Abstract
RNA interference is a cellular process of gene silencing in which small duplexes of RNA specifically target a homologous sequence for cleavage by cellular ribonucleases. The introduction of ≈22-nt small interfering RNAs (siRNAs) into mammalian cells can specifically silence cellular mRNAs without induction of the nonspecific IFN responses that are activated by longer RNA duplexes. We investigate in this article whether siRNAs can also silence the expression of the cytoplasmically replicating hepatitis C virus (HCV) RNAs by using a replicon system that supports robust HCV replication, but not the production of infectious virions. We report the efficient silencing of both cellular lamin A/C and HCV RNAs in Huh-7 hepatoma cell lines supporting HCV replication. Silencing of HCV RNAs was dose dependent and specific, inasmuch as two HCV variants that differ by 3 nt within the target sequence were only silenced by the exact homologous sequence for each. siRNAs designed to target HCV RNA triggered an exponential decrease in HCV RNA, resulting in an 80-fold decrease in HCV RNA after 4 days. The introduction of siRNAs into cells with established HCV replication cured >98% of these cells of detectable HCV antigen and replication-competent HCV RNAs. These data support the principle of siRNA-based HCV antiviral therapy.
Keywords: RNA interference‖antiviral agents
Hepatitis C virus (HCV) replication occurs in the cytoplasm and is associated with membranes that may be derived from the endoplasmic reticulum (1, 2). The genomic HCV RNA is translated to produce an ≈3,000-aa polypeptide that is processed into at least 10 proteins. The nonstructural (NS) proteins 3, 4A, 4B, 5A, and 5B form a replicase complex that promotes transcription of a genomic (−) strand intermediate. This serves as a template for production of (+) strands that are either translated or packaged into virions as genomic RNAs (reviewed in ref. 3). Although HCV is notoriously difficult to grow in vitro, systems have been developed that sustain efficient replication of HCV in cell culture. These are replicon-based strategies in which the HCV internal ribosome entry site drives expression of the neomycin phosphotransferase gene, whereas a heterologous internal ribosome entry site from encephalomyocarditis virus promotes translation of the HCV polypeptide (4, 5). Initially, subgenomic replicons that expressed only NS protein were constructed; however, replicons have since been constructed to express the entire HCV polyprotein (6, 7). These are useful for dissection of HCV replication and protein function; however, the production of infectious virus in vitro has yet to be documented.
In addition to molecular studies of HCV replication, replicons provide an excellent system to evaluate HCV antiviral agents in cell culture (reviewed in ref. 8). HCV is a major public health problem, with 170 million chronically infected people throughout the world (9). Chronically infected individuals are a reservoir for new infections as well as being at risk for progression to cirrhosis and hepatocellular carcinoma. Current therapeutic regimens have limited efficacy against certain HCV genotypes (10, 11). Although new antiviral agents are in development, an important lesson from the therapy of other viral infections, such as HIV, is that multiple drug targets may be needed to limit the emergence of drug-resistant variants.
Cellular mechanisms of gene silencing by targeting RNA transcripts exist in both plants and animals, and the molecular machinery seems to be ancient and highly conserved (reviewed in refs. 12 and 13). In Drosophila, larger double-stranded RNAs are processed into short ≈22-mers by an RNase III-like enzyme, dicer (14). These small interfering RNAs (siRNAs) become unwound and associate with an activated RNA-induced silencing complex (15). The single-stranded siRNA then acts as a guide to substrate selection, leading to the cleavage by dicer of a homologous target RNA molecule (16).
Although initial studies of RNA interference (RNAi) focused on cellular mRNA targets, evidence suggests that it may also target viral RNAs. The related process of posttranscriptional gene silencing is a well-documented antiviral system in plants (17–19), and RNAi has recently been shown to have antiviral function in animal cells (20). These processes silence the expression of transposons, repetitive elements, and viruses (reviewed in ref. 21). Viral inhibitors of RNAi have been isolated from plant viruses (22–24) and also the animal virus, flock house virus (20, 25). However, it has not been shown whether RNAi is an antiviral defense in mammals or, conversely, whether viruses that infect mammals express functions that inhibit RNAi. The goal of this study was to assess the efficacy of RNAi against HCV RNAs in a robust cell culture replication system. We show that siRNAs efficiently eliminate replicating HCV RNAs from human hepatoma cells.
Materials and Methods
Cells and HCV Replicons.
Huh-7.5 cells are a subline derived from Huh-7 hepatoma cells (46). They are highly permissive for the initiation of HCV replication. Huh-7.5 cell populations containing the following HCV replicons were used. HCV-Con1 is a full-length genotype 1b replicon with the highly adaptive serine to isoleucine substitution at amino acid 2204 of the HCV polypeptide. This has been referred to previously as Con1/Fl-neo(S2204I) (46). Pol− is a replication-defective derivative of HCV-Con1 that contains a GDD to AAG mutation in the NS5B polymerase (5). Cells were maintained in DMEM supplemented with nonessential amino acids and 10% FBS. Cells containing replicons are maintained in the above media plus 0.75 mg/ml G418.
Construction of the HCV-C/LB Replicon.
HCV-C/LB is a chimeric HCV genotype 1b replicon consisting of sequences from Con1 and an independent 1b isolate called LB (S. Gagneten and S. Feinstone, personal communication). Briefly, a full-length HCV cDNA was cloned into pRS424 by using conventional RT-PCR methods from plasma of a patient chronically infected with HCV. A consensus clone was generated by comparison and repairing of sequences from several independent clones and is termed LB (kindly provided by S. Feinstone, U.S. Food and Drug Administration, Rockville, MD). HCV-C/LB was generated by replacement of the NsiI–BglII fragment, containing part of NS3, 4A, 4B, 5A, and 5B from Con1/SG-neo (5, 25a), with the corresponding fragment from LB. HCV-C/LB contains the adaptive S2204I mutation.
RNAi Assay.
siRNAs were designed as follows, and for each, the sense-strand sequence is described (a complementary oligonucleotide was synthesized for each). Lamin A/C siRNA (siLAM), 5′-aacuggacuuccagaagaacaTT; irrelevant siRNA (siIRR), 5′-aaggacuuccagaagaacaucTT; HCV siRNA (siHCV), 5′-aaccucaaagaaaaaccaaacTT; HCV-Con1 siRNA (siCon1), 5′-aaggugcuuguggauauuuugTT; and HCV-C/LB siRNA (siC/LB), 5′-aaggcgcuuguggacauucugTT. Chemically synthesized RNA oligos were annealed, deprotected, and desalted as recommended by the manufacturer (Dharmacon, Lafayette, CO). Varying amounts (50–4,000 pmol) of RNA duplexes in annealing buffer (100 mM potassium acetate/30 mM Hepes-KOH, pH 7.4/2 mM magnesium acetate) were electroporated with 2.5 × 106 Huh7 cells in 0.4 ml of PBS, pH 7.4. Electroporation conditions were 5 pulses of 900 V with 1-s intervals on a BTX electroporator. Cells were plated and maintained in appropriate conditions. The HCV-specific siRNAs described above were all effective at silencing HCV expression, and no other sequences have been tested.
RNA Quantitation.
Total RNA was harvested with Trizol and purified as recommended by the manufacturer (Invitrogen); 0.75 μg of DNaseI-treated total RNA was reverse-transcribed with Superscript II (Invitrogen) for 1 h at 42°C. cDNA synthesis was primed with either the HCV-specific primer 5′-cacccctgctccataacc for HCV cDNA or oligo(dT) for GAPDH cDNA. The reverse transcriptase was heat-inactivated at 80°C for 20 min; 1/20 of the cDNA mix was mixed with an equal volume of 2× Sybr green master mix (Applied Biosystems). PCR was performed with primers flanking the siCon1/siC/LB site in NS4B with 5′-cgctgcttctgctttcg and 5′-cacccctgctccataacc. Values were normalized to that of GAPDH, which was amplified with primers 5′-caagctgtgggcaaggt and 5′-ggaaggccatgccagtga. PCR conditions were as follows: 50°C, 2 min; 95°C, 10 min (95°C, 15 s; 60°C, 1 min) × 40 cycles; 95°C, 15 s; 60°C, 15 s; and 95°C, 15 s. Results were analyzed with SDS 2.0 software from Applied Biosystems.
Results
Efficient Silencing of Lamin A/C in Huh-7.5 Cells That Contain Replicating HCV-Con1 RNAs.
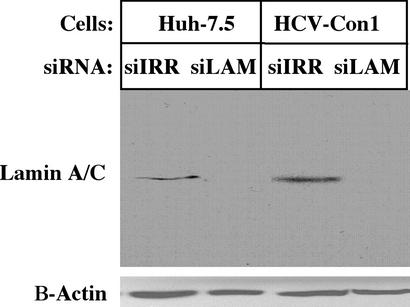
Because HCV replication in cell culture is limited to Huh-7 cells and their derivatives, we first verified that siRNAs could silence the expression of a host gene in this cellular environment. For this purpose, lamin A/C was chosen as a model, as silencing by lamin A/C siRNAs in mammalian cells has been reported (26). RNA duplexes specific for lamin A/C (siLAM) or irrelevant siRNA duplexes (siIRR) were transfected into Huh-7.5 cells under optimized conditions. Two days later, cells were harvested and examined for lamin A/C protein levels (Fig. 1). Lamin A/C protein was undetectable after electroporation of Lamin A/C siRNA, whereas the irrelevant siIRR had no effect. β-Actin protein levels were unchanged. Because some plant and animal viruses express functions that inhibit RNAi, we also tested the efficiency of lamin A/C silencing in Huh-7.5 cells containing full-length replicating HCV RNAs (HCV-Con1). The experiment was repeated as above, and lamin A/C expression was silenced in cells containing the replicating HCV-Con1 RNAs (Fig. 1). We conclude that a host gene can be efficiently silenced in Huh-7.5 cells after introduction of siRNAs and that HCV functions expressed by full-length replicons do not abrogate this process.
Figure 1.
Efficient siRNA-mediated silencing of lamin A/C expression in Huh-7.5 cells containing HCV-Con1 replicons. Naïve Huh-7.5 cells and Huh-7.5 cells containing replicating HCV-Con1 RNAs were electroporated with 1,000 pmol of either siIRR or lamin A/C siRNAs (siLAM) and plated. Two days later, protein lysates were electrophoresed on SDS/10% PAGE gels, transferred to nitrocellulose, blocked, and probed with rabbit polyclonal anti-lamin A antiserum 2032 (Cell Signaling). This was followed with secondary goat anti-rabbit IgG conjugated to horseradish peroxidase. Lamin A/C was detected by chemiluminescence. The nitrocellulose was then blocked and reacted to mouse anti-B-actin (Sigma), followed by goat anti-mouse conjugated to horseradish peroxidase. β-Actin was detected by chemiluminescence.
The Silencing of HCV Antigen Expression in Huh-7.5 Cells Is Dose-Dependent and Sequence-Specific.
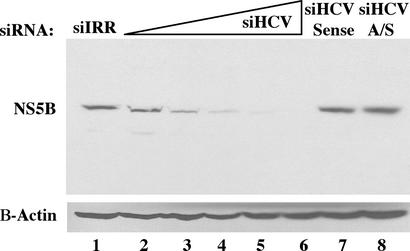
We next tested whether HCV antigen expression could be silenced by using HCV-specific siRNAs. An siRNA duplex, siHCV, which targets the 5′ core region just downstream of the ribosomal toe-print straddling the initiator methionine, was designed (27). The integrity of this region is associated with optimal translation of the HCV polypeptide, and its sequence is maintained in all replicons used in this study (28–30). Huh-7.5 cells containing HCV-Con1 replicons were electroporated with increasing concentrations of siHCV and plated in the absence of G418 selection for 4 days. Protein lysates were made, and immunoblots were performed with mAbs specific for the HCV NS5B. NS5B levels decreased after electroporation of 50 pmol of siHCV, and this decrease continued in a dose-dependent fashion (Fig. 2). Electroporation of 4,000 pmol of siHCV resulted in NS5B levels that were below the sensitivity of detection, whereas 4,000 pmol of siIRR had no effect. Silencing was optimal with an siRNA duplex, as electroporation of either the sense or antisense strand siHCV alone did not greatly affect NS5B expression.
Figure 2.
Dose-dependent silencing of NS5B expression by HCV-specific siRNAs. Huh-7.5 cells containing replicating HCV-Con1 RNAs were electroporated with the appropriate siRNA, plated, and maintained for 4 days. Protein lysates were electrophoresed on SDS/10% PAGE gels, transferred to nitrocellulose, probed with mouse monoclonal anti-NS5B, 5B.3B.1.5.3 (45), and processed as in Fig. 1. The nitrocellulose filter was then reprobed with mouse anti-β-actin as in Fig. 1. The siRNAs used are as follows: lane 1, 4,000 pmol siIRR; lane 2, 50 pmol siHCV duplex; lane 3, 150 pmol siHCV; lane 4, 450 pmol siHCV; lane 5, 1,350 pmol siHCV; lane 6, 4,000 pmol siHCV; lane 7, 4,000 pmol sense strand-only siHCV RNA; and lane 8, 4,000 pmol antisense strand-only siHCV RNA.
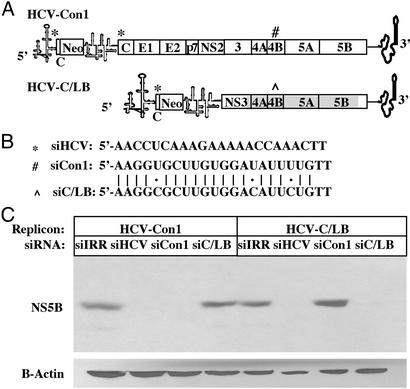
We next tested the specificity of HCV silencing by comparing the ability to silence two related replicons, the aforementioned HCV-Con1 and a replicon termed HCV-C/LB (described in Materials and Methods and in Fig. 3A). HCV-C/LB is a chimeric replicon in which Con1 sequences, including part of NS3, all of NS4A, 4B, 5A, and part of NS5B, were replaced with the corresponding region from the genotype 1b LB strain. HCV-Con1 and HCV-C/LB have similar levels of replication and protein expression (A.G. and C.M.R., unpublished data). Two siRNA duplexes differing at only three nucleotide positions in the same position of the NS4B gene were designed for each replicon (Fig. 3B). The siRNA duplex termed siCon1 is homologous to the HCV-Con1 sequence, whereas the siRNA duplex termed siC/LB is specific to the HCV-C/LB sequence. The siHCV is homologous to sequences present in both replicons. Huh-7.5 cells containing either the HCV-Con1 replicon or the HCV-C/LB replicon were electroporated with siIRR, siHCV, siCon1, and siC/LB and maintained for 4 days in the absence of selection. siHCV effectively silenced NS5B expression in both replicon-containing cell lines, whereas the irrelevant siIRR had no effect (Fig. 3C). The siCon1 silenced NS5B expression in the HCV-Con1 cells but not in the HCV-C/LB cells. The converse was true of siC/LB. Taken together, these results show that both replicon sequences can be silenced after introduction of homologous siRNAs but are resistant to silencing from siRNAs that differ in 3 of 21 nucleotide positions. The sequence specificity of silencing confirms that reduced HCV antigen expression is not resulting from a nonspecific activation of the IFN system by the double-stranded siRNAs. This is in agreement with other findings that double-stranded RNAs <30 nt do not induce IFN responses (31, 32).
Figure 3.
HCV genotype-specific silencing of NS5B expression by siRNAs. (A) HCV-Con1 and HCV-C/LB replicon structures. The shaded box in HCV-C/LB NS3, 4A, 4B, 5A, and 5B highlights the region in which HCV-Con1 sequences were replaced with the corresponding sequences from the LB isolate. *, #, and HCV-Con1 or ^ denote sites of siHCV, siCon1, and siC/LB homology, respectively. (B) Sequences for siHCV, siCon1, and siC/LB. siHCV sequence, located in the core, is conserved in both strains, whereas the siCon1 and siC/LB, located in NS4B, differ by 3 nt. (C) Huh7.5 cells containing either HCV-Con1 or HCV-C/LB replicons were electroporated with either 4 or 20 nmol, respectively, of siIRR, siHCV, siCon1, or siC/LB, plated, and maintained for 4 days. Immunoblots were performed as in Fig. 2.
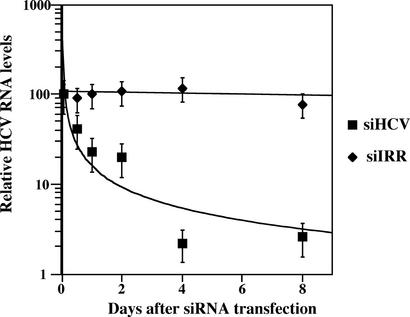
The effect of siRNA silencing on HCV RNA levels was next investigated by using real-time RT-PCR. Huh-7.5 cells with replicating HCV-Con1 were electroporated with either siIRR or siCon1, and total RNAs were harvested at 12, 24, 48, 96, and 192 h after electroporation. RT-PCR on total RNA was performed with HCV-specific primers flanking the expected site of RNA cleavage, and cDNAs were quantitated. These values were normalized to GAPDH levels, and the relative HCV/GAPDH ratio at time = 0 was then normalized to 100 (Fig. 4). The introduction of siIRR and maintenance of cells in the absence of selection had little effect on relative HCV RNA levels. This continued high level of HCV replication in the absence of selection is similar to previously reported results in which HCV replicon RNAs were detected for at least 10 months after the removal of selective pressure (33). Introduction of siCon1 into Con1 cells triggered an exponential decrease in HCV RNAs. This decrease was evident within 12 h of siHCV transfection. Four days after transfection, the total HCV RNA levels had dropped 80-fold, and this level was still maintained after 8 days.
Figure 4.
siRNA triggered a decrease in HCV RNAs. HCV-Con1 cells were electroporated with 4 nmol of either siIRR or siCon1 and plated, and total RNA was harvested 12, 24, 48, 96, and 192 h after electroporation. RNA was reverse-transcribed with either oligo(dT) or HCV-specific primers, and RNA levels were quantified with real-time PCR by using primers that flank the siRNA target sequence as described in Materials and Methods. HCV RNA values were normalized first to GAPDH RNA values and then to the ratio of HCV/GAPDH immediately after electroporation of siRNAs (t = 0). The ratio of HCV/GAPDH at t = 0 is given the relative value of 100, and subsequent time point values reflect changes in the HCV/GAPDH ratio.
siRNA-Mediated Curing of Persistently Replicating HCV-Con1 RNAs Occurs in >98% of Cells.
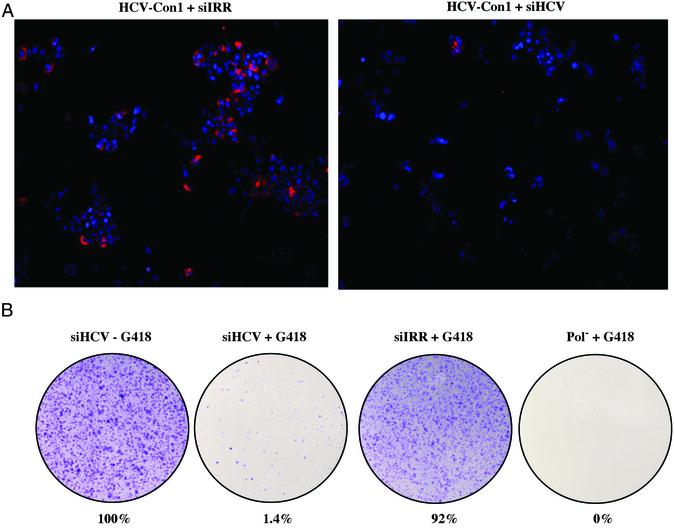
Fig. 4 shows that silencing resulted in ≈2-log decrease in HCV RNA levels. It was not clear whether this reflected a general reduction in all cells or whether a small percentage of cells maintained HCV RNA and protein expression, whereas the vast majority were “cured” of HCV. We next examined the silencing of stably replicating HCV RNAs on a single-cell level in two experiments: (i) NS5A expression by immunofluorescence and (ii) the formation of G418-resistant colonies. In the first experiment, Huh-7.5 cells containing HCV-Con1 were electroporated with either siIRR or siHCV, plated, and maintained for a total of 5 days in the absence of G418 selection. Cells were washed, fixed, probed for NS5A expression (red), counterstained with Hoechst dye to label the nuclei (blue), and viewed by microscopy (Fig. 5A). Cells treated with siIRR showed typical reticular NS5A staining in the cytoplasm of ≈40% of cells, consistent with previous observations (2). In cells treated with siHCV, NS5A staining could be detected in only <1% of cells. This strong fluorescence in a small percentage of cells is consistent with siRNA clearance of HCV below detectable levels in the majority of cells, with a small subset of cells maintaining HCV antigen expression.
Figure 5.
Curing of Huh-7.5 cells containing replicating HCV-Con1 RNAs with siRNAs. (A) HCV-Con1 cells were electroporated with 4 nmol siIRR or siHCV, plated, and maintained for 3 days. Cells were then trypsinized, plated in eight-well chamber slides, and maintained for 2 days. Cells were washed, fixed in methanol at −80°C, and probed with anti-NS5A mAb (Maine Biotechnology, Portland, ME). Cells were then reacted with goat anti-mouse IgG conjugated to Rhodamine for 1 h followed by Hoechst dye for 20 min. Slides were mounted, viewed by microscopy with a Nikon TE200, and captured with spot ccd software. (B) HCV-Con1 cells were electroporated with 4 nmol siHCV or siIRR, plated, and maintained for 1 week. Cells were then trypsinized, and 105 cells were plated in 100-mm2 dishes and maintained either in the presence or absence of 0.75 mg/ml G418 for 10 days. Cells were washed, fixed in 7% paraformaldehyde, stained with crystal violet, and counted. Values were normalized to cells electroporated with HCV siRNAs and maintained in the absence of G418. As a control, naive Huh7.5 cells were electroporated with Pol−, an HCV-Con1 replication defective mutant, and maintained as above in the presence of G418. Representative data are from three independent experiments.
To test this hypothesis functionally, we next assessed the efficacy of siHCV in curing Huh-7.5 cells of replicating full-length HCV. The ability to form G418-resistant colonies depends on the persistent expression of neomycin phosphotransferase from replicating HCV RNAs. As such, the formation of G418-resistant foci is an indirect measure of HCV replication competence at a single cell level. Huh-7.5 cells containing HCV-Con1 RNAs were electroporated with either siIRR or siHCV, plated, and maintained in the absence of G418 selection. After 1 week, cells were trypsinized and 105 cells were plated in duplicate 100-mm2 dishes and maintained with or without G418 selection for 10 days. Cells were then washed, fixed, stained with crystal violet, and counted. As a control, naïve Huh-7.5 cells were electroporated with a polymerase-defective mutant replicon (Pol−) that is replication-incompetent (5). The siHCV-transfected cells maintained in the absence of selection were normalized to 100% (Fig. 5B). Cells treated with siHCV formed G418-resistant colonies in only 1.4% of cells. In contrast, cells electroporated with siIRR formed G418-resistant colonies in 92% of cells. These results support the conclusion that HCV-specific siRNAs mediate the clearance of replicating HCV RNAs in >98% of cells to levels below the sensitivity of detection. It is unclear whether the remaining G418-resistant cells (i) received insufficient amounts of siRNAs, (ii) contain HCV replicon escape mutants that are resistant to siRNAs, or (iii) have cellular defects in the RNAi machinery.
Discussion
The data in this study suggest that siRNAs can elicit an anti-HCV response in cell culture. Cells harboring actively replicating HCV were sensitive to siRNA targeting of both cellular (lamin A/C) and viral (HCV) transcripts. We have yet to find evidence that HCV expresses functions that inhibit silencing after the introduction of siRNAs in cell culture. While this manuscript was in preparation, it was reported that both HIV and poliovirus are sensitive to RNAi in cell culture (34–38). This finding lends credence to the principle of siRNAs as a general antiviral and also questions whether RNAi is a natural host defense against viruses. The sensitivity of these viruses to siRNAs suggests that mammalian RNAi does not exert evolutionary antiviral pressure in nature, at least in the case of these three viruses. However, it remains possible that these viruses may inhibit the processing of larger dsRNAs into ≈22-mer siRNAs in vivo, a step that was bypassed by the introduction of chemically synthesized siRNAs in these experiments. The general activity of the RNAi machinery in mammals is of considerable interest. It has been shown that transgene silencing can be induced by the introduction of chemically synthesized siRNAs into adult mouse liver (39). This finding, in combination with the results presented in this study, suggests that the RNAi machinery in mammals may be activated to perform anti-HCV functions even if that is not their natural function. Mice that contain chimeric mouse/human livers support HCV infection and replication (40) and may provide a model to test the efficacy of siRNAs against HCV in vivo, in a fully infectious system.
The specific silencing by RNAi without induction of nonspecific IFN responses is attractive from a therapeutic standpoint. However, the specificity of silencing distinct HCV genotype 1b sequences (Fig. 3) highlights the possibility of reduced efficacy against viral escape mutants. In this study, target sequences that differed by 3 nt from the siRNA could not be silenced. In another study, poliovirus escape mutants rapidly emerge after siRNA treatment (35). One advantage of HCV as a model for using RNAi as an antiviral agent is that HCV replication is not as robust as poliovirus. The average HCV RNA copy number in HCV-Con1 cells is ≈9,000 (P. Balfe, K. J. Blight, P. Lopez, C.M.R., and J. A. McKeating, unpublished data), whereas poliovirus copy number is ≈100,000 per cell.
The targeted HCV sequences in this study are not completely conserved in different HCV genotypes. They are contained in the coding region, with no predicted RNA structure that would restrict genetic variation at the nucleotide level. In essence, the sites were chosen precisely for their sequence variation so that we could test the specificity of silencing different replicons. Given the proof of principle presented in this article, a logical next step would be the identification of optimal target sequences for therapeutic evaluation. Highly conserved nucleotide sequences exist within the 5′ and 3′ UTRs in addition to certain parts of the coding region within core and NS5B. One can also envision a degenerate siRNA set against an invariable 7-aa sequence, which is common in the HCV genome. Multiple siRNAs targeting evolutionarily restricted HCV RNA sequences may limit the emergence of escape mutants. Recently developed stable RNAi silencing vectors (37, 41–44) can be used to generate cell lines that constitutively express high levels of siRNAs that target HCV. These cell lines could be used to address the question of viral escape in cell culture, as caveats associated with the transfection efficiency of chemically synthesized siRNAs into cells are avoided.
Variations of these stable RNAi silencing vectors may also function in gene therapy wherein numerous modes exist for expressing therapeutic RNAs, such as antisense RNAs or ribozymes. A goal would be the prevention of productive HCV infection by siRNA expression in liver cells. This would be particularly useful in the transplant setting wherein newly transplanted livers are highly susceptible to reinfection with HCV.
The mechanism of anti-HCV activity is of interest. The nascent HCV polyprotein promotes association of the polysomal genome RNA with the rough endoplasmic reticulum. Proposed sites of HCV RNA replication are membranous webs composed of endoplasmic reticulum or endoplasmic reticulum-derived membranes (1, 2). It remains to be determined whether polysomal RNAs and RNAs in the replication complex, as either templates or nascent products, are similarly accessible to RNAi-mediated cleavage. Alternatively, efficient cleavage of HCV mRNA may be sufficient to clear cells of replicating RNA if one or more of the replicase components are limiting.
In addition to therapeutic potential, these data highlight the use of RNAi as a molecular tool for dissecting gene function and significance. The effective silencing of lamin A/C in HCV-replicating cells reveals the potential of RNAi as a tool to investigate HCV–host interactions, and in particular, the significance of the many reported interactions for HCV replication.
Acknowledgments
We acknowledge Paola Concari and The Rockefeller University gene array facility for assistance with real-time PCR, Steve Feinstone for his generous gift of the HCV genotype 1b LB strain, and Darius Moradpour for the mouse monoclonal anti-NS5B, 5B.3B.1.5.3. We thank Joseph Marcotrigiano, Julie Stacey, and Holly Hanson for critical reading of the manuscript. This work was supported in part by Public Health Service Grants CA57973 and AI40034 and the Greenberg Medical Research Institute. G.R. is supported by postdoctoral fellowship American Cancer Society Grant PF-02-016-01-MBC. A.G. is supported by a postdoctoral fellowship grant from the Cancer Research Institute.
Abbreviations
- RNAi
RNA interference
- siRNAs
small interfering RNAs
- HCV
hepatitis C virus
- NS
nonstructural
- siIRR
irrelevant siRNA
- siHCV
HCV siRNA
- siCon1
HCV-Con1 siRNA
- siC/LB
HCV-C/LB siRNA
Note Added in Proof.
While this manuscript was in review, it was reported that phosphorylated single-stranded antisense siRNAs are as efficient as double-stranded RNAs in silencing (47). We have repeated the experiment shown in Fig. 2 and shown (unpublished data) that in vitro phosphorylated antisense siRNAs alone efficiently silenced NS5B expression.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Egger D, Wolk B, Gosert R, Bianchi L, Blum H E, Moradpour D, Bienz K. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mottola G, Cardinali G, Ceccacci A, Trozzi C, Bartholomew L, Torrisi M R, Pedrazzini E, Bonatti S, Migliaccio G. Virology. 2002;293:31–43. doi: 10.1006/viro.2001.1229. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach B D, Rice C M. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 1. Philadelphia: Lippincott; 2001. pp. 991–1041. [Google Scholar]
- 4.Lohmann V, Korner F, Koch J O, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 5.Blight K J, Kolykhalov A A, Rice C M. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 6.Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, Strand D, Bartenschlager R. J Virol. 2002;76:4008–4021. doi: 10.1128/JVI.76.8.4008-4021.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda M, Yi M, Li K, Lemon S M. J Virol. 2002;76:2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randall G, Rice C M. Curr Opin Infect Dis. 2001;14:743–747. doi: 10.1097/00001432-200112000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Anonymous. Wkly Epidemiol Rec. 1997;72:341–344. [Google Scholar]
- 10.Zeuzem S, Feinman S V, Rasenack J, Heathcote E J, Lai M Y, Gane E, O'Grady J, Reichen J, Diago M, Lin A, et al. N Engl J Med. 2000;343:1666–1672. doi: 10.1056/NEJM200012073432301. [DOI] [PubMed] [Google Scholar]
- 11.Heathcote E J, Shiffman M L, Cooksley W G, Dusheiko G M, Lee S S, Balart L, Reindollar R, Reddy R K, Wright T L, Lin A, et al. N Engl J Med. 2000;343:1673–1680. doi: 10.1056/NEJM200012073432302. [DOI] [PubMed] [Google Scholar]
- 12.Bosher J M, Labouesse M. Nat Cell Biol. 2000;2:E31–E36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- 13.Hannon G J. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Nykanen A, Haley B, Zamore P D. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 16.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty W G, Lindbo J A, Smith H A, Parks T D, Swaney S, Proebsting W M. Mol Plant Microbe Interact. 1994;7:544–552. [PubMed] [Google Scholar]
- 18.Ruiz M T, Voinnet O, Baulcombe D C. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Kaff N S, Covey S N, Kreike M M, Page A M, Pinder R, Dale P J. Science. 1998;279:2113–2115. doi: 10.1126/science.279.5359.2113. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Li W X, Ding S W. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 21.Voinnet O, Pinto Y M, Baulcombe D C. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D C. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Li W X, Ding S W. Curr Opin Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 24.Voinnet O, Lederer C, Baulcombe D C. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach B D, Rice C M. Mol Cell. 2002;9:925–927. doi: 10.1016/s1097-2765(02)00539-7. [DOI] [PubMed] [Google Scholar]
- 26.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 27.Kolupaeva V G, Pestova T V, Hellen C U. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda M, Ping L H, Rijnbrand R C, Amphlett E, Clarke B, Rowlands D, Lemon S M. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 29.Lu H H, Wimmer E. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minks M A, West D K, Benvin S, Baglioni C. J Biol Chem. 1979;254:10180–10183. [PubMed] [Google Scholar]
- 32.Manche L, Green S R, Schmedt C, Mathews M B. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. J Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coburn G A, Cullen B R. J Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gitlin L, Karelsky S, Andino R. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 36.Jacque J M, Triques K, Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee N S, Dohjima T, Bauer G, Li H, Li M J, Ehsani A, Salvaterra P, Rossi J. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 38.Novina C D, Murray M F, Dykxhoorn D M, Beresford P J, Riess J, Lee S K, Collman R G, Lieberman J, Shankar P, Sharp P A. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 39.McCaffrey A P, Meuse L, Pham T T, Conklin D S, Hannon G J, Kay M A. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 40.Mercer D F, Schiller D E, Elliott J F, Douglas D N, Hao C, Rinfret A, Addison W R, Fischer K P, Churchill T A, Lakey J R, et al. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 41.Brummelkamp T R, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 42.McManus M T, Petersen C P, Haines B B, Chen J, Sharp P A. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paddison P J, Caudy A A, Bernstein E, Hannon G J, Conklin D S. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester W C. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moradpour D, Bieck E, Hugle T, Wels W, Wu J Z, Hong Z, Blum H E, Bartenschlager R. J Biol Chem. 2002;277:593–601. doi: 10.1074/jbc.M108748200. [DOI] [PubMed] [Google Scholar]
- 46.Blight K J, McKeating J A, Rice C M. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]