Abstract
The sooty mangabey (SM) (Cercocebus atys) is the natural host of a simian immunodeficiency virus, termed SIVsm, which gave rise to human immunodeficiency virus type 2. Data on the geographic distribution, prevalence, and genetic diversity of SIVsm in the wild remains limited. To address this issue, noninvasive strategies based on screening SM fecal and urine specimens for SIVsm-specific antibodies and virion RNA (vRNA) were developed, and the results were correlated with viral loads in plasma. Twenty-three SIVsm-infected and 27 uninfected SMs were evaluated. Time-matched urine, fecal and plasma samples were collected over a 2-month period from 16 captive naturally infected SMs. The remaining 7 infected and 27 uninfected SMs were sampled once. Each specimen was subjected to enhanced chemiluminescence-Western blot analysis and nested reverse transcriptase (RT) PCR. The results showed that urine was highly sensitive (96%) and specific (100%) for detection of SIVsm antibodies, while fecal detection was much less sensitive (16%). Conversely, vRNA detection was more sensitive in feces (50%) than in urine (2%) samples. Fecal-vRNA detection correlated with viral loads in plasma (P < 0.002). SMs with detectable fecal vRNA had a mean viral load in plasma of 458,006 copies/ml, while those with undetectable fecal vRNA had a mean viral load in plasma of 29,428 copies/ml. Moreover, for every log increase in the viral load in plasma, the odds of detecting virus in fecal samples increased 87-fold. Genetic diversity of SIVsm in the SM colony was characterized by sequencing partial gag (846 bp) and gp43 (439 bp) fragments. Surprisingly, four new SIVsm lineages were identified, two of which were initially detected by fecal RT-PCR. This study documents the suitability of noninvasive methods for the detection and molecular characterization of new SIV variants. These assays will be useful for studying the phylogeny and epidemiology of SIVsm infections in the wild, and they hold promise as tools for investigating natural SIV infections in endangered nonhuman primates.
Phylogenetic analyses of full-length genomic sequences have identified six major lineages of simian immunodeficiency viruses (SIVs) (9, 21). SIVsm, a virus isolated from captive (14, 23, 31, 37), as well as wild (7), sooty mangabeys (SMs), forms a single phylogenetic lineage of diverse viruses which also includes SIVmac from captive macaques and human immunodeficiency virus type 2 (HIV-2). SMs are distributed in coastal West Africa from Senegal to Ivory Coast, which is coincident with the center of HIV-2 endemicity, where all of the divergent HIV-2 and SIVsm variants have been identified (6, 7, 16, 31, 43, 50). SMs are kept as pets or hunted for food (31); thus, there are plausible routes for zoonotic transmission. Moreover, nonsterile injections in the past may have contributed to the emergence of lentiviruses in the human population (12). All of these arguments support the hypothesis that HIV-2 arose through cross-species transmission of SIVsm from SMs (16, 17, 21, 23, 31). However, out of the seven HIV-2 lineages (which were previously called subtypes but are now referred to as groups according to the latest nomenclature proposal [B. T. Foley, personal communication]), only groups A and B are responsible for the HIV-2 epidemic (11, 16, 21), with no closely related simian counterpart having been discovered for either. The remaining five HIV-2 groups (C to G) are represented by single viruses identified in healthy individuals from Liberia (group D), Sierra Leone (groups E and F), and Ivory Coast (group G) (6, 7, 16, 50, 61). These countries are within the present-day (and historical) range of the SM in West Africa.
A full account of SIVsm diversity is required in order to understand the origins of the various HIV-2 lineages, as well as the conditions that led to their initial introduction into humans and subsequent spread (or lack thereof) in the new host. This is an important aspect of AIDS research, because >30 different species of African nonhuman primates are now known to harbor SIV (21). Numerous nonhuman primate lentiviruses have been discovered in recent years (2, 8-10, 18, 32, 40, 42, 53, 54), and a major concern is that these viruses have the potential to cross over to humans. Many SIVs grow in vitro in human cells (1, 2, 18, 22, 41, 45), and serological evidence for SIVmnd type 2 in humans has been reported (53). The prevalence of SIVs in the different species of nonhuman primates has been reported to range between 5 and 40% (7, 42, 52). However, these values are likely to be an underestimate (42). A study of SIVagm prevalence in wild grivet monkeys revealed infection rates of up to 90% in sexually active adults (27, 44). Thus, to determine the prevalence of lentiviral infection in the different species of nonhuman African primates and to evaluate the risk for cross-species transmission to humans, samples from wild primates are required (21, 42). However, this approach is problematic because wild monkeys are difficult to sample. Moreover, most of these species are highly endangered, indicating that blood or other tissues are generally not available. The utility of fecal and urine samples for detecting SIVcpz-specific antibodies and virion RNA (vRNA) and the use of these noninvasive approaches to detect and characterize SIVcpz infection in wild chimpanzees have been reported (49). Urine and stool specimens have also been used to document HIV-1 antibodies and RNA in infected children and adults (20, 58, 59, 62). However, none of these reports has examined the systemic viral load as a predictor of noninvasive vRNA detection, nor have SMs been studied by this approach.
In this study, we have applied the noninvasive SIV detection approaches to a new host species, the SM, to investigate the utility of this approach for (i) determining SIVsm prevalence and (ii) investigating SIVsm diversity in the wild. Both are critical to understand HIV-2 emergence. The sensitivity and specificity of antibody and vRNA detection in urine and fecal samples were determined, which confirmed urine Western blot analysis as the most sensitive noninvasive test for identifying SIVsm-infected SMs. Moreover, an acceptable sensitivity was found for vRNA detection in fecal samples, and fecal vRNA and systemic viral loads were strongly correlated. Finally, the noninvasive approaches prompted a comprehensive analysis of the genetic diversity of SIVsm in the Tulane National Primate Research Center (TNPRC) SM colony. Surprisingly, four new SIVsm lineages were uncovered that had previously gone unrecognized.
MATERIALS AND METHODS
Animals.
A total of 50 SMs were included in this study. Twenty-one naturally SIVsm-infected and four uninfected animals were housed at TNPRC. The TNPRC colony was originally established in 1980 with animals from the Yerkes National Primate Research Center (YNPRC) except for one monkey (G932), which was brought to TNPRC from New Iberia, La. Two SIVsm-infected and 23 non-SIVsm-infected SMs were housed at YNPRC. All SIVsm-infected animals were housed in individual cages. The research at both facilities complied with all relevant federal guidelines and institutional policies. The identification numbers, ages, sexes, and weights of the animals are presented in Table 1. Ages ranged from 3.3 to 24.8 years for the SIVsm-positive group (17 males and 6 females) and from 1.1 to 25.2 years for the SIVsm-negative group (18 males and 9 females).
TABLE 1.
Viral load, CD4 and CD8 cell numbers, and vital statistics of SMs in TNPRC and YNPRC colonies
| I.D. code | Monkey colonya | Age (yr) | Wt (kg) | Sexb | SIVsm statusc | Viral load (mean)d | No. of CD4 cells (mean) | No. of CD8 cells (mean) | CD4/CD8 cell ratio (mean) |
|---|---|---|---|---|---|---|---|---|---|
| A023 | Tu | 24.8 | 8.7 | M | + | 48,085f | 612f | 3,027f | 0.20f |
| D087 | Tu | 22.8 | 7.1 | M | + | <500f | 1,675f | 3,758f | 0.45f |
| D174 | Tu | 20.1 | 10.6 | M | + | 97,780 | 179 | 1,627 | 0.11 |
| D177 | Tu | 19.8 | 9.9 | M | + | 49,680 | 790 | 704 | 1.12 |
| E038 | Tu | 18.8 | 5.9 | F | + | NDe | ND | ND | ND |
| E039 | Tu | 18.2 | 12.1 | M | + | 14,878 | 506 | 688 | 0.74 |
| E041 | Tu | 19.2 | 10.3 | M | + | 591,198 | 784 | 1,117 | 0.70 |
| F098 | Tu | 18.2 | 12.5 | M | + | <500 | NRg | 547 | ND |
| G932 | Tu | 19.2 | 11.0 | M | + | 43,290f | 758f | 571f | 1.32f |
| M918 | Tu | 10.1 | 10.9 | M | + | ND | 830f | 1,042f | 0.80f |
| M922 | Tu | 12.3 | 7.6 | F | + | 22,364 | 480 | 734 | 0.65 |
| M923 | Tu | 13.0 | 7.3 | F | + | 1,083,620 | 788 | 546 | 1.44 |
| M927 | Tu | 11.7 | 6.7 | F | + | ND | 307 | 255 | 1.20 |
| M930 | Tu | 12.1 | 11.9 | M | + | 42,520 | 307 | 1,007 | 0.30 |
| M933 | Tu | 12.1 | 10.8 | M | + | 97,641 | 681 | 1,134 | 0.60 |
| M935 | Tu | 13.2 | 12.4 | M | + | 825,128 | 776 | 584 | 1.33 |
| M940 | Tu | 12.1 | 12.8 | M | + | 1,395 | 641 | 1,339 | 0.48 |
| M946 | Tu | 10.3 | 5.8 | F | + | 54,517 | 1,089 | 995 | 1.09 |
| M947 | Tu | 11.9 | 11.6 | M | + | 56,622 | 354 | 460 | 0.77 |
| M949 | Tu | 11.2 | 13.2 | M | + | 20,996 | 698 | 1,262 | 0.55 |
| M951 | Tu | 16.7 | 11.1 | M | + | 25,143 | 358 | 1,039 | 0.34 |
| F105 | Tu | 17.1 | 9.8 | M | − | 929 | 991 | 0.94 | |
| A039 | Tu | 22.6 | 11.5 | M | − | 384 | 973 | 0.39 | |
| M937 | Tu | 14.7 | 13.0 | M | − | 684 | 1,330 | 0.51 | |
| C215 | Tu | 25.2 | 6.5 | F | − | 569 | 7,058 | 0.08 | |
| FWk | Ye | 3.3 | 8.4 | F | + | ND | 1,727f | 1,666f | 1.04f |
| FNg | Ye | 16.3 | 9.8 | M | + | ND | 582f | 1,151f | 0.51f |
| FKu | Ye | 5.7 | 8.6 | M | − | ND | ND | ND | |
| FWn | Ye | 10.1 | 6.5 | F | − | ND | ND | ND | |
| FSo | Ye | 9.3 | 11.0 | M | − | ND | ND | ND | |
| FUr | Ye | 7.3 | 6.1 | F | − | 1,021f | 1,210f | 0.84f | |
| FOs | Ye | 7.1 | 7.5 | F | − | ND | ND | ND | |
| FRo | Ye | 9.4 | 6.0 | F | − | ND | ND | ND | |
| FQk | Ye | 12.4 | 7.2 | F | − | ND | ND | ND | |
| FFk | Ye | 13.1 | 8.3 | F | − | ND | ND | ND | |
| FYl | Ye | 11.3 | 6.4 | F | − | 1,970f | 1,054f | 1.87f | |
| FWj | Ye | 13.1 | 9.4 | M | − | ND | ND | ND | |
| FGv | Ye | 4.9 | 8.8 | F | − | ND | ND | ND | |
| FWo | Ye | 9.3 | 11.2 | M | − | 1,214f | 1,448f | 0.84f | |
| FKq | Ye | 8.3 | 12.4 | M | − | 1,249 | 1,761 | 0.71 | |
| FSs | Ye | 7.1 | 9.9 | M | − | ND | ND | ND | |
| FQp | Ye | 9.1 | 12.5 | M | − | ND | ND | ND | |
| FRq | Ye | 8.2 | 13.1 | M | − | 1,675f | 3,460f | 0.48f | |
| FPr | Ye | 7.3 | 12.2 | M | − | ND | ND | ND | |
| FRs | Ye | 7.1 | 11.3 | M | − | ND | ND | ND | |
| FKt | Ye | 6.4 | 10.9 | M | − | ND | ND | ND | |
| FMp | Ye | 9.2 | 13.5 | M | − | ND | ND | ND | |
| FUo | Ye | 9.3 | 11.4 | M | − | 1,227f | 1,187f | 0.65f | |
| FWy | Ye | 1.2 | NDe | M | − | ND | ND | ND | |
| FYy | Ye | 1.1 | ND | M | − | ND | ND | ND |
Tu, Tulane colony; Ye, Yerkes colony.
F, female; M, male.
+, positive; −, negative.
Mean, average of four data points.
ND, not done.
Single data point.
NR, no reaction with CD4 antibodies (SK3/B-D, 7E14/Exalpha, OkT4/Ortho, OKT4A/Ortho, and M-T477/Pharmingen [CD4 clone/manufacturer]).
Sample collection.
Plasma, urine and fecal samples were collected from each animal on the same day. For 16 of the 21 SIVsm-infected SMs from TNPRC, repeat samples (four per animal) were collected every 2 weeks. Urine and feces were collected in the morning prior to cage cleaning. Samples were placed in individual sterile tubes and were immediately frozen at −80°C until they were used.
Quantification of plasma vRNA.
Plasma was separated from EDTA-treated whole blood by centrifugation at 1,200 × g for 10 min and stored as 1-ml aliquots at −80°C. Quantification of SIVsm RNA in plasma was done at Bayer Reference Testing Laboratory (Emeryville, Calif.) by the most recent version of the SIV branched-DNA (bDNA, version 3.0) assay. This version was found to be applicable to all previously known variants of SIVmac and SIVsm and the limit of detection was more than 500 copies/ml (J. Booth, E. Sawyer, E. McNelley, D. Tayama, C. Wingfield, D. Cox, and K. Leung, 18th Ann. Symp. Nonhuman Primate Models AIDS, abstr. 129, 2000).
Determination of CD4+- and CD8+-T-lymphocyte subsets by flow cytometry.
Lymphocytes from peripheral blood were stained for analysis on a Becton Dickinson (San Jose, Calif.) FACScan flow cytometer with the following monoclonal antibodies; anti-human CD4-allophycocyanin (clone SK3), anti-human CD8-peridinin-chlorophyll (clone SK1), and anti-human CD3-fluorescein isothiocyanate (clone SP34) (Becton Dickinson Immunocytometry System). Data were analyzed using Cell Quest software (Becton Dickinson).
Western blot analysis.
Enhanced chemiluminescence (ECL)-immunoblot analysis was performed as described previously (49) using commercially available SIVmac strips (Zeptometrix, Buffalo, N.Y.). A 33% suspension of feces was prepared with 1× sample buffer containing 10 mM phosphate-buffered saline (PBS) (pH 7.4), 0.05% (wt/vol) Tween 20, 2.5 mM EDTA, 0.1% (wt/vol) NaN3, 0.1% (wt/vol) bovine serum albumin, and 1% (wt/vol) IGEPAL detergent (Sigma, St. Louis, Mo.); vortexed; and then centrifuged twice (13,800 × g for 25 min followed by 4,000 × g for 10 min at 4°C) to remove solid debris. The resulting clarified supernatant (1 ml) was incubated with the Western blotting strips. Urine samples (0.9 ml) were mixed with 100 μl of 10× sample buffer and used directly for immunoblotting. Plasma samples were diluted 1:100 in blocking buffer which included 1× sample buffer and 5% nonfat dry milk.
The SIV immunoblots were initially hydrated in PBS-T for 10 min and then incubated with blocking buffer for 1 h at room temperature. Fecal, urine, or plasma samples (1 ml) were incubated with the strips overnight at 4°C on an orbital shaker. The strips were washed three times in PBS-T for 10 min each time and then incubated for 1 h at room temperature with 1:1,000 goat anti-rhesus immunoglobulin G antibodies conjugated to horseradish peroxidase (Southern Biotechnology, Birmingham, Ala.). Following three washes in PBS-T for 10 min each time, the strips were developed by ECL-Western blotting detection reagents (AP Biotech, Piscataway, N.J.) according to the manufacturer's instructions. The strips were exposed to Kodak X-OMAT film for 20 s, 30 s, 1 min, and 5 min. Blots exhibiting a gp140 band alone or in combination with other virus-specific bands, or reactivities of any three structural proteins in the absence of a gp140 band, were scored positive. Blots exhibiting no reactivity were scored negative, while blots exhibiting reactivities other than the ones described above were scored indeterminate.
Nucleic acid extractions.
Total RNA was extracted from fecal samples using the RNAqueous-Midi kit (Ambion, Austin, Tex.). Briefly, 6 ml of lysis-binding solution was added to 0.5 g of fecal sample and vortexed vigorously until the sample was thoroughly homogenized. The suspension was clarified by centrifugation (16,000 × g; 3 min), and an equal volume of 64% ethanol was added. The solution was passed through a glass fiber filter unit to bind nucleic acids and washed three times with wash buffer. The nucleic acids were eluted (1 ml), with RNA subsequently preferentially precipitated with LiCl and spun. The resulting pellet was washed once with cold 70% ethanol, air dried, and then resuspended in 50 μl of RNase-free water. Urine pellets (1 ml) were obtained by centrifugation at 45,000 × g for 1.5 h at 4°C. Total nucleic acids were extracted using the NucliSens HIV-1 QT kit (Organon-Teknika, Boxtel, The Netherlands), and eluted in 50 μl of RNase-free water. DNA was also extracted from the peripheral blood mononuclear cells (PBMCs) of 17 SIVsm-infected SMs from TNPRC using the QIAamp kit (Qiagen, Valencia, Calif.).
RT-PCR.
Diagnostic reverse transcriptase (RT) PCR was initially performed using pol primers that were designed to amplify divergent strains of SIV from the six different primate lentivirus lineages. This pol primer set amplifies a 330-bp fragment in the integrase region of pol, using the outer primers pol-F1 (5′-CCA GCN CAC AAA GGN ATA GGA GG-3′) and pol-R1 (5′-ACB ACY GCN CCT TCH CCT TTC-3′) and the inner primers pol-F2 (5′-GCA AGT GGA TAC TTA GAA GCA GAA GT-3′) and pol-R2 (5′-CCC AAT CCC CCC TTT TCT TTT AAA ATT-3′). Subsequently, SIVsm-specific pol primers were designed using SIVsm consensus sequences to amplify a 425-bp fragment. These were SM-POL-F1 (5′-AAT GCC ANC ARA AAG GAG AAG C-3′) and SM-POL-R1 (5′-ACT GCT CCT TCC CCT TTC CA-3′) in the first round and SM-POL-F2 (5′-ATA CAT GGR CAR GTA AAT GCA GA-3′) and SM-POL-R2 (5′-TCC TCC CCT TCT TTT AAA ATT CAT-3′) in the second round of PCR.
For cDNA synthesis, an RT-PCR master mixture consisting of 1× buffer II (Perkin-Elmer, La Jolla, Calif.), 5 mM MgCl2, 1 mM deoxynucleoside triphosphate, 5 mM dithiothreitol, 20 pmol of reverse-transcription primer (pol-R1 or SM-POL-R1), 20 U of RNase inhibitor (Promega, Madison, Wis.), and 100 U of Superscript RT II (Gibco-BRL, Rockville, Md.) was prepared. Ten microliters of this master mixture was combined with 10 μl of fecal or urine nucleic acid extracts and incubated for 1 h at 42°C to allow for cDNA synthesis. The PCR was performed in a volume of 50 μl containing 1× buffer II (Roche Molecular Biochemicals, Indianapolis, Ind.), 2.0 mM MgCl2, 0.4 mM deoxynucleoside triphosphate, 10 pmol of outer primers (F1-R1), 1.25 U of Taq polymerase (Roche), and 10 μl of cDNA. The thermocycling profile included denaturation at 94°C for 2 min, followed by 45 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 30 s, and extension at 72°C for 1 min, with an additional extension of 72°C for 10 min. Two microliters of the first-round PCR product was used for nested PCR amplification with the inner primers (F2-R2) under the same thermocycling parameters, except that the annealing temperature was changed to 50°C.
To characterize the genetic diversity of SIVsm at TNPRC, nested PCR was also performed to amplify gag and gp43 sequences from primary PBMC DNA (6). The SM-gag primer set amplifies an 846-bp fragment, and the SM-gp43 primer set amplifies a 439-bp fragment. All PCR products were purified (Qiagen) and directly sequenced using the inner primers in an ABI automated DNA sequencer.
Phylogenetic analysis.
The pol, gag, and env sequences from newly characterized SIVsm strains were aligned with HIV-2, SIVsm, and SIVmac reference sequences from the Los Alamos National Laboratory HIV Sequence Database (http://hiv-web.lanl.gov) using the CLUSTAL X program (55). The alignment was manually adjusted, and poorly aligned regions were excluded. Gap-containing sites were removed prior to analysis. Pairwise evolutionary distances were calculated with the DNADIST program of the Phylip package (13) using a Kimura two-parameter model of nucleotide substitutions. All phylogenetic trees were constructed by the neighbor-joining method (48) in the Phylip package, and the reproducibility of the branching orders was estimated by 1,000 bootstraps.
Statistical methods.
Statistical methods were used to estimate the sensitivities and specificities of the fecal and urine Western blot assays, as well as fecal and urine vRNA detection tests. Statistical methods were also employed to correlate viral loads in plasma and feces and to determine the vRNA level in plasma that was predictive (with 95% probability) of a positive fecal-vRNA test result.
Sensitivity and specificity.
Test sensitivities and specificities were estimated using results from all available specimens. Because tests of the same type (e.g., urine Western blotting) were performed on serial specimens from the same SM, the results were corrected for correlated data sets. Since there were no indeterminate test results for any of the specimens, the data set consisted of binary data (positive and negative test results). The most common approach to fitting a binary outcome measure to data sets is to use a logistic regression model. Since we could not assume zero correlation between the repeated measures for specimens from the same SM, we estimated the parameters of a logistic regression model with correlated data using the generalized estimating equations described by Ronco and Biggeri (47). The dichotomous test results were the outcome, and the true infection status of the SM was the predictor. Once the logistic regression model was fitted, sensitivities and specificities were estimated using the following equations: sensitivity = exp (β1 + β2)/1 + exp (β1 + β2) and specificity = 1/exp (β1), where β1 represents the intercept and β2 represents the slope in the logistic regression model. Ninety-five percent confidence intervals (CI) for sensitivity and specificity were calculated by substituting the upper and lower limits of the 95% CI of the beta estimates (part of the output) in the formula above.
Association between viral load in plasma and fecal-vRNA detection.
Measurements of the viral load in plasma, as well as fecal-vRNA detection results, were available for each SM for each of four different time points. This data set was again fitted to a repeated-measures logistic regression model. The fecal-vRNA test results were the outcome, and the data on the viral load in plasma from the different time points were the predictors. We also included a variable to indicate the four time points as a predictor in the model. The results indicated that the viral load (but not the number of times that the test was repeated) was a significant predictor of vRNA detection (P = 0.0004). The viral-load value that predicted a positive fecal-vRNA test result with 95% confidence and the odds ratio (OR) of the probability of a positive test for each unit increase in the viral load were again estimated using a logistic regression model. The OR was calculated using the following formula: OR = exp (β), where β is the parameter estimate for the log viral load from the logistic regression model. The viral-load value required to predict with 95% probability a positive fecal-vRNA test result was calculated using the following formula: P = 1/1 + exp −[β0 + β1 (viral load) + β2 (time = 1) + β3 (time = 2) + β4 (time = 3)], where P represents the predicted probability fixed at 95%; β1 represents the parameter estimate for the viral load from the model; and β2, β3, and β4 represent the remaining beta values for time points 1, 2, and 3 (26).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are accessible under GenBank accession numbers AY158968 to AY158984 and AY159604 to AY159629.
RESULTS
Animal vital statistics and baseline T-lymphocyte numbers.
The age, weight, sex, SIVsm infection status, and systemic viral load, as well as CD4 and CD8 cell counts and ratios, for each animal are listed in Table 1. Eighty-four percent (21 of 25) of SMs at TNPRC are naturally infected with SIVsm. The SM infection statuses of all animals were confirmed by serology (using a commercial HIV-2 enzyme-linked immunosorbent assay) and by PCR amplification of SIVsm viral sequences from uncultured mangabey PBMC DNA. Serial samples (collected four times bimonthly) were available for 16 of the 21 SIVsm-infected SMs from TNPRC. Samples from SMs at the YNPRC were collected once and served as negative controls for noninvasive urine and fecal-antibody detection.
CD4+-T-cell numbers were measured in a subset of the SMs studied. Consistent with a previous report (4), the number of peripheral blood CD4+ T cells correlated negatively with the ages of the SMs in the uninfected group (r = −0.49; P = 0.029), whereas no such correlation was observed in the SIVsm-infected group (P = 0.95). There were no significant differences between the absolute numbers of CD4 cells in SIVsm-negative (16 samples; age, 16.1 ± 4.6 years) and -positive (19 samples; age, 15.6 ± 4.4 years) animals aged 10 years or older, although the CD4 cell numbers of SIVsm-infected mangabeys were slightly lower, yet still in the normal range (934 ± 439 versus 651 ± 335 cells/μl; P = 0.06). Comparison of CD4 cells in the SIVsm-infected and uninfected groups was not done for animals <10 years of age because only one SIVsm-infected SM was <10 years of age. One interesting exception was F098, a clinically asymptomatic animal that had no detectable CD4+ cells despite the use of different CD4 monoclonal antibodies for staining (L. Chakrabarti, unpublished data). The reason for this lack of CD4+ staining is not known. Similarly, no significant differences (P = 0.07) were found between the CD8+-T-cell counts of SIVsm-positive (1,158 ± 867 cells/μl) and SIVsm-negative (2,026 ± 1,806 cells/μl) animals aged 10 years or older.
Noninvasive detection of SIVsm-specific antibody by ECL-Western blot analysis.
Ultrasensitive Western blot analysis was employed to detect SIVsm-specific antibodies in urine and stools, since antibodies are present in these samples at lower concentrations than in serum or plasma. Using SIVmac1A11-coated immunoblotting strips (Zeptometrix) and ECL detection methods, anti-SIVsm antibodies were detected in 59 of 59 plasma samples from infected SMs (100% sensitivity) but in none of 24 plasma samples from uninfected SMs (100% specificity). Anti-gp140 antibodies were detected in plasma samples from SIVsm-infected mangabeys at dilutions of up to 1:10,000, which contrasts with results observed with commercial HIV-1 strips, which detected HIV-1 gp160 reactivity in plasma samples from HIV-1-infected individuals diluted up to 1:1,000,000 (B. Ling and P. Marx, unpublished results). Thus, these data indicated that the sensitivity of the SIVmac strips was 2 orders of magnitude lower than that of the Food and Drug Administration-approved HIV-1 strips. These differences in sensitivity may be due to only partial cross-reactivity of antibodies directed against genetically highly divergent viruses. An alternative explanation is that naturally infected animals have lower antibody titers to specific viral antigens (Gag) than humans and macaques (1, 38). Finally, it is also possible that the HIV-1- and SIVmac-coated test strips contain different amounts of virus-specific antigens.
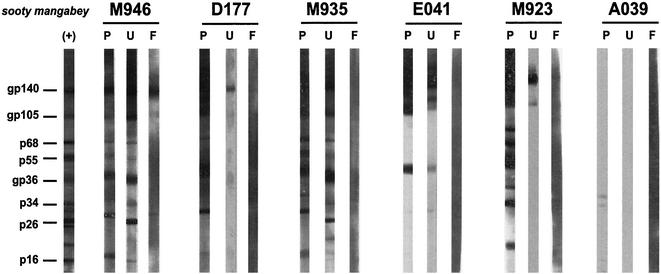
SIVsm-specific antibodies were detected in fecal and urine samples from SIVsm-infected, but not from uninfected, mangabeys (Fig. 1, M946 and A039, respectively). Statistical analyses were used to compute the sensitivity and specificity of urine and fecal-antibody detection, accounting for correlated sample sets (83 urine samples were obtained from 43 animals, and 91 fecal samples were obtained from 42 animals). The results are shown in Table 2. The urine Western blot test was highly sensitive for detecting virus-specific antibodies (96% sensitivity), while the fecal Western blot assay was less sensitive (16%). The availability of triplet plasma, urine, and fecal samples collected on the same date allowed a qualitative comparison of their immunoblot profiles. As shown in Fig. 1, antibody reactivities in the plasma were usually directed toward all SIV antigens. Urine antibody reactivities ranged from multiple- to single-band reactivities. Antibody-positive fecal samples usually exhibited only a single gp140 band. Moreover, only 10 of 64 fecal samples were reactive. These data suggest that the abundance of virus-specific antibodies in SMs decreases from plasma to urine to feces and that the antibodies with the highest titers are directed toward the viral envelope glycoprotein (gp140).
FIG. 1.
Detection of SIVsm-specific antibodies in plasma (P), urine (U), and fecal (F) samples from naturally SIVsm-infected SMs by ECL-Western blotting. Samples from each animal were collected on the same day. (+), positive control from the Western blotting kit (Zeptometrix). Samples were scored as positive in the presence of a gp140 band alone or in combination with other virus-specific bands or, in the absence of a gp140 band, with reactivities of any three structural proteins. M946, D177, M935, E041, and M923 are derived from SIVsm-infected mangabeys, while A039 represents a negative control.
TABLE 2.
Sensitivities and specificities of noninvasive SIVsm diagnostic assaysa
| Assay | SIVsm-positive SMs
|
SIVsm-negative SMs
|
||||
|---|---|---|---|---|---|---|
| Individuals (no. SIV+/no. tested) | Samples (no. SIV+/ no. tested) | Sensitivity (95% CI) (%) | Individuals (no. SIV+/no. tested) | Samples (no. SIV+/ no. tested) | Specificity (95% CI) (%) | |
| Antibody detection | ||||||
| Urine | 18/19 | 54/56 | 96 (87-99) | 0/24 | 0/27 | 100 (38-100) |
| Fecal | 7/18 | 10/64 | 16 (9-27) | 0/26 | 0/29 | 100 (64-100) |
| Viral-RNA detection | ||||||
| Urine | 1/18 | 1/53 | 2 (0-12) | 0/24 | 0/27 | 100 (80-100) |
| Fecal | 13/17 | 28/60 | 50 (32-69) | 0/26 | 0/29 | 100 (76-100) |
Sensitivity and specificity calculations were done using all samples tested rather than individuals and were corrected for correlated sample sets.
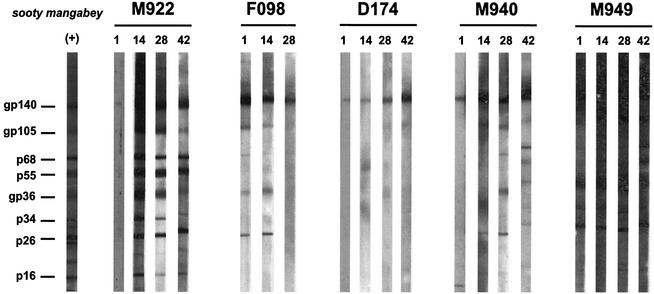
We also tested the reproducibility of the urine Western blot test by analyzing repeat samples collected from the same individual at different times. Figure 2 shows that the antibody reactivities are reproducible when different samples from the same mangabey are compared. For example, all four urine samples from animal D174 exhibited a single gp140 band, while all four urine samples from M949 exhibited a broad banding pattern. However, there were exceptions. For example, M922 urine collected on day 1 exhibited only a gp140 band, while broader reactivities were observed at later time points. This degree of variability is not unexpected given the nature of the sample. Moreover, in no instance did variability in the banding pattern influence the SIVsm-positive or -negative diagnosis.
FIG. 2.
Detection of SIVsm-specific antibodies in sequential urine samples from the same mangabey collected on days 1, 14, 28, and 42. (+), positive control from the Western blotting kit (Zeptometrix).
Viral loads in plasma in naturally SIV-infected SMs.
vRNA loads in plasma were quantified by an improved version of the SIV bDNA assay (Bayer Reference Testing Laboratory). The probe set for this assay was redesigned to maximize reactivity with variants of the SIVmac251 and SIVsmH4 groups of viruses (Booth et al., 18th Ann. Symp. Nonhuman Primate Models AIDS). Therefore, the new bDNA assay should also detect more divergent SIVsm strains. We assessed the reliability of the assay by testing four identical sets of double-blinded samples on two different dates. There were no significant differences between the values in the paired samples (data not shown).
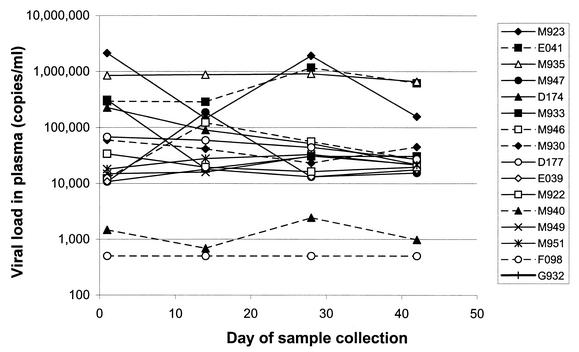
As shown in Fig. 3, viral-load levels in SMs ranged from <500 to 2 × 106 copies/ml, similar to values reported in other studies and for other species of African nonhuman primates (5, 19, 28, 39, 46). Low viral loads were detected for M940 (2,444 copies/ml), while the viral load for F098 was below the detection limit of the assay (500 copies/ml). In a subset of SMs, sequential plasma samples were analyzed (Fig. 3). There was a positive correlation for a specific mangabey's viral load from one time to the next (r = 0.75; P < 0.01), indicating that (i) a viral-load set point exists for individual SMs, and (ii) a single viral-load measurement should be representative of the set point for a given SM.
FIG. 3.
SIVsm viral loads in plasma in naturally infected SMs in the time period during which blood, urine, and feces were collected. vRNA levels in plasma were determined using the bDNA assay (Booth et al., 18th Ann. Symp. Nonhuman Primate Models AIDS). The values in SMs ranged from <500 to 2 × 106 copies/ml.
vRNA detection in fecal samples correlates with viral load in plasma.
The presence of vRNA in fecal and urine samples of SIVsm-infected SMs was determined using methods previously developed to detect HIV-1 and SIVcpz vRNAs in urine and fecal samples from experimentally and naturally infected chimpanzees (49). Samples from 18 SIVsm-infected and 26 uninfected SMs were included in this study. The results are summarized in Table 2. Similar to previous results with chimpanzees, vRNA detection in urine was largely negative, with 1 of 59 urine samples yielding an amplification product (M949). By contrast, 28 of 60 fecal specimens from SIVsm-infected mangabeys were RT-PCR positive, indicating a sensitivity of fecal-vRNA detection of 50% (Table 2). No sequences were amplified from 27 urine and 29 fecal samples from 26 uninfected mangabeys.
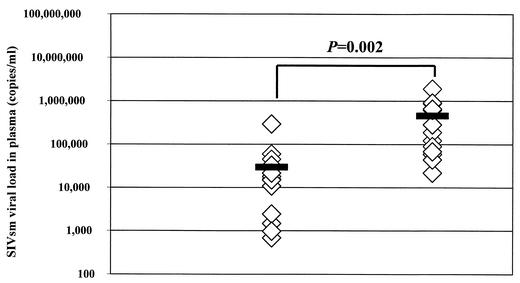
The availability of corresponding data on the viral loads in plasma for many of the fecal samples prompted us to investigate whether there was a correlation of the systemic viral burden and fecal-vRNA detection. Interestingly, SMs with detectable fecal vRNA had a mean viral load in plasma of 458,006 copies/ml, while those with undetectable fecal vRNA had a mean viral load in plasma of 29,428 copies/ml. This difference in means was highly significant (P = 0.002) (Fig. 4). The OR for this correlation was computed, revealing that for every log increase in the viral load in plasma, the odds of detecting virus in fecal samples increased 87-fold. The data set also allowed an estimation of the relation between viral-load levels in plasma and the detectability of fecal vRNA. Thus, if an SM had a viral load in plasma of ≤6,693 copies/ml, the probability of a fecal-vRNA-negative test result was >95%. In contrast, SMs with viral loads in plasma of ≥138,909 copies/ml had a >95% probability of having a positive fecal-vRNA test result.
FIG. 4.
Dependence of fecal-vRNA detection on viral load in plasma. SMs with undetectable fecal vRNA had a mean viral load in plasma (solid bar) of 29,428 copies/ml, whereas SMs with detectable fecal vRNA had a mean level of 458,006 copies/ml. This difference was statistically significant (P = 0.002).
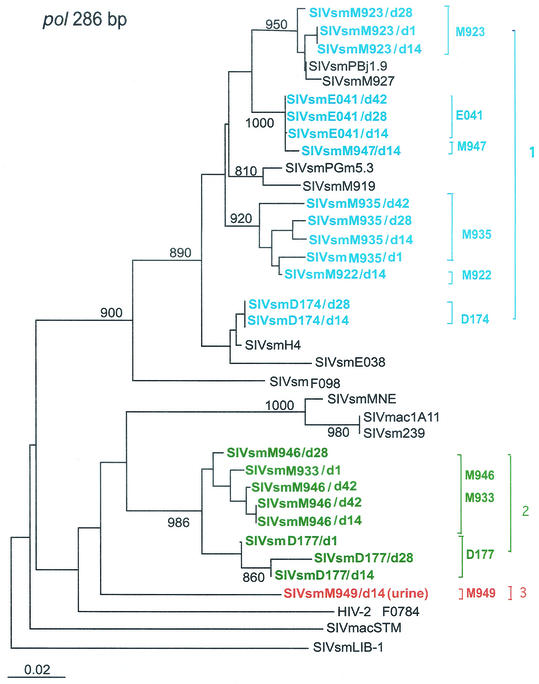
To confirm the authenticity of the pol PCR products, we subjected all fecal amplification products to sequencing and phylogenetic analyses. Figure 5 shows that amplification products from the same mangabey (e.g., sequential samples from M923, E041, D174, and D177) tended to cluster together. Some SMs, such as M946 and M933, had very closely related viruses that could not be distinguished in the short pol fragment. The most interesting result, however, was the unexpected branching of some SIVsm strains. Most RNA sequences clustered within the SIVsmPBj/SIVsmH4 group of viruses, which is not surprising given the origin of SIVsmH4 in the TNPRC colony of SMs (15, 23). However, some sequences fell outside the previously established SIVsm and SIVmac clusters. Notably, sequences from M946-M933, D177, and M949 seemed highly divergent from all other SIVsm and SIVmac strains in this short pol region (Fig. 5).
FIG. 5.
Phylogenetic analysis of feces- and urine-derived SIVsm sequences. Partial pol nucleotide sequences (286 bp) amplified by RT-PCR from fecal samples from naturally SIVsm-infected SMs (blue, green, and red) were compared with HIV-2 subtypes and SIVsm reference sequences from the HIV Sequence Database (29). The results suggested a minimum of three divergent SIVsm lineages in the sooty mangabeys studied, including two (green and red) not previously known to infect the Tulane SM colony. Phylogenetic trees were estimated by the neighbor-joining method. The reliability was estimated from 1,000 bootstrap replicates; bootstrap values on the branches with less than 800 replicates were not shown. The bar indicates 0.02 amino acid substitutions per site.
Four new divergent SIVsm lineages in a colony of SMs at TNPRC.
To assess the degree of genetic diversity of SIVsm in the Tulane SM colony, we amplified gag and env sequences from PBMC DNAs of 16 naturally SIVsm-infected SMs. High-molecular-weight DNA was extracted from PBMCs using a QIAamp DNA Mini Kit (Qiagen), and nested PCR was performed using previously reported gag and env gp43 region primers (6) to allow comparison with known HIV-2 and SIVsm strains. All amplification products were sequenced, revealing the existence of previously unknown SIVsm lineages.
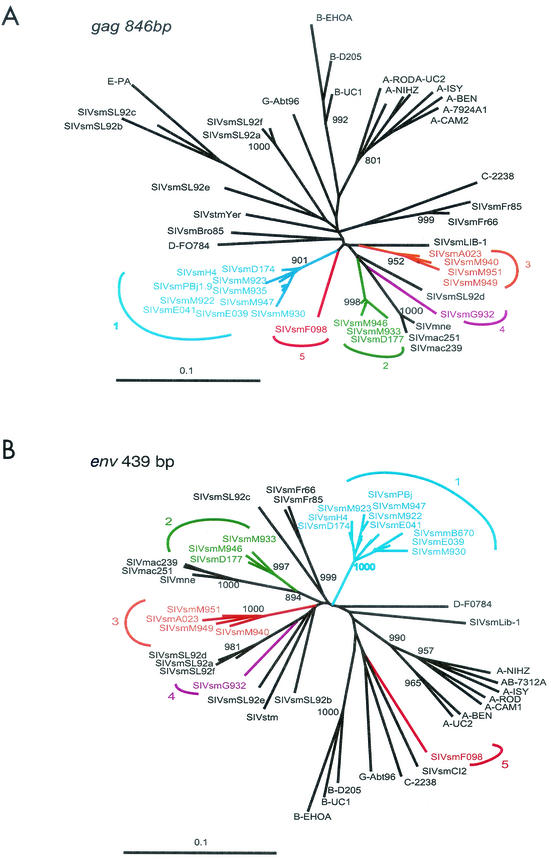
As shown in Fig. 6, a total of five lineages of SIVsm exist in SMs at TNPRC. Half of the newly derived viruses (8 of 16) fell within the previously identified SIVsmPBj/SIVsmH4 group, now designated lineage 1. However, the other half fell into one of four new groups (lineages 2 to 5) that were roughly equidistant from each other and different from all previously identified groups (Table 3). In the env fragment, intralineage genetic distances were <6%, whereas interlineage nucleotide sequence distances were >13%, with the greatest degree of divergence seen for viruses of the fifth lineage, which differed from all other strains by 23 to 25% (Table 3).
FIG. 6.
Detection of four new SIVsm lineages in Tulane SMs. Lineage 1 (blue) contains previously reported strains (http://hiv-web.lanl.gov). Lineages 2 to 5 (green, orange, purple, and red) are newly discovered. The trees are based on gag sequences (A) and env gp43 sequences (B). The phylogenetic trees were estimated by the neighbor-joining method. The reliability was estimated from 1,000 bootstrap replicates; bootstrap values on the branches of <80% are not shown. Bootstrap values on the branches with less than 800 replicates were not shown. The bar indicates 0.02 amino acid substitutions per site.
TABLE 3.
env and gag genetic distances of the five different lineages of TNPRC sooty manabeys
| Gene | SIVmac251 | SIVsmH4 | Lineage 1 | Lineage 2 | Lineage 3 | Lineage 4 | Lineage 5 |
|---|---|---|---|---|---|---|---|
| env | |||||||
| SIVmac251 | |||||||
| SIVsmH4 | 16.84 | ||||||
| Lineage 1 | 17.62 ± 0.72 | 3.29 ± 1.07 | 4.31 ± 1.49 | ||||
| Lineage 2 | 12.73 ± 0.39 | 13.56 ± 0.35 | 13.18 ± 0.72 | 3.91 ± 1.12 | |||
| Lineage 3 | 17.7 ± 0.94 | 14.49 ± 0.88 | 14.43 ± 1.08 | 13.11 ± 1.06 | 5.35 ± 0.53 | ||
| Lineage 4 | 17.49 | 16.45 | 16.24 ± 0.48 | 14.6 ± 0.81 | 14.13 ± 1.51 | ||
| Lineage 5 | 26.06 | 22.82 | 23.04 ± 0.77 | 23.48 ± 0.29 | 24.13 ± 0.36 | 24.95 | |
| gag | |||||||
| SIVmac251 | |||||||
| SIVsmH4 | 16.84 | ||||||
| Lineage 1 | 13.48 ± 0.61 | 5.98 ± 2.17 | 5.23 ± 2.32 | ||||
| Lineage 2 | 12.16 ± 0.57 | 12.98 ± 0.4 | 11.31 ± 1.27 | 3.35 ± 1.34 | |||
| Lineage 3 | 17.7 ± 0.94 | 13.61 ± 0.85 | 14.37 ± 1.09 | 13.75 ± 0.96 | 3.66 ± 0.70 | ||
| Lineage 4 | 10.97 | 13.38 | 11.85 ± 1.03 | 12.81 ± 0.42 | 15.05 ± 0.67 | ||
| Lineage 5 | 16.15 | 13.42 | 13.57 ± 1.06 | 15.18 ± 0.48 | 17.40 ± 0.58 | 15.83 |
A similar pattern was observed for the gag gene analyses, with 3 to 5% intralineage genetic distance and 13 to 18% interlineage nucleotide genetic distance. Again, SIVsmF098, the representative of the fifth lineage, was the most divergent virus among all strains. F098 was brought to TNPRC from YNPRC as a juvenile at the age of 3 years. SIVsmG932 formed an independent lineage in both the gag and env trees. G932 is 30 years old and was transferred from New Iberia to TNPRC in 1982. No additional information is available concerning its African origin. Thus, these divergent SIVsm strains were probably introduced to the United States via mangabey importation and were acquired by the infected SMs in their native habitat in West Africa. None of the new SIVsm lineages was particularly closely related to any of the known HIV-2 groups. Thus, SIVsmSL92b and SIVsmSL92c (Fig. 6) (6) remain the closest SIVsm relatives to an HIV-2 group.
The viruses found in fecal and urine samples (lineages 1 to 3) were consistent with the lineages found in peripheral blood of the same animals (Fig. 6). We were not able to recover representatives of lineages 4 (SIVsmG932) and 5 (SIVsmF098) in fecal samples. This is not surprising, since these two monkeys also had low viral loads in plasma by the bDNA assay (Table 1).
Having identified highly divergent SIVsm strains among the SMs analyzed, we considered the possibilities that the relatively lower viral loads obtained for animals which carried divergent viruses was due to a greater mismatch of the probes used for bDNA analysis and that similarly negative fecal RT-PCR amplifications were caused by primer mismatch. To address these questions, new primers were designed for the divergent viruses and used for fecal amplification. Importantly, this did not increase the frequency of recovering fecal vRNA, indicating that genetic diversity was not the reason for the negative RT-PCR results. The same primers were also used for RT-PCR from plasma for those animals which had negative fecal vRNA, and in these cases, amplification products were always obtained. Nevertheless, the correlation of the viral load in plasma and fecal-vRNA detection was recalculated for lineage 1 alone. The mean viral load in plasma for fecal-vRNA-positive specimens was 606,113 copies/ml, while the mean for fecal-vRNA-negative samples was 42,162 copies/ml. This difference remained statistically highly significant (P = 0.001), indicating that the viral load in plasma-fecal-vRNA correlation holds even after accounting for the SIVsm lineages for which bDNA may have underestimated the true viral load.
DISCUSSION
African nonhuman primates are the reservoirs for all known simian lentiviruses. Strong phylogenetic and geographical evidence shows that HIV-1 and HIV-2 are derived from SIVcpz and SIVsm lineages, respectively. However, the details of when, where, and how HIV emerged are not yet known. Further study of the lentiviruses in nonhuman primates is important for providing insights into the origins and emergence of HIV in humans.
This study used recently developed techniques for analysis of urine and feces in SMs for SIVsm detection and characterization (49). Two colonies of captive SMs were included in this study: the SM colony at the TNPRC (containing mostly SIVsm-infected monkeys) for sensitivity evaluations and the SMs housed at YNPRC (mostly SIVsm seronegative) for specificity evaluations. The monkeys at TNPRC originated from YNPRC in the 1980s, with the exception of G932, which was transferred from New Iberia, La., to TNPRC in 1982. The countries of origin for the infected SMs in this study are unknown. It has long been believed that the viruses infecting both Yerkes and Tulane SMs were very closely related, because it has been assumed that most animals acquired their infections in captivity. However, our results show that this is clearly not the case. Rather, the high degree of SIVsm diversity seen in the Tulane colony indicates that many of the founder animals of the Yerkes colony must have already been infected prior to their exportation from West Africa.
The viral loads in the plasma of these naturally infected SMs ranged from <500 to 2 × 106 copies/ml. When animals were retested for 42 days during the study, the viral-load levels in plasma were relatively unchanged. The viral loads were similar to the set points observed in pathogenic SIV infections of rhesus macaques. The major difference is that SMs remain healthy even with high viral loads while high set points in SIV-infected rhesus monkeys and HIV-1-infected humans predict progression to AIDS (30, 34). These observations, together with those of other investigators, thus indicate fundamental differences in the immune responses of SMs versus macaques and humans, which exhibit a negative correlation between CD4 counts and the viral load in plasma (5, 28, 30, 33, 34, 46). The question of how mangabeys can maintain high CD4+-T-cell levels despite high levels of viral loads in plasma is still unanswered, but most recent data indicate that nonpathogenic SIVsm infection is characterized by limiting bystander immunopathology and preserved regenerative capacity (3; M. B. Feinberg, personal communication). Also, similar observations were reported for other African nonhuman primate species naturally infected with SIVs (19, 38, 39).
Our study also demonstrates the feasibility and utility of noninvasive approaches to detect and molecularly characterize divergent SIV strains in endangered primate populations. ECL-Western blotting was used to detect SIVsm-specific antibodies in fecal and urine samples, and vRNA was successfully amplified primarily from fecal samples by RT-PCR. The analysis of fecal and urine samples from SMs of known infection status allowed us to calculate and compare the sensitivities and specificities of these tests (this was done by correcting for correlated sample sets). These data demonstrated that the sensitivity of antibody detection was significantly greater in urine (96%; CI, 87 to 99%) than in feces (16%; CI, 9 to 27%). By contrast, the sensitivity of vRNA detection was much greater in feces (50%; CI, 32 to 69%) than in urine (2%; CI, 0 to 12%). Thus, a combination of urine and fecal analyses would be most useful for field studies, with urine antibody determinations allowing prevalence determinations and fecal-vRNA amplification allowing molecular confirmation and phylogenetic analyses.
Previous studies of chimpanzee plasma, fecal, and urine samples indicated that ECL-based immunoblotting could detect even low concentrations of antibodies (10−7 dilutions of plasma samples from infected individuals still yielded positive results) using commercially available Western blotting strips. Our study shows that the sensitivity of commercially available SIV strips is 2 orders of magnitude lower than that of HIV-1 strips. This may compromise field studies, where samples will be collected under less ideal conditions. Moreover, a greater viral diversity might influence the sensitivity of antibody detection, which has been shown, for example, for HIV-1 group O (51). In order to overcome such limitations, the sensitivity of available SIVsm strips will have to be improved, possibly by including genetically engineered antigens and peptides and a cocktail of antigens from more divergent strains. One should note that antibody concentration in urine might be highly variable, as urine density varies significantly at different times of the day. The urine sampling was standardized in our study, but such conditions are difficult to reproduce in the wild. Moreover, even under standardized conditions, antibody concentrations seemed to vary in urine samples from the same SM (M940 [Fig. 2]).
One important outcome of our study is the demonstration that systemic and fecal viral loads are correlated and that our ability to detect and amplify vRNA from fecal samples depends on a high viral load in plasma. Although our studies did not yield a particular threshold value, the data indicated that viral loads equal to or less than ∼7,000 copies/ml had a 95% or greater probability to yield a negative result. Conversely, a positive amplification could be expected with >95% probability for fecal samples from mangabeys with >140,000 copies/ml. The dependence of fecal-vRNA detection on the viral load in plasma is interesting, since viral-load levels in plasma may differ in different species. Analysis of additional naturally infected primate species will be necessary to determine to what extent fecal-RNA detection can be used for noninvasive molecular epidemiological studies of SIV infection.
To date, SIVsm is the most diverse group of SIVs found in a single monkey subspecies (http://hiv-web.lanl.gov). This finding is probably related to the high number of isolates available. Our study reemphasizes the need for comprehensive testing: the more viruses analyzed, the more diversity emerges for nonhuman primate lentiviruses. The remarkable diversity among SIVsm strains in captive SMs, along with a high prevalence in the wild, strongly suggests that the founders of the original colony in the United States were infected when imported from Africa with divergent viruses circulating in their home range. This situation was also observed in a colony of mandrills in Gabon, where two of the founders were infected with two different viruses (53). Both viruses were isolated in 1988 (56), but only one was characterized (57). Ten years later, the second one was shown to be a different virus (53).
In summary, our study shows that highly divergent SIVs can infect a single colony of captive primates, indicating that carefully conducted genetic studies are required to understand the diversity and evolution of primate lentiviruses, their potential for cross-species transmission, and the origins of HIV. Noninvasive investigations may prove to be an extremely useful tool for these studies.
Acknowledgments
Binhua Ling and Mario L. Santiago contributed equally to this work.
This work was supported by funds from grants RO1 AI 44596, RO1 AI 50529, P51 RR000164, and P30 AI 27767 from the National Institutes of Health.
We thank Yingying Li, Cynthia Rodenburg, Megan Mefford, and Lynn Fresh for expert technical assistance; Louis Martin, Calvin Lanclos, and Julie Bruhn for flow cytometry analyses; and Theresa Secrist for administrative support. We also thank the veterinary and animal care staff of TNPRC for their service and expertise.
REFERENCES
- 1.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J. P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. St. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bostik, P., A. E. Mayne, F. Villinger, K. P. Greenberg, J. D. Powell, and A. A. Ansari. 2001. Relative resistance in the development of T cell anergy in CD4+ T cells from simian immunodeficiency virus disease-resistant sooty mangabeys. J. Immunol. 166:506-516. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Age-dependent changes in T cell homeostasis and SIV load in sooty mangabeys. J. Med. Primatol. 29:158-165. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, D. D. Ho, L. Zhang, and P. A. Marx. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., P. Telfer, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewley, J. P., J. C. Lewis, D. W. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A. M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damond, F., C. Apetrei, D. L. Robertson, S. Souquiere, A. Lepretre, S. Matheron, J. C. Plantier, F. Brun-Vezinet, and F. Simon. 2001. Variability of human immunodeficiency virus type 2 (HIV-2) infection patients living in France. Virology 280:19-30. [DOI] [PubMed] [Google Scholar]
- 12.Drucker, E., P. G. Alcabes, and P. A. Marx. 2001. The injection century: massive unsterile injections and the emergence of human pathogens. Lancet 358:1989-1992. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1993. PHYLIP (phylogeny interference package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 14.Fultz, P. N., H. M. Mcclure, D. C. Anderson, R. B. Swenson, R. Anand, and A. Srinivasan. 1986. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys). Proc. Natl. Acad. Sci. USA 83:5286-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fultz, P. N., H. M. McClure, D. C. Anderson, and W. M. Switzer. 1989. Identification and biologic characterization of an acutely lethal variant of simian immunodeficiency virus from sooty mangabeys (SIV/SMM). AIDS Res. Hum. Retrovir. 5:397-409. [DOI] [PubMed] [Google Scholar]
- 16.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, F., L. Yue, A. T. White, P. G. Pappas, J. Barchue, A. P. Hanson, B. M. Greene, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 18.Georges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Dubreuil, Z. Chen, S. M. Smith, A. Georges, F. Gao, B. H. Hahn, and P. A. Marx. 1998. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein, S., I. Ourmanov, C. R. Brown, B. E. Beer, W. R. Elkins, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2000. Wide range of viral load in healthy African green monkeys naturally infected with simian immunodeficiency virus. J. Virol. 74:11744-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottfried, T. D., J. C. Sturge, and H. B. Urnovitz. 1999. A urine test system for HIV-1 antibodies. Am. Clin. Lab. 18:4.. [PubMed] [Google Scholar]
- 21.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 24.Holland, C. A., J. H. Ellenberg, C. M. Wilson, S. D. Douglas, D. C. Futterman, L. A. Kingsley, A. B. Moscicki, et al. 2000. Relationship of CD4+ T cell counts and HIV type 1 viral loads in untreated, infected adolescents. AIDS Res. Hum. Retrovir. 16:959-963. [DOI] [PubMed] [Google Scholar]
- 25.Holznagel, E., S. Norley, S. Holzammer, C. Coulibaly, and R. Kurth. 2002. Immunological changes in simian immunodeficiency virus (SIV(agm))-infected African green monkeys (AGM): expanded cytotoxic T lymphocyte, natural killer and B cell subsets in the natural host of SIV(agm). J. Gen. Virol. 83:631-640. [DOI] [PubMed] [Google Scholar]
- 26.Hosmer, D. W., Jr., and S. Lemeshaw. 1989. Applied logistic regression. John Wiley & Sons, Inc., New York, N.Y.
- 27.Jolly, C. J., J. E. Phillips-Conroy, T. R. Turner, S. Broussard, and J. S. Allan. 1996. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops). J. Med. Primatol. 25:78-83. [DOI] [PubMed] [Google Scholar]
- 28.Kaur, A., R. M. Grant, R. E. Means, H. Mcclure, M. Feinberg, and R. P. Johnson. 1998. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J. Virol. 72:9597-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinksy (ed.). 2001. Human retroviruses and AIDS 2000: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 30.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marx, P. A., Y. Li, N. W. Lerche, S. Sutjipto, A. Gettie, J. A. Yee, B. H. Brotman, A. M. Prince, A. Hanson, R. G. Webster, and R. Desrosiers. 1991. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type 2 from a west African pet sooty mangabey. J. Virol. 65:4480-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCormack, G. P., and J. P. Clewley. 2002. The application of molecular phylogenetics to the analysis of viral genome diversity and evolution. Rev. Med. Virol. 12:221-238. [DOI] [PubMed] [Google Scholar]
- 33.Mellors, J. W. 1998. Viral-load tests provide valuable answers. Sci. Am. 279:90-93. [DOI] [PubMed] [Google Scholar]
- 34.Mellors, J. W., C. R. Rinaldo, P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 35.Miura, T., J. Sakuragi, M. Kawamura, M. Fukasawa, E. N. Moriyama, T. Gojobori, K. Ishikawa, J. A. Mingle, V. B. Nettey, H. Akari, M. Enami, H. Tsujimoto, and M. Hayami. 1990. Establishment of a phylogenetic survey system for AIDS-related lentiviruses and demonstration of a new HIV-2 subgroup. AIDS 4:1257-1261. [DOI] [PubMed] [Google Scholar]
- 36.Mohamed, O. A., R. Ashley, A. Goldstein, J. McElrath, J. Dalessio, and L. Corey. 1994. Detection of rectal antibodies to HIV-1 by a sensitive chemiluminescent Western blot immunodetection method. J. Acquir. Immune Defic. Syndr. 7:375-380. [PubMed] [Google Scholar]
- 37.Murphey-Corb, M., L. N. Martin, S. R. Rangan, G. B. Baskin, B. J. Gormus, R. H. Wolf, W. A. Andes, M. West, and R. C. Montelaro. 1986. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature 321:435-437. [DOI] [PubMed] [Google Scholar]
- 38.Norley, S., B. Beer, S. Holzammer, J. zur Megede, and R. Kurth. 1999. Why are the natural hosts of SIV resistant to AIDS? Immunol. Lett. 66:47-52. [DOI] [PubMed] [Google Scholar]
- 39.Onanga, R., C. Kornfeld, I. Pandrea, J. Estaquier, S. Souquiere, P. Rouquet, V. Poaty-Mavoungou, O. Bourry, S. M'Boup, F. Barre-Sinoussi, F. Simon, C. Apetrei, P. Roques, and M. C. Muller-Trutwin. 2002. High levels of viral replication contrast with only transient changes in CD4+ and CD8+ cell numbers during the early phase of experimental infection with simian immunodeficiency virus SIVmnd-1 in Mandrillus sphinx. J. Virol. 76:10256-10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osterhaus, A. D., N. Pedersen, G. Van Amerongen, M. T. Frankenhuis, M. Marthas, E. Reay, T. M. Rose, J. Pamungkas, and M. L. Bosch. 1999. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin). Virology 260:116-124. [DOI] [PubMed] [Google Scholar]
- 41.Owen, S. M., S. Masciotra, F. Novembre, J. Yee, W. M. Switzer, M. Ostyula, and R. B. Lal. 2000. Simian immunodeficiency viruses of diverse origin can use CXCR4 as a coreceptor for entry into human cells. J. Virol. 74:5702-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peeters, M., W. Janssens, K. Fransen, J. Brandful, L. Heyndrickx, K. Koffi, E. Delaporte, P. Piot, G. M. Gershy-Damet, and G. Van Der Groen. 1994. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Cote d'Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res. Hum. Retrovir. 10:1289-1294. [DOI] [PubMed] [Google Scholar]
- 44.Phillips-Conroy, J. E., C. J. Jolly, B. Petros, J. S. Allan, and R. C. Desrosiers. 1994. Sexual transmission of SIVagm in wild grivet monkeys. J. Med. Primatol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 45.Poaty-Mavoungou, V., R. Onanga, I. Bedjabaga, and E. Mavoungou. 2001. Simian immunodeficiency virus from mandrill (Mandrillus sphinx) SIVmnd experimentally infects human and nonhuman primate cells. Microb. Infect. 3:599-610. [DOI] [PubMed] [Google Scholar]
- 46.Rey-Cuille, M. A., J. L. Berthier, M. C. Bomsel-Demontoy, Y. Chaduc, L. Montagnier, A. G. Hovanessian, and L. A. Chakrabarti. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronco, G., and A. Biggeri. 1999. Estimating sensitivity and specificity when repeated tests are performed on the same subject. J. Epidemiol. Biostat. 4:329-336. [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465.. [DOI] [PubMed] [Google Scholar]
- 50.Schim Van Der Loeff, M. F., and P. Aaby. 1999. Towards a better understanding of the epidemiology of HIV-2. AIDS 13(Suppl. A):S69-S84. [PubMed] [Google Scholar]
- 51.Simon, F., T. D. Ly, A. Baillou-Beaufils, V. Fauveau, J. De Saint-Martin, I. Loussert-Ajaka, M. L. Chaix, S. Saragosti, A. M. Courouce, D. Ingrand, C. Janot, and F. Brun-Vezinet. 1994. Sensitivity of screening kits for anti-HIV-1 subtype O antibodies. AIDS 8:1628-1629. [DOI] [PubMed] [Google Scholar]
- 52.Simon, F., S. Souquiere, F. Damond, A. Kfutwah, M. Makuwa, E. Leroy, P. Rouquet, J. L. Berthier, J. Rigoulet, A. Lecu, P. T. Telfer, I. Pandrea, J. C. Plantier, F. Barre-Sinoussi, P. Roques, M. C. Muller-Trutwin, and C. Apetrei. 2001. Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res. Hum. Retrovir. 17:937-952. [DOI] [PubMed] [Google Scholar]
- 53.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takehisa, J., Y. Harada, N. Ndembi, I. Mboudjeka, Y. Taniguchi, C. Ngansop, S. Kuate, L. Zekeng, K. Ibuki, T. Shimada, B. Bikandou, Y. Yamaguchi-Kabata, T. Miura, M. Ikeda, H. Ichimura, L. Kaptue, and M. Hayami. 2001. Natural infection of wild-born mandrills (Mandrillus sphinx) with two different types of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 17:1143-1154. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsujimoto, H., R. W. Cooper, T. Kodama, M. Fukasawa, T. Miura, Y. Ohta, K. Ishikawa, M. Nakai, E. Frost, G. E. Roelants, J. Roffi, and M. Hayami. 1988. Isolation and characterization of simian immunodeficiency virus from mandrills in Africa and its relationship to other human and simian immunodeficiency viruses. J. Virol. 62:4044-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]
- 58.Urnovitz, H. B., J. C. Sturge, T. D. Gottfried, and W. H. Murphy. 1999. Urine antibody tests: new insights into the dynamics of HIV-1 infection. Clin. Chem. 45:1602-1613. [PubMed] [Google Scholar]
- 59.Van Der Hoek, L., R. Boom, J. Goudsmit, F. Snijders, and C. J. Sol. 1995. Isolation of human immunodeficiency virus type 1 (HIV-1) RNA from feces by a simple method and difference between HIV-1 subpopulations in feces and serum. J. Clin. Microbiol. 33:581-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolfheim, J. H. 1983. Primates of the world. University of Washington, Seattle.
- 61.Yamaguchi, J., S. G. Devare, and C. A. Brennan. 2000. Identification of a new HIV-2. subtype based on phylogenetic analysis of full-length genomic sequence. AIDS Res. Hum. Retrovir. 16:925-930. [DOI] [PubMed] [Google Scholar]
- 62.Yolken, R. H., S. Li, J. Perman, and R. Viscidi. 1991. Persistent diarrhea and fecal shedding of retroviral nucleic acids in children infected with human immunodeficiency virus. J. Infect. Dis. 164:61-66. [DOI] [PubMed] [Google Scholar]