficiently as wild-type HIV-1 despite differences in the requirements for cellular proteins in TAR recognition. In the HIV-1 Tat-TAR interaction, a heterodimeric complex between Tat and cyclin T1 is needed to recognize the bulge and loop of TAR (5, 11, 23, 25, 34, 45, 46, 56), whereas in the BIV Tat-TAR interaction, the arginine-rich domain of Tat adopts a β-hairpin conformation upon binding to the bulge region and generates a high-affinity complex on its own (14, 15, 43, 57). Although cyclin T1 does not appear to participate directly in BIV TAR recognition, it presumably still interacts with the Tat activation domain, allowing Cdk9 recruitment, transcriptional activation, and replication (2, 6).
The JDV Tat RNA-binding domain is especially interesting in that it is able to bind to both HIV-1 and BIV TARs in the two different binding modes (49). Here we have shown that the JDV Tat domain also supports viral replication through both TAR elements, consistent with a previous report showing that the full-length JDV Tat protein can substitute for HIV-1 Tat in viral replication (12). We have also shown that a hybrid TAR composed of the HIV-1 loop and BIV bulge region (H/B TAR) that is able to bind the HIV-1 and BIV domains in the two binding modes (50) supports replication with any of the Tat proteins. The generation of replication-competent viruses with mixed binding modes suggests that Tat-TAR interactions with rather different structural characteristics can readily evolve in a viral setting via multifunctional intermediates. In addition, the finding that HIV-1 TAR can be recognized by the BIV domain by mutation of a single loop nucleotide to create an extra base pair (the L6 mutant) (50), resulting in replication-competent viruses (Fig. 6), suggests that other simple pathways can be used to evolve new interactions that require only one or a few mutations and do not involve multifunctional intermediates. The selective pressure for the Tat-TAR interaction, the high mutation rate of HIV-1, and the relatively small changes needed to create new peptide-RNA interactions with the arginine-rich motif (20) apparently can provide multiple pathways to coevolve an RNA-protein interaction, as also emphasized by previous studies of TAR and Tat mutants (3, 33, 41, 52).
The substitution of the HIV-1 Tat-TAR interaction with the other lentiviral interactions described here suggests that a viral replication system may be used to select for novel Tat-TAR binding interactions. Indeed, we have been utilizing this strategy to identify TAR binding peptides from combinatorial libraries (unpublished data), with the intent of finding high-affinity binders that might ultimately be used to disrupt the Tat-TAR complex. Berkhout and coworkers (40, 53) have shown that there are both minimal and maximal levels of activation that can support viral replication, suggesting that such viral selection systems may require fine-tuning to identify the desired molecules. Nevertheless, the structural diversity of the lentiviral Tat-TAR systems provides an interesting opportunity to explore the evolution and possible utility of peptide-RNA interactions.
Acknowledgments
We thank Mark Feinberg for providing the R7/3 proviral plasmids and for helpful discussions; Mervi Detorio and Maureen Oliveira of the Wainberg laboratory for technical assistance; Valerie Calabro, Chandreyee Das, Steve Landt, and members of the Frankel laboratory for helpful discussions; and Raul Andino, Valerie Calabro, and Chandreyee Das for comments on the manuscript.
This research was supported by NIH grant RO1 AI29135 (A.D.F.) and by the Canadian Institutes of Health Research (M.A.W.).
REFERENCES
- 1.Aboul-ela, F., J. Karn, and G. Varani. 1995. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 253:313-332. [DOI] [PubMed] [Google Scholar]
- 2.Barboric, M., R. Taube, K. Fujinaga, and B. M. Peterlin. 2000. Binding of Tat to TAR and recruitment of positive transcription elongation factor b occur independently in bovine immunodeficiency virus. J. Virol. 74:6039-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout, B., and B. Klaver. 1993. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 21:5020-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., H. L. Wiegand, P. D. Bieniasz, and B. R. Cullen. 2000. Functional differences between human and bovine immunodeficiency virus Tat transcription factors. J. Virol. 74:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calnan, B. J., S. Biancalana, D. Hudson, and A. D. Frankel. 1991. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 5:201-210. [DOI] [PubMed] [Google Scholar]
- 8.Calnan, B. J., B. Tidor, S. Biancalana, D. Hudson, and A. D. Frankel. 1991. Arginine-mediated RNA recognition: the arginine fork. Science 252:1167-1171. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, S., S. A. Nadin-Davis, Y. Wannemuehler, and J. A. Roth. 1993. Identification of transactivation-response sequences in the long terminal repeat of bovine immunodeficiency-like virus. J. Virol. 67:4399-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chadwick, B. J., R. J. Coelen, G. E. Wilcox, L. M. Sammels, and G. Kertayadnya. 1995. Nucleotide sequence of Jembrana disease virus: a bovine lentivirus associated with an acute disease syndrome. J. Gen. Virol. 76:1637-1650. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., Y. Fong, and Q. Zhou. 1999. Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc. Natl. Acad. Sci. USA 96:2728-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H., J. He, S. Fong, G. Wilcox, and C. Wood. 2000. Jembrana disease virus Tat can regulate human immunodeficiency virus (HIV) long terminal repeat-directed gene expression and can substitute for HIV Tat in viral replication. J. Virol. 74:2703-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., G. Wilcox, G. Kertayadnya, and C. Wood. 1999. Characterization of the Jembrana disease virus tat gene and the cis- and trans-regulatory elements in its long terminal repeats. J. Virol. 73:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, L., and A. D. Frankel. 1994. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry 33:2708-2715. [DOI] [PubMed] [Google Scholar]
- 15.Chen, L., and A. D. Frankel. 1995. A peptide interaction in the major groove of RNA resembles protein interactions in the minor groove of DNA. Proc. Natl. Acad. Sci. USA 92:5077-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cujec, T. P., H. Okamoto, K. Fujinaga, J. Meyer, H. Chamberlin, D. O. Morgan, and B. M. Peterlin. 1997. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg, M. B., D. Baltimore, and A. D. Frankel. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, S., and E. C. Holland. 1988. HIV-1 tat trans-activation requires the loop sequence within tar. Nature 334:165-167. [DOI] [PubMed] [Google Scholar]
- 19.Fong, S. E., J. D. Greenwood, J. C. Williamson, D. Derse, L. A. Pallansch, T. Copeland, L. Rasmussen, A. Mentzer, K. Nagashima, G. Tobin, and M. A. Gonda. 1997. Bovine immunodeficiency virus tat gene: cloning of two distinct cDNAs and identification, characterization, and immunolocalization of the tat gene products. Virology 233:339-357. [DOI] [PubMed] [Google Scholar]
- 20.Frankel, A. D. 2000. Fitting peptides into the RNA world. Curr. Opin. Struct. Biol. 10:332-340. [DOI] [PubMed] [Google Scholar]
- 21.Frankel, A. D., and C. O. Pabo. 1988. Cellular uptake of the Tat protein from human immunodeficiency virus. Cell 55:1189-1193. [DOI] [PubMed] [Google Scholar]
- 22.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Martinez, L. F., G. Mavankal, J. M. Neveu, W. S. Lane, D. Ivanov, and R. B. Gaynor. 1997. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 16:2836-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gold, M. O., X. Yang, C. H. Herrmann, and A. P. Rice. 1998. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J. Virol. 72:4448-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonda, M. A., D. G. Luther, S. E. Fong, and G. J. Tobin. 1994. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 32:155-181. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 31.Kameoka, M., L. Rong, M. Gotte, C. Liang, R. S. Russell, and M. A. Wainberg. 2001. Role for human immunodeficiency virus type 1 Tat protein in suppression of viral reverse transcriptase activity during late stages of viral replication. J. Virol. 75:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 33.Klaver, B., and B. Berkhout. 1994. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 13:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 35.Laspia, M. F., A. P. Rice, and M. B. Mathews. 1989. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59:283-292. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Z. Q., D. Sheridan, and C. Wood. 1992. Identification and characterization of the bovine immunodeficiency-like virus tat gene. J. Virol. 66:5137-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long, K. S., and D. M. Crothers. 1999. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry 38:10059-10069. [DOI] [PubMed] [Google Scholar]
- 38.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marciniak, R. A., and P. A. Sharp. 1991. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 10:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marzio, G., M. Vink, K. Verhoef, A. de Ronde, and B. Berkhout. 2002. Efficient human immunodeficiency virus replication requires a fine-tuned level of transcription. J. Virol. 76:3084-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuveut, C., and K. T. Jeang. 1996. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J. Virol. 70:5572-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 43.Puglisi, J. D., L. Chen, S. Blanchard, and A. D. Frankel. 1995. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 270:1200-1203. [DOI] [PubMed] [Google Scholar]
- 44.Puglisi, J. D., R. Tan, B. J. Calnan, A. D. Frankel, and J. R. Williamson. 1992. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257:76-80. [DOI] [PubMed] [Google Scholar]
- 45.Richter, S., H. Cao, and T. M. Rana. 2002. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human cyclin T1-Tat-TAR ternary complex formation. Biochemistry 41:6391-6397. [DOI] [PubMed] [Google Scholar]
- 46.Richter, S., Y. H. Ping, and T. M. Rana. 2002. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosen, C. A., J. G. Sodroski, and W. A. Haseltine. 1985. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell 41:813-823. [DOI] [PubMed] [Google Scholar]
- 48.Roy, S., U. Delling, C. H. Chen, C. A. Rosen, and N. Sonenberg. 1990. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 4:1365-1373. [DOI] [PubMed] [Google Scholar]
- 49.Smith, C. A., V. Calabro, and A. D. Frankel. 2000. An RNA-binding chameleon. Mol. Cell 6:1067-1076. [DOI] [PubMed] [Google Scholar]
- 50.Smith, C. A., S. Crotty, Y. Harada, and A. D. Frankel. 1998. Altering the context of an RNA bulge switches the binding specificities of two viral Tat proteins. Biochemistry 37:10808-10814. [DOI] [PubMed] [Google Scholar]
- 51.Tao, J., and A. D. Frankel. 1993. Electrostatic interactions modulate the RNA-binding and transactivation specificities of the human immunodeficiency virus and simian immunodeficiency virus Tat proteins. Proc. Natl. Acad. Sci. USA 90:1571-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verhoef, K., and B. Berkhout. 1999. A second-site mutation that restores replication of a Tat-defective human immunodeficiency virus. J. Virol. 73:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verhoef, K., M. Koper, and B. Berkhout. 1997. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology 237:228-236. [DOI] [PubMed] [Google Scholar]
- 54.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193-198. [DOI] [PubMed] [Google Scholar]
- 55.Weeks, K. M., and D. M. Crothers. 1991. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell 66:577-588. [DOI] [PubMed] [Google Scholar]
- 56.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 57.Ye, X., R. A. Kumar, and D. J. Patel. 1995. Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem. Biol. 2:827-840. [DOI] [PubMed] [Google Scholar]