Abstract
Sterol 27-hydroxylase (CYP27A1) is required for bile acid synthesis by both the classical and alternate pathways. Cyp27a1−/− mice exhibit a dramatic increase in the activity of cytochrome P450 3A (CYP3A), which catalyzes side-chain hydroxylations of bile acid intermediates, thereby facilitating their excretion in the bile and urine. We examine the role of the nuclear xenobiotic receptor PXR (pregnane X receptor) in this process. We demonstrate that expression of Cyp3a11 and other established PXR target genes is increased in the Cyp27a1−/− mice. WhenCyp27a1−/− mice are fed a diet containing either cholic acid or chenodeoxycholic acid, expression of CYP7A1, which catalyzes the rate-limiting step in bile acid biosynthesis, is strongly suppressed. In parallel, the induction of Cyp3a11 observed in these mice is reversed, suggesting that bile acid intermediates serve as PXR activators. In support of this hypothesis, three potentially toxic sterols (7α-hydroxy-4-cholesten-3-one, 5β-cholestan-3α,7α,12α-triol, and 4-cholesten-3-one), including two that are known to accumulate in Cyp27a1−/− mice, are efficacious activators of mouse PXR. All three compounds are more potent activators of mouse PXR than of human PXR, which may explain in part why humans who lack functional CYP27A1 do not display a corresponding increase in CYP3A activity and are stricken with the disease cerebrotendinous xanthomatosis. Taken together, these results reveal the existence of a feedforward regulatory loop by which potentially toxic bile acid intermediates activate PXR and induce their own metabolism. In addition, this study demonstrates that animal models with alterations in gene expression can be used to identify endogenous ligands for orphan nuclear receptors.
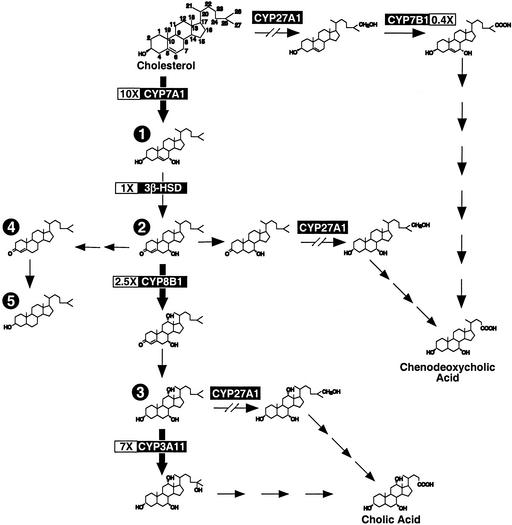
The conversion of cholesterol to bile acids occurs exclusively in the liver. This process involves the translocation of cholesterol and intermediates through various compartments of the cell, where they encounter a wide array of enzymes necessary for their ultimate conversion to the primary C24 bile acids, chenodeoxycholic acid (CDCA) and cholic acid (CA). Two principal pathways of bile acid biosynthesis are known. The classic or neutral pathway involves modification of the sterol ring structure followed by hydroxylation and cleavage of the side chain, whereas in the alternate or acidic pathway, side-chain hydroxylation precedes the ring modifications (Fig. 1). In both pathways, sterol 27-hydroxylase (CYP27A1), a mitochondrial cytochrome P450 enzyme, plays a key role.
Figure 1.
Hepatic mRNA expression levels of several enzymes involved in bile acid biosynthesis are altered in the Cyp27a1−/− mouse. Relative hepatic RNA levels were determined by Northern analysis from male, mixed-strain, wild-type and Cyp27a1−/− mice fed a standard rodent diet. The values shown in boxes represent the mean ± SEM of the fold change in mRNA levels (n = 6 mice per genotype). Bile acid intermediates that would be predicted to accumulate upstream of CYP3A11 because of these changes in enzyme levels include the following: 1, 7α-hydroxycholesterol (5-cholesten-3β,7α-diol); 2, 7α-hydroxy-4-cholesten-3-one (4-cholesten-7α-ol-3-one); 3, 5β-cholestan-3α,7α,12α-triol; 4, 4-cholesten-3-one; and 5, cholestanol (5α-cholestan-3β-ol).
In humans, mutations in the CYP27A1 gene are responsible for the rare genetic disorder cerebrotendinous xanthomatosis (CTX) (1, 2). CTX is characterized by the reduced formation of normal bile acids, the appearance of glucuronidated 25-hydroxylated bile alcohols in serum and urine, and elevated cholesterol and cholestanol levels in nearly all tissues (3). Clinical features in CTX patients include tendon xanthomas, mental retardation, cerebellar ataxia, cataracts, and peripheral neuropathy (4). Interestingly, disruption of the Cyp27a1 gene in mice does not phenocopy the human disease (5). Cyp27a1−/− mice reproduce only a few of the biochemical changes seen in human CTX. In particular, there are only trace levels of 25-hydroxylated bile alcohols, far less than those detected in humans (6, 7), and cholesterol biosynthesis is enhanced in various organs (8). Also in contrast to humans, these mice do not show evidence of xanthomas, accumulation of excessive cholesterol or cholestanol, or neural dysfunction. In fact, the phenotype of Cyp27a1−/− mice with respect to bile acid depletion is more benign than that for mice lacking the gene for cholesterol 7α-hydroxylase (Cyp7a1), deletion of which affects only the classic pathway of bile acid biosynthesis (9).
Interestingly, Cyp27a1−/− mice display a marked increase in the activity of CYP3A, an enzyme that can hydroxylate the side chain of sterols and bile acid intermediates (7). These metabolites can be either excreted from the body in the bile or urine or converted to CA. Notably, CYP3A activity does not appear to be increased in human CTX, suggesting a potential explanation for the dichotomy in CYP27A1−/− phenotypes between mice and humans (7, 10).
Expression of CYP3A is regulated in both species by the orphan nuclear pregnane X receptor (PXR), which is activated by a broad spectrum of xenobiotics including some prescription drugs and herbal remedies (reviewed in ref. 11). PXR also is activated in cell-based reporter assays by a variety of chemicals produced by the body (endobiotics), including steroid hormones and bile acids; however, the concentrations of these chemicals required to activate PXR appear to be higher than those that occur in vivo and, to date, it has remained unclear whether PXR is activated by endobiotics under physiological or pathophysiological conditions.
In this report, we provide evidence that PXR is activated in the Cyp27a1−/− mouse by the accumulation of several potentially toxic bile acid intermediates. Concordant with the differential activation of CYP3A in mouse versus human, several of these intermediates are less potent activators of human PXR (also called SXR). These results demonstrate that PXR can be activated by endobiotics under certain pathophysiological conditions and they emphasize the role of PXR as a sensor of toxic lipids. Moreover, because activation of PXR may ultimately lead to detoxification of these intermediates, our results suggest that PXR agonists may be of therapeutic potential in alleviating symptoms associated with the accumulation of hepatotoxic steroids in human CTX.
Materials and Methods
Materials.
5-Cholesten-3β 7α-diol (7α-OH-cholesterol), 4-cholesten-7α-ol-3-one (7α-OH-4-cholesten-3-one), and 4-cholesten-3-one were purchased from Steraloids (Newport, RI). Pregnenolone 16α-carbonitrile (PCN; 5-pregnen-3β-ol-20-one-16α-carbonitrile), CDCA, CA, lithocholic acid, and TCPOBOP {1,4-bis[2-(3,5-dichloropyridyloxyl)]benzene} were obtained from Sigma. 5β-cholestan-3α,7α,12α-triol (GW614417) was custom synthesized by Steraloids.
Animals and Diets.
Sterol 27-hydroxylase-deficient mice (Cyp27a1−/−; ref. 5) were maintained on a mixed-strain background, as were gender- and age-matched wild-type controls. Mice were housed in a temperature-controlled facility with 12-h light/dark cycles, and tissue samples were taken at the end of the dark cycle when mice were fed. Mice were given ad libitum a cereal-based powdered rodent diet (no. 7001, Teklad, Madison, WI) that contains 0.02% (wt/wt) cholesterol and 4% (wt/wt) fat. This diet was supplemented with 0.1% or 0.2% (wt/wt) CA or CDCA as indicated. At the end of a feeding study, mice were anesthetized and exsanguinated through the ascending vena cava, and tissues were harvested and flash-frozen in liquid nitrogen. All experiments were approved by the Institutional Animal Care and Research Advisory Committee at University of Texas Southwestern Medical Center.
Northern Analysis.
Total RNA was isolated from the liver tissue of individual mice, and poly(A+) RNA was purified from the RNA of individual mice (Figs. 1 and 2) or from a pool containing equal quantities of RNA from the five to seven mice per group (Fig. 3), by using oligo(dT) columns (Amersham Pharmacia Biotech). Five micrograms of mRNA was fractionated on a 1% agarose/formaldehyde gel and transferred to a membrane (ZetaProbe, Bio-Rad). Hybridizations were performed by using 32P-labeled cDNA probes for mouse PXR (12), β-actin, CYP7A1, CYP7B1, CYP27A1, and 3β-hydroxy-Δ5-C27-steroid oxidoreductase (3βHSD), from David Russell, University of Texas Southwestern Medical Center (13, 14). Probes for other mouse genes were generated by RT-PCR using the following primers (GenBank accession numbers are provided in parentheses): constitutive androstane receptor (CAR) (AF009327) 5′-GGAATTCGGAATG and 5′-GCGGATGGCCTCAA; cytochrome P450 2B10 (CYP2B10) (NM_009998.1) 5′-GGCTTCTTGCTACTCTTAGC and 5′-ATTTCCACCAGCTGTCTCAG; cytochrome P450 3A11 (CYP3A11) (NM_007818.1) 5′-TTTTCTGTCTTCACAAACCGG and 5′-CAAACCTCATGCCAATGCAG; sterol 12α-hydroxylase (CYP8B1) (AF090317) 5′-GGGTACCAGTCTGTAGATGG and 5′-AGTCTCTGGTGGAAGAGACG; multidrug resistance-associated protein 2 (MRP2) (AF227274) 5′-GACTCTGACAACTTGAATGGGACC and 5′-CCTTCTCTCCGCTCTTGATGTTAC; organic anion transporter polypeptide (OATP2) (NM_030687) 5′-CATGTGCATATGTATCCAAATCAC and 5′-CCTCATCACAGCTTAGTTTTCCGT; and small heterodimer partner (SHP) (L76567) 5′-AAGGATCCGCTGGGAAGAAACAGGAACAAG and 5′-CTAGCTAGCTGGAGGCACCAGACTCCATTC. All cDNA probes that were generated by PCR were verified by sequencing. The relative expression levels of RNA were quantified by PhosphorImager (Molecular Dynamics), and standardized against β-actin controls.
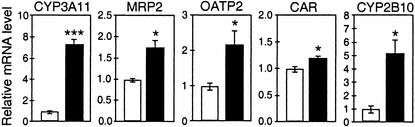
Figure 2.
PXR target genes show enhanced hepatic expression in the Cyp27a1−/− mouse. mRNA was isolated from the livers of chow-fed, male, wild-type and Cyp27a1−/− mice of a mixed-strain background (C57BL/6:129Sv). Northern analysis was performed, quantified by PhosphorImager, standardized against β-actin, and mathematically adjusted to yield a value of 1 for the wild-type group (white bars). The relative expression values (black bars) are depicted as the mean and SEM for each genotype (n = 6), and asterisks denote statistically significant results as determined by Student's t test (*, P < 0.05; ***, P < 0.0001).
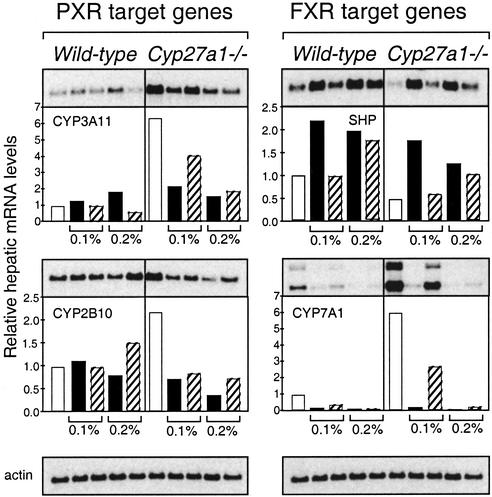
Figure 3.
Bile acid feeding relieves the elevated hepatic CYP3A11 mRNA levels seen in the Cyp27a1−/− mouse. Male, mixed-strain wild-type and Cyp27a1−/− mice were fed normal chow (white bars) or chow diets containing CA (black bars) or CDCA (hatched bars) at 0.1% or 0.2% (wt/wt) for 16 days. Total RNA was isolated from individual livers, and equivalent amounts from each animal (n = 5–7) were pooled in groups for mRNA purification. Northern analyses were performed on the pooled mRNA (shown as blots in the upper part of the panel for each mRNA), quantified by PhosphorImager, and standardized against β-actin (bottom blots), and the results are shown as histograms.
Cell Reporter Assay.
CV1 cells were transfected as described (15, 16) with an expression plasmid to supply a nuclear hormone receptor, a reporter plasmid containing the appropriate DNA response element, and a β-galactosidase- or secreted placental alkaline phosphatase-expression vector to control for transfection efficiency. The reporter plasmids used were XREM-luc (PXR; ref. 17), FXREIBABPx3-tk-luc [farnesoid X receptor (FXR); ref. 18], DR3ratCYP3A1x3-tk-luc (CAR; M. Makishima and D.J.M., unpublished data), and mSPPx3-tk-luc [VDR (vitamin D receptor); ref. 19]. Ligands were dissolved in DMSO and cells were exposed to ligands or vehicle at 0.1% vol/vol for 36–40 h.
Fluorescence Polarization Assay.
The mouse PXR ligand-binding domain (residues 105–431) was fused to the C terminus of glutathione S-transferase (GST), expressed in Escherichia coli, and purified on glutathione-coated beads. A fluorescein-labeled peptide (1 nM, with amino acid sequence ILRKLLQE) was incubated with purified GST-mPXR (≈1 μM) and candidate ligands in 100 μl of buffer (150 mM NaCl/10 mM potassium phosphate/2 mM CHAPS/2 mM EDTA/1 mM DTT, pH 7.3) in a black polypropylene 96-well plate on a shaker for 1 h (20). Ligand-dependent recruitment of the coactivator peptide was measured as an increase in fluorescence polarization by using an LJL analyst (LJL Biosystems, Sunnyvale, CA).
Statistical Analysis.
GraphPad prism computer software was used to perform all statistical analyses (GraphPad, San Diego). Experimental results comparing two groups were analyzed by the Student's t test to establish significant differences between a treatment group and the chow-fed wild-type control group. Multiple groups were analyzed by one-way ANOVA followed by the Newman–Keuls multiple comparison test. Statistical significance was declared if the calculated P value was <0.05. All values are represented as means ± SEM.
Results and Discussion
In a previous analysis of genes involved in bile acid synthesis, we showed that expression of cholesterol 7α-hydroxylase (CYP7A1) and CYP8B1 is increased and that oxysterol 7α-hydroxylase (CYP7B1) expression is decreased in the livers of Cyp27a1−/− mice compared with their wild-type counterparts (ref. 8 and Fig. 1). Other groups have recently demonstrated that the enzymatic activity of CYP3A11, which serves as a 5β-cholestan-3α,7α,12α-triol hydroxylase, is markedly up-regulated in the livers of Cyp27a1−/− mice (7, 21).
To determine whether PXR signaling is activated in the Cyp27a1−/− mice, we examined the expression level of Cyp3a11, a well-documented PXR target gene (17, 22, 23). As shown in two independent experiments, CYP3A11 expression was increased 7- to 10-fold in the Cyp27a1−/− mice compared with wild-type mice (Figs. 1 and 2). Hepatic mRNA expression of several other PXR target genes (23–25) was also increased in Cyp27a1−/− mice. MRP2 (ABCC2) was increased ≈1.7-fold, OATP2 (SLC21A5) was increased ≈2-fold, and CYP2B10 was increased >4-fold in the Cyp27a1−/− mice. Expression of the nuclear receptor CAR, which is regulated by PXR (23), was modestly but significantly increased. PXR expression was unaffected in the Cyp27a1−/− mice (data not shown). The pronounced up-regulation of several PXR target genes strongly suggests increased levels of one or more PXR ligands in Cyp27a1−/− mice.
We next examined whether the induction of PXR target genes in Cyp27a1−/− mice is reversed under conditions in which the production of bile acid intermediates is suppressed. Accordingly, Cyp27a1−/− and wild-type mice were fed a diet supplemented with either 0.1% or 0.2% CA or CDCA (the daily equivalent of 1–2 times the bile acid pool size) to suppress Cyp7a1. As expected, the addition of these bile acids to the diet caused an induction in the expression of the orphan receptor SHP, which is known to be regulated by the bile acid receptor FXR, and a corresponding suppression of Cyp7a1 (Fig. 3). We note that the lower dose of CDCA suppressed Cyp7a1 without increasing Shp expression, which may be explained by the existence of other SHP-independent mechanisms whereby bile acids repress Cyp7a1 (26, 27). At these doses, CA or CDCA had little or no effect on the expression of the PXR target genes Cyp3a11 or Cyp2b10 in wild-type mice. However, both bile acids efficiently suppressed the expression of these genes in Cyp27a1−/− mice. These data suggest that one or more bile acid intermediates are responsible for the elevated expression of PXR target genes in Cyp27a1−/− mice.
We next determined whether bile acid intermediates that are elevated in the Cyp27a1−/− mice serve as PXR agonists. Studies by Honda et al. (6) showed marked increases in the concentration of several bile acid precursors in hepatic microsomes from Cyp27a1−/− mice as compared with wild-type mice, including >10-fold increases in the concentrations of 7α-hydroxycholesterol (1) and 7α-hydroxy-4-cholesten-3-one (2), and an 80-fold increase in the concentration of 5β-cholestan-3α,7α,12α-triol (3) (Fig. 1). Cholestanol (5) levels were only modestly increased in these hepatic microsome preparations. The concentration of the putative intermediate 4-cholesten-3-one (4) was not determined (28, 29). We tested whether these compounds could activate PXR, CAR, FXR, or VDR in cell-based reporter assays (Fig. 4A), because these receptors have been implicated in CYP3A regulation (11, 18, 23). Both the mouse and human PXR were included in these studies because of the marked species differences in the ligand activation profiles (30).
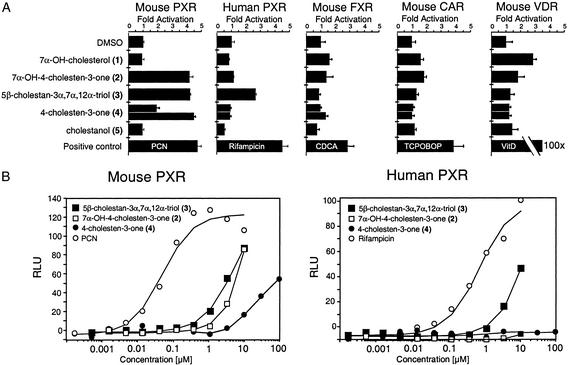
Figure 4.
Identification of bile acid intermediates that activate PXR. (A) CV-1 cells were cotransfected with mouse or human PXR and the XREM-luciferase reporter (17), mouse FXR and the FXREIBABPx3-tk-luciferase reporter (34), mouse CAR and DR3rat CYP3A1-tk-luciferase, or mouse VDR and mSPPx3-tk-luc (19). Cells were exposed to various ligands, all provided at 10 μM, except 4-cholesten-3-one (lower bar of the pair; 33 μM), CDCA (100 μM), TCPOBOP (0.5 μM), and 1,25-dihydroxyvitamin D3 (100 nM) for ≈40 h before assaying for luciferase activity. Transfection efficiency was corrected by analysis of β-galactosidase activity because of the cotransfection of a constitutive lacZ expression reporter. See Fig. 1 for the structures of bile acid intermediates. (B) Dose–response analysis of 5β-cholestan-3α,7α,12α-triol and 7α-OH-4-cholesten-3-one activation of mouse PXR and human PXR. Transfection experiments were performed as in A, except secreted placental alkaline phosphatase was used to correct for transfection activity and results are expressed as relative luciferase units (RLU).
Expression plasmids for the various receptors were transfected into CV-1 cells with luciferase reporter plasmids driven by cognate response elements. The cells were then exposed to 10 μM concentrations of 7α-hydroxycholesterol (1), 7α-hydroxy- 4-cholesten-3-one (2), 5β-cholestan-3α,7α,12α-triol (3), 4- cholesten-3-one (4), or cholestanol (5). Rifampicin (10 μM), PCN (10 μM), TCPOBOP (0.5 μM), CDCA (100 μM), and 1,25-dihydroxyvitamin D3 (100 nM) were included as positive controls for human PXR, mouse PXR, mouse CAR, mouse FXR, and mouse VDR, respectively.
Neither cholestanol (5) nor 7α-hydroxycholesterol (1) activated the human or mouse PXR. Notably, 7α-hydroxy-4-cholesten-3-one and 4-cholesten-3-one (compounds 2 and 4; Fig. 1) activated mouse PXR but did not activate human PXR at concentrations of 10–33 μM. In contrast, 5β-cholestan-3α,7α,12α-triol (3) activated both the mouse and human PXR. 7α-Hydroxy-4-cholesten-3-one, 5β-cholestan-3α,7α,12α-triol, and 4-cholesten-3-one (compounds 2, 3, and 4) had no activity on mouse or human FXR, CAR, VDR, liver X receptor α, or PPARα (Fig. 4A; data not shown). Similar results were obtained when these assays were performed with GAL4-receptor chimeras in which the DNA-binding domain of yeast GAL4 was fused to the ligand-binding domain of mouse or human PXR, thereby eliminating any species-specific effect of gene reporter context (data not shown). Full dose–response analysis showed that the triol (3) was slightly more potent on mouse PXR (half-maximal effective concentration, EC50 = 2.5 μM) than human PXR (EC50 = 5 μM) (Fig. 4B). Both 7α-hydroxy-4-cholesten-3-one (2) and 5β-cholestan-3α, 7α, 12α-triol (3) are present in the livers of Cyp27a1−/− mice at low micromolar concentrations (6), which is consistent with a role for these compounds as endogenous PXR agonists. The increased sensitivity of mouse PXR to 7α-hydroxy-4-cholesten-3-one (2), 4-cholesten-3-one (4), and 5β-cholestan-3α,7α,12α-triol (3) may explain in part why CYP3A expression is induced in Cyp27a1−/− mice but not in human CTX.
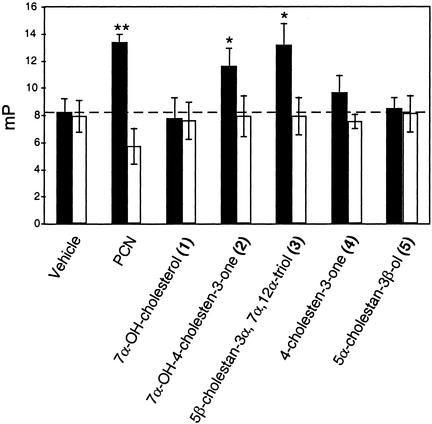
We tested whether these bile acid intermediates could bind directly to mouse PXR by using a fluorescence polarization assay (FPA) that detects ligand-dependent interactions between the ligand-binding domain of PXR and a fluorescein-labeled peptide derived from the steroid receptor coactivator 1 (SRC-1; ref. 20). The mouse PXR agonist PCN was included as a positive control. The results (Fig. 5) confirmed the findings obtained in the cell-based reporter assay and showed that 7α-hydroxy-4-cholesten-3-one (2), 4-cholesten-3-one (4), and 5β-cholestan-3α,7α,12α-triol (3) bound directly to mouse PXR.
Figure 5.
5β-Cholestan-3α,7α,12α-triol and 7α-hydroxy-4-cholesten-3-one directly bind mouse PXR as determined by fluorescence polarization assay. A fluorescein-tagged SRC1 peptide (ILRKLLQE) was incubated with bacterially expressed, purified GST-mPXR (black bars) or GST (white bars) in the presence of vehicle (DMSO), PCN (10 μM), or bile acid intermediates at 10 μM. Ligand-induced recruitment of the fluorescein-tagged SRC1 peptide was monitored by an increase in millipolarization fluorescence units (mP). Data shown represent the mean ± SEM of six samples.
In summary, our data indicate that mouse PXR is activated by a class of potentially toxic endobiotics, namely bile acid precursors. The relationship between bile acid synthesis and activation of PXR is further supported by the coordinate regulation of Cyp7a1 and Cyp3a11 under a variety of conditions. Thus, inhibition of bile acid biosynthesis after dietary manipulation (low-dose bile acid feeding) or gene disruption (Cyp7a1−/−) results in the repression of Cyp3a11 expression (Table 1). High-dose bile acid feeding results in the up-regulation of Cyp3a11, perhaps through the formation of the secondary bile acid, lithocholic acid, that can also activate PXR (22, 27, 31, 32). Conversely, Cyp3a11 expression is stimulated when Cyp7a1 expression is increased either pharmacologically (high-cholesterol diet, liver X receptor agonist) or through gene disruption (Cyp27a1−/−; Fxr−/−) (Table 1). The only mouse strains that do not display this coordinate regulation of Cyp7a1 and Cyp3a11 are the Pxr knockout and the liver-conditional Rxrα knockout (31), which are predicted outcomes because PXR signaling is lost in both strains.
Table 1.
Relationships between CYP7A1/CYP27A1 expression and CYP3A11
| Mouse model | mRNA expression
|
Ref. | ||
|---|---|---|---|---|
| CYP7A1 | CYP27A1 | CYP3A11 | ||
| Dietary manipulations | ||||
| 2% cholesterol feeding (7 d) | ↑ 2× | NC | ↑ ≈50% | J.J.R., unpublished |
| LXR agonist feeding (7 d) | ↑ 6× | NC | ↑ 2× | J.J.R., unpublished |
| Knockout mice | ||||
| Cyp27a1−/− | ↑ 10× | Absent | ↑ 10× | Current study |
| Cyp7a1−/− | Absent | NC | ↓ ≈50% | J.J.R., unpublished |
| Fxr−/− | ↑ 2× | ↓ ≈75% | ↑ | 31 |
| Pxr−/− | ↓ ≈50% | NC | ↑ 4× | 32 |
| Liver conditional Rxrα−/− | ↑ | ? | ↓ | 33 |
The hepatic mRNA expression levels for these three genes are expressed relative to wild-type, chow-fed mice.
References are given for studies that measured both CYP7A1 and CYP3A11. NC, no change; LXR, liver X receptor; ?, not determined.
The induction of CYP3A11 activity by bile acid precursors is likely to represent a host defense mechanism for solubilizing and removing these potentially harmful chemicals from the body. Notably, a corresponding induction of CYP3A enzyme activity does not appear to occur in human CTX (7). Our data support the hypothesis that this difference may be due to the fact that the human PXR is not as sensitive as the mouse receptor to the three identified bile acid intermediates. Future studies with Cyp27a1−/− mice lacking PXR or engineered to contain only the human PXR gene will be invaluable in assessing the relative contribution of this pathway to the elimination of bile acid precursors. Consistent with this hypothesis, mouse and human PXR have remarkably different specificities for xenobiotic ligands, which explains many of the differences in drug metabolism between the two species (11). With regard to CTX, the sensitivity of mouse (but not human) PXR to compound 4 is of particular interest. Compound 4 is a precursor to cholestanol (5) (Fig. 1; refs. 28 and 29), which accumulates to toxic levels in CTX patients but not in Cyp27a1−/− mice. These results suggest that differential activation of mouse PXR by compound 4 would prevent accumulation of toxic levels of cholestanol by inducing its catabolism through CYP3A11 or another PXR-inducible enzyme. Future studies on the substrate specificity of these metabolites by CYP3A or other PXR target gene products should address this issue. Because human CYP3A4 can also catalyze the hydroxylation of bile acid precursors (21), potent human PXR agonists that increase CYP3A4 activity may prove useful in the treatment of disease in which a toxic accumulation of bile acid intermediates occurs.
In closing, the finding that Cyp27a1−/− mice have altered expression of PXR target genes was integral to the discovery of bile acid precursors as endogenous PXR ligands. This approach to hunting for nuclear receptor ligands may become more common as gene expression profiles are characterized in additional mouse strains.
Acknowledgments
We thank Dr. Stephen D. Turley, Stephen Osterman, and Amanda Fletcher for assistance in design and execution of animal studies, and Dr. David Russell for review of the manuscript. K.C.G. and M.U. are Associates and D.J.M. is an Investigator of the Howard Hughes Medical Institute. This work was supported by funds to D.J.M. from the Howard Hughes Medical Institute and the Robert A. Welch Foundation.
Abbreviations
- CA
cholic acid
- CAR
constitutive androstane receptor (NR1I3)
- CDCA
chenodeoxycholic acid
- CTX
cerebrotendinous xanthomatosis
- CYP2B10
cytochrome P450 2B10
- CYP3A11
cytochrome P450 3A11
- CYP8B1
sterol 12α-hydroxylase
- CYP27A1
sterol 27-hydroxylase
- FXR
farnesoid X receptor (NR1H4)
- PCN
pregnenolone 16α-carbonitrile
- PXR
pregnane X receptor (NR1I2)
- SHP
small heterodimer partner (NR0B2)
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxyl)]benzene
- VDR
vitamin D receptor
References
- 1.Cali J J, Hsieh C L, Francke U, Russell D W. J Biol Chem. 1991;266:7779–7783. [PMC free article] [PubMed] [Google Scholar]
- 2.Björkhem I, Leitersdorf E. Trends Endocrinol Metab. 2000;11:180–183. doi: 10.1016/s1043-2760(00)00255-1. [DOI] [PubMed] [Google Scholar]
- 3.Björkhem I, Boberg K M, Leitersdorf E. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 2961–2988. [Google Scholar]
- 4.Salen G, Shefer S, Berginer V M. In: Metabolic Basis of Inherited Disease. Stanbury J B, Wyngaarden J B, Fredrickson D S, Brown M S, Goldstein J L, editors. New York: McGraw–Hill; 1981. pp. 713–730. [Google Scholar]
- 5.Rosen H, Reshef A, Maeda N, Lippoldt A, Shpizen S, Triger L, Eggertsen G, Björkhem I, Leitersdorf E. J Biol Chem. 1998;273:14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
- 6.Honda A, Salen G, Matsuzaki Y, Batta A K, Xu G, Leitersdorf E, Tint G S, Erickson S K, Tanaka N, Shefer S. J Lipid Res. 2001;42:291–300. [PubMed] [Google Scholar]
- 7.Honda A, Salen G, Matsuzaki Y, Batta A K, Xu G, Leitersdorf E, Tint G S, Erickson S K, Tanaka N, Shefer S. J Biol Chem. 2001;276:34579–34585. doi: 10.1074/jbc.M103025200. [DOI] [PubMed] [Google Scholar]
- 8.Repa J J, Lund E G, Horton J D, Leitersdorf E, Russell D W, Dietschy J M, Turley S D. J Biol Chem. 2000;275:39685–39692. doi: 10.1074/jbc.M007653200. [DOI] [PubMed] [Google Scholar]
- 9.Ishibashi S, Schwarz M, Frykman P K, Herz J, Russell D W. J Biol Chem. 1996;271:18017–18023. doi: 10.1074/jbc.271.30.18017. [DOI] [PubMed] [Google Scholar]
- 10.Salen G, Shefer S, Tint G S, Nicolau G, Dayal B, Batta A K. J Clin Invest. 1985;76:744–751. doi: 10.1172/JCI112030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin B, Redinbo M R, Kliewer S A. Annu Rev Pharmacol Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- 12.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterstrom R H, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz M, Russell D W, Dietschy J M, Turley S D. J Lipid Res. 1998;39:1833–1843. [PubMed] [Google Scholar]
- 14.Schwarz M, Wright A C, Davis D L, Nazer H, Björkhem I, Russell D W. J Clin Invest. 2000;106:1175–1184. doi: 10.1172/JCI10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones S A, Moore L B, Shenk J L, Wisely G B, Hamilton G A, McKee D D, Tomkinson N C, LeCluyse E L, Lambert M H, Willson T M, et al. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 16.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin B, Hodgson E, Liddle C. Mol Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 18.Makishima M, Lu T T, Xie W, Whitfield G K, Domoto H, Evans R M, Haussler M R, Mangelsdorf D J. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 19.Noda M, Vogel R L, Craig A M, Prahl J, DeLuca H F, Denhardt D T. Proc Natl Acad Sci USA. 1990;87:9995–9999. doi: 10.1073/pnas.87.24.9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz J R, Tu H, Luk A, Repa J J, Medina J C, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf D J, et al. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furster C, Wikvall K. Biochim Biophys Acta. 1999;1437:46–52. doi: 10.1016/s0005-2760(98)00175-1. [DOI] [PubMed] [Google Scholar]
- 22.Xie W, Radominska-Pandya A, Shi Y, Simon C M, Nelson M C, Ong E S, Waxman D J, Evans R M. Proc Natl Acad Sci USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maglich J M, Stoltz C M, Goodwin B, Hawkins-Brown D, Moore J T, Kliewer S A. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 24.Xie W, Barwick J L, Simon C M, Pierce A M, Safe S, Blumberg B, Guzelian P S, Evans R M. Genes Dev. 2000;14:3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smirlis D, Muangmoonchai R, Edwards M, Phillips I R, Shephard E A. J Biol Chem. 2001;276:12822–12826. doi: 10.1074/jbc.M005930200. [DOI] [PubMed] [Google Scholar]
- 26.Kerr T A, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell D W, Schwarz M. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Lee Y K, Bundman D, Han Y, Thevananther S, Kim C S, Chua S S, Wei P, Heyman R A, Karin M, Moore D D. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 28.Björkhem I, Karlmar K E. Biochim Biophys Acta. 1974;337:129–131. doi: 10.1016/0005-2760(74)90046-0. [DOI] [PubMed] [Google Scholar]
- 29.Skrede S, Björkhem I, Buchmann M S, Hopen G, Fausa O. J Clin Invest. 1985;75:448–455. doi: 10.1172/JCI111719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore L B, Maglich J M, McKee D D, Wisely B, Willson T M, Kliewer S A, Lambert M H, Moore J T. Mol Endocrinol. 2002;16:977–986. doi: 10.1210/mend.16.5.0828. [DOI] [PubMed] [Google Scholar]
- 31.Schuetz E G, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim R B, Ramachandran V, Komoroski B J, et al. J Biol Chem. 2001;276:39411–39418. doi: 10.1074/jbc.M106340200. [DOI] [PubMed] [Google Scholar]
- 32.Staudinger J L, Goodwin B, Jones S A, Hawkins-Brown D, MacKenzie K I, LaTour A, Liu Y, Klaassen C D, Brown K K, Reinhard J, et al. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y J, An D, Cai Y, Repa J J, Hung-Po Chen T, Flores M, Postic C, Magnuson M A, Chen J, Chien K R, et al. Mol Cell Biol. 2000;20:4436–4444. doi: 10.1128/mcb.20.12.4436-4444.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grober J, Zaghini I, Fujii H, Jones S A, Kliewer S A, Willson T M, Ono T, Besnard P. J Biol Chem. 1999;274:29749–29754. doi: 10.1074/jbc.274.42.29749. [DOI] [PubMed] [Google Scholar]