Abstract
Several studies have suggested the involvement of cutaneous human papillomaviruses (HPVs) in the development of nonmelanoma skin cancers. Here we have characterized the in vitro properties of E7 proteins of three cutaneous HPV types, 10, 20, and 38, which are frequently detected in skin specimens. We show that HPV38 E7 is able to inactivate the tumor suppressor pRb and induces loss of G1/S transition control, a key event in carcinogenesis. In contrast, HPV10 and HPV20 E7 proteins do not display these in vitro transforming activities. We also show that the two early proteins E6 and E7 of HPV38 are sufficient to corrupt the cell cycle and senescence programs in primary cells, inducing active and long-lasting proliferation of primary human keratinocytes, the natural host cells. Our study shows that E6 and E7 of this cutaneous HPV type have transforming activity in primary human cells, suggesting a role for HPV38 infection in skin carcinogenesis. In further support of such a role, we detected HPV38 DNA in approximately 50% of nonmelanoma skin cancers, but only in 10% of healthy skin specimens (P < 0.001).
Nonmelanoma skin cancer is the most frequently occurring malignancy in the Caucasian population (34, 38, 47). Although these cancers have a good prognosis and are not normally associated with mortality, an increasing incidence of other invasive cancers and cancer mortality following nonmelanoma skin cancers has been reported (17, 24, 28, 29). Several lines of evidence suggest the involvement of an infective agent in the etiology of this condition. Patients suffering from a rare genetic immune suppression termed epidermodysplasia verruciformis and individuals under long-lasting immunosuppression are prone to develop these cancers (21, 22, 30, 37). Epidermodysplasia verruciformis patients are highly susceptible to human papillomavirus (HPV) infections by a specific subgroup of cutaneous HPVs, the so-called epidermodysplasia verruciformis types (e.g., HPV5 and HPV8), that lead to extensive verrucosis of confluent flat warts (22, 30, 37). In approximately 30% of cases, the HPV lesions develop into multifocal squamous cell carcinomas.
Supporting the infectious role of cutaneous HPV types in the tumorigenesis of nonmelanoma skin cancers is the fact that other members of the papillomavirus family are clearly oncogenic (55). Indeed, clinical, epidemiological, and molecular data have demonstrated that mucosal high-risk HPV types (e.g., high-risk HPV16 and HPV18) are the etiological agents of anogenital cancers as well as a subgroup of head and neck cancers (55). The early region of these HPV types encodes two oncoproteins, E6 and E7, which associate with and neutralize the cellular tumor suppressors p53 and retinoblastoma (pRb), respectively (32, 36).
Independent studies suggest that cutaneous HPV types may also be involved in the development of squamous cell carcinoma and basal cell carcinomas in the general population (6, 7, 14, 43). These indications are based only on studies assessing viral DNA presence in skin tumors by PCR, which has revealed the presence of a large number of different HPV types, both in healthy skin and in tumors. In addition, the transforming properties of the majority of the cutaneous HPV types, which represent a criterion to evaluate the potential in vivo oncogenicity, have been poorly investigated.
We have characterized several transforming properties of three cutaneous HPV types, 10, 20, and 38. HPV20 and HPV38 are epidermodysplasia verruciformis types and have frequently been detected in skin specimens in non-epidermodysplasia verruciformis patients (5, 10), while HPV10, a common cutaneous HPV type, is associated with benign lesions (27).
In this study, we present compelling evidence that E7 of HPV38 but not of HPV10 and HPV20 induces loss of G1/S control. We also demonstrate that HPV38 E6 and E7 are sufficient to corrupt the cell cycle and senescence programs in primary cells. Finally, we found that HPV38 is highly prevalent in nonmelanoma skin cancers. Our data suggest a role for HPV38 infection in skin carcinogenesis.
MATERIALS AND METHODS
Retroviral expression vectors.
The retroviral vectors pBabe-puro and pBabe-neo were described previously (35), while pLXSN was obtained from Clontech (Palo Alto, Calif.). The open reading frames of HPV10, -16, -20, and -38 E7 were amplified by PCR to introduce ecoRI and SalI sites upstream and downstream, respectively, of the different genes. In addition to the restriction enzyme site, the primers comprise the complementary sequence of the first or the last six codons of each gene. The viral genes were cloned in pBabe-neo with or without the hemagglutinin (HA) tag sequence at the 5′ end. H-ras V12 and HPV16 E6/E7 or HPV38 E6/E7 were cloned in pBabe-puro and pLXSN, respectively, using the procedure described above. The pBabe-puro p16INK4a construct was kindly provided by Bruno Amati (DNAX Research Institute, Palo Alto, Calif.).
Cell culture.
NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 4.5 g of glucose per liter, supplemented with 10% calf serum (NIH 3T3). Bosc23, Phoenix, and HaCaT were grown in DMEM supplemented with 10% fetal calf serum. Primary human oral fibroblasts were isolated from oral mucosa (49, 50) and cultured in the medium described above. Primary human keratinocytes were isolated from skin of adult individuals as previously described (8) and grown together with NIH 3T3 feeder layers in FAD medium containing 3 parts Ham's F12, 1 part DMEM, 5% fetal calf serum, insulin (5 μg/ml), epidermal growth factor (10 ng/ml), cholera toxin (8.4 ng/ml), adenine (24 μg/ml), and hydrocortisone (0.4 μg/ml). Feeder layers were prepared by irradiating NIH 3T3 (137Cs; 80 Gy).
Retroviral infections.
High-titer retroviral supernatants (>5 × 106 IU/ml) were generated by transient transfection of Bosc23 cells (ecotropic viruses) or Phoenix cells (amphotropic viruses) and used to infect rodent or human cells, respectively, as described previously (40). After infection, NIH 3T3 cells were selected in 1 mg of G418 per ml (7 to 8 days) or 2.0 μg of puromycin per ml (48 h). Primary oral fibroblasts were selected in 0.5 mg of G418 (6 to 7 days) or 0.5 μg of puromycin (4 to 5 days) per ml, while primary human keratinocytes were selected in 0.02 mg of G418 per ml (2 to 3 days). Coexpression of the viral genes and p16INK4a or ras was achieved by consecutive retroviral infection as previously described (18, 42).
Determination of cellular parameters.
For the fluorescence-activated cell sorting (FACS) analysis, cells, after retroviral infection, were selected for 48 h, harvested by trypsinization, washed with phosphate-buffered saline (PBS), and fixed in 80% methanol at −20°C for 30 min. Approximately 106 cells were washed in PBS and resuspended in 500 μl of PBS containing RNase A (0.1 mg/ml). After incubation (30 min at 37°C), propidium iodide was added (5 μl of a 50-μg/ml solution). Analysis by flow cytometry was performed with a Becton Dickinson FACSort. Cell cycle profiles were determined by using the Modfit cell cycle analysis software package.
Soft agar assays were performed by seeding cells in 35-mm-diameter dishes onto an underlayer of 0.5% low-gelling agarose in growth medium. Cells were plated in medium containing 0.28% SeaPlaque agarose and fed every 4 to 5 days with growth medium. Colonies were allowed to grow for 2 to 4 weeks and scored microscopically.
For the determination of population doublings, cells were cultured in 75-cm2 flasks and trypsinized when they reached approximately 80% confluence (3 × 106 to 4 × 106 cells/ml). Population doublings were calculated, taking into consideration the number of passages and the split ratio.
For senescence-associated β-galactosidase staining, senescence-associated β-galactosidase activity (15) was determined as described elsewhere (42).
Immunoblot analysis and antibodies.
Total cellular extracts were prepared in lysis buffer as described previously (18). Cell extracts were fractionated by electrophoresis on a sodium dodecyl sulfate-polyacrylamide gel. Immunoblot analysis was performed with the following antibodies: anti-HA tag (MMS-101R, Babco, Richmond, Calif.); anti-pRb (14001A; Pharmingen, San Diego, Calif.); anti-β-tubulin (clone TUB2.1, Sigma, Deisenhofen, Germany); and anti-p16INK4a (kindly provided by Gordon Peters, Imperial Cancer Research Fund, London, United Kingdom).
GST pulldown assay.
Glutathione S-transferase (GST) fusion protein synthesis, HaCaT protein extracts, and the GST pulldown assay were performed as described previously (1).
Telomerase assay.
Cell lysis and telomerase assays were performed with the TRAPeze kit (Intergen Company, Oxford, United Kingdom); 50 ng of total protein extract was used for each assay, each with or without RNase inactivation (RNase- and DNase-free; Roche, Mannheim, Germany). Products were separated in nondenaturing 10% polyacrylamide gels, visualized by autoradiography and phosphorimager scanning (Fujifilm Bas-1500), and quantified with TINA 2.0 software.
RT-PCR analysis.
Total RNA was extracted from mammalian cells with the RNeasy kit (Qiagen GmbH, Germany), adding a DNase I treatment to prevent cellular DNA contamination in the PCR. cDNA was synthesized with the First Strand cDNA synthesis kit (MBI Fermentas, Heidelberg, Germany) with a random hexamer primer and the Moloney murine leukemia virus reverse transcriptase.
Detection of HPV38 DNA in skin specimens by PCR.
Actinic keratosis, basal cell carcinomas, and squamous cell carcinomas were obtained from Istituto di Dermatologia, Facoltà di Medicina, Università di Bari, Bari, Italy. Breast skin from individuals of the same geographical region was used for healthy skin controls. Paraffin blocks were randomly mixed, and the subsequent analysis was blinded to the clinical data. DNA extraction was performed as described previously (54). Empty paraffin blocks and distilled water were included within the samples as negative controls. Hot start PCR was performed with Platinum Supermix (Life Technologies, Rockville, Md.).
To check the quality of all DNA samples, a fragment of the β-globin gene was amplified by PCR. The primers used annealed in the 38E6 open reading frame, 5′-TGGAACTTACCAAAACCTCAAAC-3′ and 5′-CCTCTTGAGTCCAGATTAACTG-3′. Southern blotting was performed as described in the application manual for filter hybridization (Roche Molecular Biochemicals, Mannheim, Germany). The analysis of HPV38 DNA in skin specimens was repeated twice, including the cutting of the paraffin blocks and DNA extraction. The data were considered only when identical results were obtained in both analyses.
Immunofluorescence.
The immunofluorescent staining was performed as previously described (53), with primary antibodies anti-pankeratin (Dako, Glostrup, Denmark) and antivimentin (Progen, Heidelberg, Germany).
In situ hybridization.
Hybridization was performed with the entire HPV38 genome (8,000 bp) as probe. Viral DNA was labeled with biotin-14-dATP by nick translation according to the manufacturer's instructions (Life Technologies, Rockville, Md.). All steps were performed according to the protocol in the Genepoint kit (Dako, Glostrup, Denmark). Counterstaining was performed in Mayer's hematoxylin, resulting in light-blue nuclei.
Statistical analysis.
For the statistical comparison of values in the different experiments of viral gene-expressing cells and controls, we used a one-sided paired t test. For each experiment, we calculated the respective P value and a statistically significant result was stated if the P value was less than 0.05.
RESULTS
HPV16 E7 and HPV38 E7 have similar biological properties.
HPV16 E7 associates with pRb and promotes its degradation via the proteasome pathway, inducing uncontrolled proliferation (reviewed in reference 36). We first determined the ability of the three E7s to associate with pRb in vitro with a GST pulldown assay. Equal amounts of GST-E7 fusion proteins (Fig. 1A) were immobilized on glutathione-Sepharose beads and incubated with cellular protein extracts of immortalized human keratinocytes (HaCaT) (9). The amount of pRb associated with the E7 proteins was determined by Western blot analysis.
FIG. 1.
In vitro ability of E7 protein from HPV types 10, 20, and 38 to associate with pRb. (A) Quantification of GST fusion proteins. The different GST-E7 fusion proteins were purified as described previously (1). Five microliters of fusion proteins immobilized on glutathione beads was applied to a polyacrylamide-sodium dodecyl sulfate gel and stained with Coomassie brilliant blue R-250 to estimate the protein concentration. (B) GST pulldown assay. HaCaT cellular extracts (600 μg) were incubated with beads containing approximately 1 μg of the different fusion proteins. After extensive washing of the beads, the amount of pRb associated with the different GST-E7 proteins was determined by immunoblot analysis with a specific anti-pRb antibody (14001A; Pharmingen). One-tenth of the total cellular extract (60 μg) used in the GST pulldown assay (input) was applied to a polyacrylamide-sodium dodecyl sulfate gel. GST, GST16E6, and GST16E7G24 (a mutant form of 16E7 that does not efficiently associate with pRb) were used as negative controls.
HPV10 and HPV20 E7s weakly associate with pRb as an HPV16 E7 mutant (C24G) (Fig. 1B), which harbors a mutation in the pRb-binding domain (amino acids 22 to 26). In contrast, HPV38 E7 binds pRb with an efficiency similar to that of HPV16 E7 (Fig. 1B). These results are in agreement with a previous report in which the pRb binding affinity of HPV10 and HPV38 E7 was determined with different recombinant proteins and assays (16). The weak interaction of HPV16 C24G with pRb (Fig. 1B) is most likely due to the pRb low-affinity binding site located in the C-terminal region of the viral protein (39).
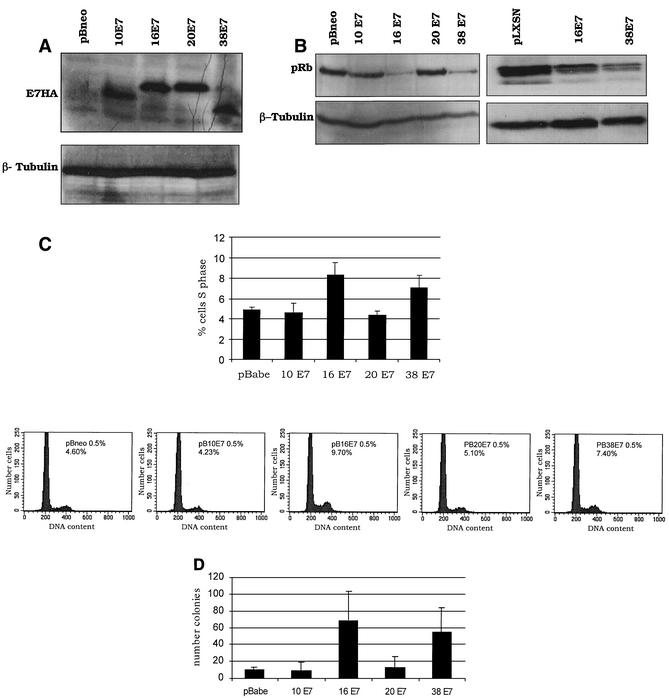
It has been demonstrated that pRb binding efficiency does not necessarily correlate with the ability to degrade the tumor suppressor and to induce cellular transformation (1, 13, 18, 19, 41). Thus, we evaluated the ability of the cutaneous E7s to promote pRb destabilization. The proteins were fused at the N terminus to the hemagglutinin (HA) epitope to allow comparison of the expression levels in rodent fibroblasts. Figure 2A shows that all the E7 proteins were expressed at similar levels. HPV16 E7- and HPV38 E7-expressing cells exhibited reduced pRb levels in comparison to control or HPV10 and HPV20 E7 cells (Fig. 2B, left panel). The ability of HPV38 E7 to induce pRb destabilization was not limited to rodent cells, but was also observed in primary human cells (Fig. 2B, right panel).
FIG. 2.
In vitro biological properties of E7 protein from HPV types 10, 20, and 38. (A) Expression of E7 proteins in NIH 3T3 cells. One hundred micrograms of protein extract was fractionated on a 12% polyacrylamide-sodium dodecyl sulfate gel, transferred to a polyvinylidene difluoride membrane, and incubated with an anti-HA-tag monoclonal antibody (MMS-101R; Babco) or a monoclonal anti-β-tubulin antibody (TUB2.1; Sigma). (B) Levels of pRb in NIH 3T3 (left panel) or primary human fibroblast (right panel) cells expressing the different E7 proteins. One hundred micrograms of protein extracts of cells expressing the different oncoproteins as indicated was applied to an 8% polyacrylamide-sodium dodecyl sulfate gel, transferred to a polyvinylidene difluoride membrane, and incubated with an anti-pRb (14001A; Pharmingen) or β-tubulin (TUB2.1; Sigma) antibody. (C) Ability of E7 proteins to stimulate S-phase entry in serum-deprived NIH 3T3 cells. Mock-infected NIH 3T3 cells (infected with empty retrovirus) or infected NIH 3T3 cells expressing the E7 proteins, as indicated in the figure, were grown in DMEM with 0.5% calf serum for 48 h. The percentage of the population in S phase was determined by flow cytometry. The results are averages of five independent experiments, and differences were statistically significant (pBabe versus HPV16 E7, P = 0.013; pBabe versus HPV38 E7, P = 0.010). The cell cycle profiles of a representative experiment are also shown (bottom panels). (D) Efficiency of the E7 proteins in inducing anchorage-independent growth. The results are averages of three independent experiments, each performed in duplicate. The difference between the values of HPV16 E7 or HPV38 E7 cells and control cells (pBabe) was statistically significant (pBabe versus HPV16 E7, P = 0.0001; pBabe versus HPV38 E7, P = 0.0004).
Consistent with their efficiency in inactivating pRb, HPV16 and HPV38 E7s were able to induce transformation of rodent cells, which acquired the ability to proliferate in the absence of mitogenic factors and to grow in an anchorage-independent manner (Fig. 2C and D). To exclude the possibility that the addition of HA to E7s alters their biological properties, all the experiments described above were also performed with cells expressing the native viral proteins. In all assays, native and HA-tagged proteins behaved similarly (data not shown).
The cyclin-dependent kinase (CDK) inhibitor p16INK4a is a negative-feedback cell cycle regulator that prevents unscheduled proliferation (45). High levels of p16INK4a lead to cell cycle arrest and/or senescence (11, 45). Loss of its expression favors tumor development in mice (44) and is a common event in human cancers (12, 25). HPV16 E7 abolishes the cell cycle-regulatory functions of p16INK4a (31). This event is intimately associated with the ability of E7 to promote pRb degradation (18).
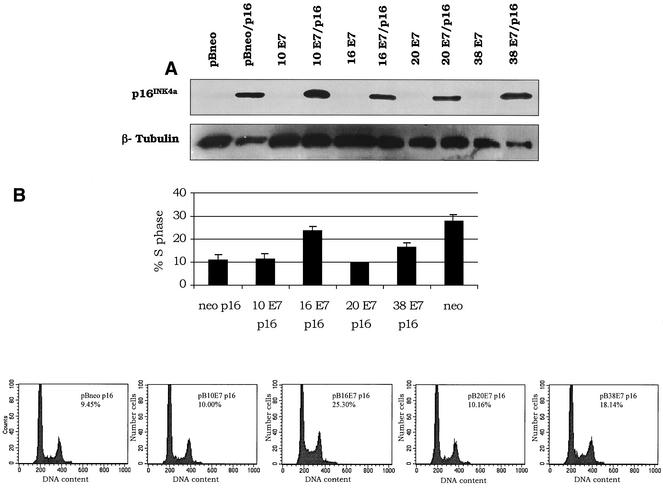
To further characterize the transforming properties of the E7 proteins, we used double retroviral infections (18) to coexpress the E7 and p16INK4a genes in NIH 3T3 cells and analyzed their cell cycle profiles. Although p16INK4a was efficiently expressed in combination with all E7 proteins tested (Fig. 3A), only those cells coexpressing HPV38 or HPV16 E7 were able to proliferate (Fig. 3B). HPV16 E7 can also alter cell cycle regulation by associating with and blocking the activity of two other CDK inhibitors, p27KIP1 and p21WAF1 (36). Both CDK inhibitors inhibit cyclin E/CDK2 activity and, analogously to p16INK4a, induce a cell cycle block in G1 phase (45). We observed that HPV38 E7, like HPV16 E7, circumvented the p21WAF1- and p27KIP1-imposed G1 arrest (data not shown).
FIG. 3.
Induction of S phase by E7 proteins in the presence of high levels of p16INK4a. (A) Expression of human p16INK4a in NIH 3T3 cells. One hundred micrograms of protein extracts of cells infected with different recombinant retroviruses (viral genes/p16INK4a) as indicated was applied to a 15% polyacrylamide-sodium dodecyl sulfate gel, transferred onto a polyvinylidene difluoride membrane, and incubated with an anti-human p16INK4a or β-tubulin (TUB2.1; Sigma) antibody. (B) Ability of E7 proteins to stimulate S-phase entry in p16INK4a-expressing NIH 3T3 cells. Double-infected cells were harvested, fixed, and stained with propidium iodide after neomycin and puromycin selection. The cell cycle profile was analyzed by flow cytometry. The data represent the means of four independent experiments. The difference between the values of HPV16 E7 or HPV38 E7 cells and control cells (pBabe) was statistically significant (pBabe versus HPV16 E7, P = 0.03; pBabe versus HPV38 E7, P = 0.03). The cell cycle profiles of a representative experiment are also shown (bottom panels).
Together, these data demonstrate that HPV38 E7 efficiently targets pRb and deregulates the G1/S transition.
HPV38 E6 and E7 alter cell cycle and senescence programs of primary cells.
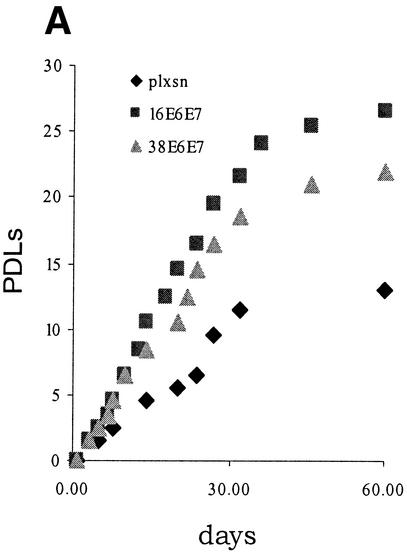
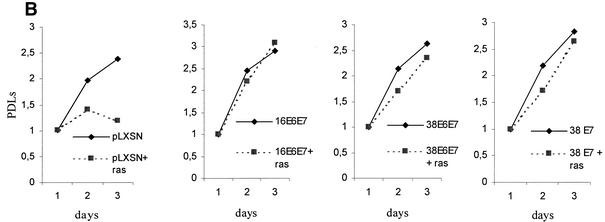
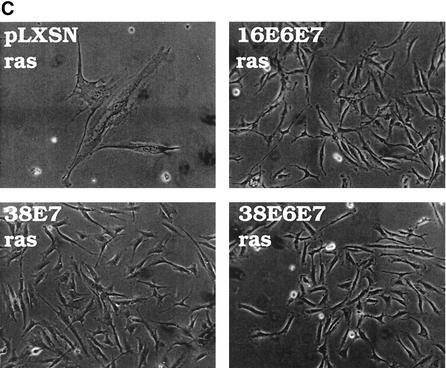
Our data show that, like HPV16 E7, HPV38 E7 has a strong transforming activity in immortalized fibroblasts. In contrast, HPV10 and HPV20 E7 proteins do not display this property. Based on these findings, HPV10 and HPV20 were not analyzed further. Studies on HPV16 have shown that the transforming activity of E7 is greatly increased in primary cells by the presence of E6 (20, 52), reflecting the natural situation in a viral infection, in which the two proteins act in concert to promote carcinogenesis. Therefore, we next analyzed the events induced by expression of the HPV38 E6 and E7 genes in primary human oral fibroblasts. After retroviral infection, the levels of E6 and E7 transcripts were determined by RT-PCR (data not shown). As shown in Fig. 4A, coexpression of both viral genes considerably stimulated proliferation of these cells. Mutations that constitutively activate Ras proteins can be found in skin cancers (reviewed in reference 3). Overexpression of activated ras in primary human fibroblasts causes accumulation of the CDK inhibitors p16INK4a, p21WAF1, and p27KIP1 (11), leading to a G1 cell cycle arrest and ultimately to premature senescence. However, this event is counteracted by expression of other oncogenes, such as adenovirus E1A (42). We determined whether HPV38 E6 and E7 were able to abolish the G1 arrest imposed by ras. Consistent with previous findings, cells expressing ras alone were not able to proliferate (Fig. 4B) and acquired a flat and enlarged morphology, characteristic of arrested cells (Fig. 4C) (42). In contrast, the cells coexpressing ras and the viral genes were able to proliferate at levels similar to those of the control cells (viral genes only) (Fig. 4B) and exhibited a transformed phenotype characterized by small and highly refractile cells (Fig. 4C). Similar results were obtained in cells expressing HPV38 E7 alone (Fig. 4C).
FIG. 4.
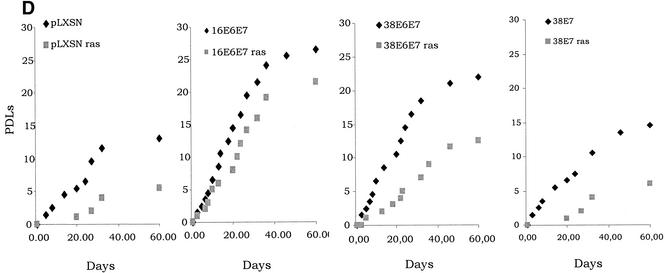
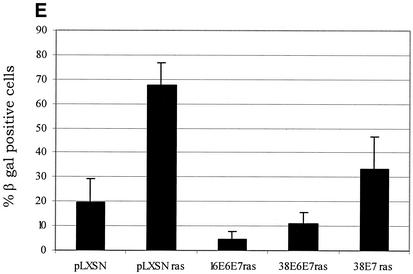
Activity of HPV38 E6 and E7 in primary human fibroblasts. (A) Long-term analysis of the growth profile of primary human fibroblasts expressing the HPV proteins. The graph represents the number of population doublings (PDL) at the indicated times. PDL 0 refers to the point at which drug selection was complete after infection with the different recombinant retroviruses. (B) Growth time course of the different cultures. After selection, growth was followed daily for 3 days by measuring the cellular protein content of cell cultures as previously described (46). The data are the means of two independent experiments, each performed in triplicate. (C) Morphology of primary human fibroblasts expressing the HPV proteins and the ras oncogene. The different cell lines (as indicated in the figure) were photographed at day 5 postselection. The same magnification was used (10×) in all the photomicrographs. (D) Long-term analysis of the growth profile of primary human fibroblasts expressing the HPV genes and ras. The graph represents the number of population doublings (PDL) at the indicated times. PDL 0 refers to the point at which drug selection was complete after retroviral infection. (E) Detection of senescent cells in the different cultures by β-galactosidase staining. Primary human oral fibroblasts were stained for β-galactosidase (pH 6) at day 30 postselection. Ten independent fields of at least 100 cells each were counted, and the percentage of stained blue cells was calculated.
To further analyze the properties of the HPV38 proteins, we followed the cultures expressing the E6 and E7 genes for another 2 months. The control cells (ras alone) remained attached to the plate but did not grow significantly, and the majority were positive for β-galactosidase at pH 6.0, a marker of senescence (15) (Fig. 4D and E). In contrast, HPV38 E6/E7-ras cells continued to proliferate, although less efficiently than their HPV16 counterparts (Fig. 4D). Interestingly, HPV38 E7 alone appears to be less efficient than HPV38 E6/E7 in promoting long proliferation of cells overexpressing ras, indicating that E6 also contributes to this event. Most importantly, few senescent cells were detected in HPV38 E6/E7 culture, while a much higher percentage of senescent cells was observed in the HPV38 E7 culture (Fig. 4E). Together, these data show that both proteins, HPV38 E6 and E7, have the ability to alter the cell cycle control of ras-expressing cells.
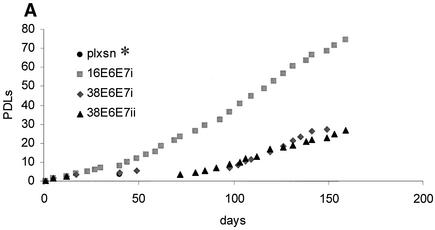
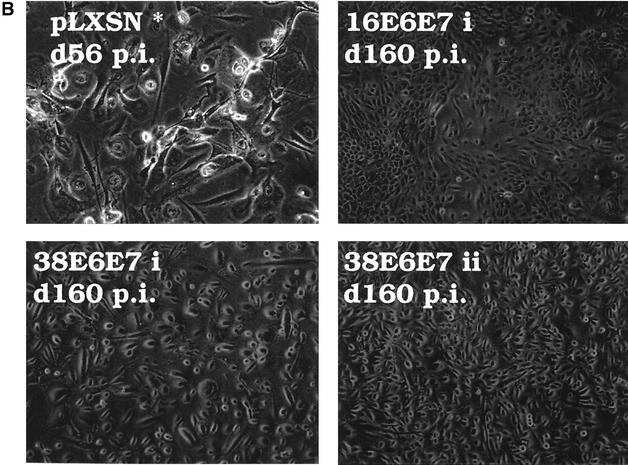
Since HPV infection and its life cycle are restricted to keratinocytes, we analyzed the effects induced by HPV38 E6 and E7 expression in primary human keratinocytes. HPV16 E6 and E7 were included as positive controls because several independent studies have demonstrated their ability to increase the life span of and immortalize these cells (55). Following infection with various recombinant amphotropic retroviruses, as indicated in Fig. 5A, keratinocytes were cultured in selective medium and their longevity was determined by monitoring growth (Fig. 5A). Control cells reached senescence after three to four doublings, while HPV16 E6 and E7 keratinocytes showed continuous proliferation (5 to 6 months, ≈80 population doublings). HPV38 E6- and E7-expressing cells grew for a few passages and then underwent a time period with no apparent growth. Thereafter, they began to proliferate (5 to 6 months, ≈30 population doublings) (Fig. 5A).
FIG. 5.

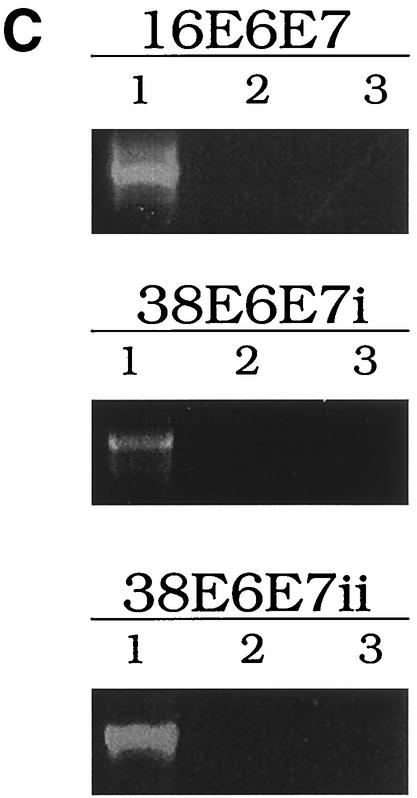
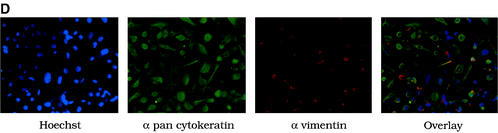
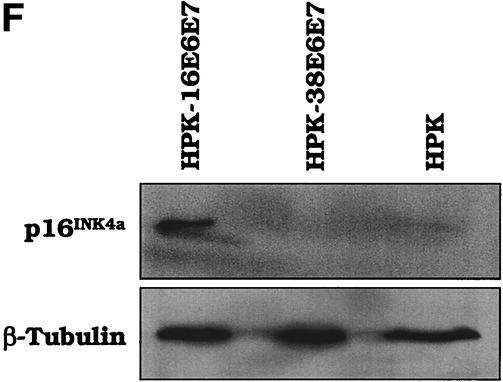
HPV38 E6 and E7 increase the life span of primary human keratinocytes. (A) Long-term analysis of the growth profile of primary human keratinocytes. The graph represents the number of population doublings (PDL) at the indicated times. PDL 0 refers to the point at which drug selection was complete after retroviral infection. After 60 days, pLXSN∗ cultures (infected with empty vector retrovirus) did not contain any viable keratinocytes. (B) Morphology of primary human keratinocytes expressing E6 and E7 of HPV16 or HPV38. The different cell cultures (as indicated in the figure) were photographed at the indicated times postinfection. After 60 days, pLXSN∗ cultures (infected with empty vector retrovirus) did not contain any viable keratinocytes. HPV38 E6/E7 keratinocytes i and ii are different cultures which were obtained in two independent experiments. (C) Determination of mRNA levels of E6 and E7 by RT-PCR. Total RNA and cDNA were prepared as described in Materials and Methods. The cDNA obtained was used as the template for PCR (lane 1). As a negative control, a PCR was performed omitting the reverse transcriptase step (lane 2) or with water as the template (lane 3). (D) Immunofluorescence of HPV38 E6/E7 keratinocytes. Cells were fixed and double stained with the pankeratin antibody (Dako; green) and the vimentin antibody (Progen; red), which stain keratinocytes and fibroblasts, respectively. DNA was stained with Hoechst 33342 blue. (E) Telomerase activity in primary human keratinocytes expressing E6 and E7 of HPV16 or HPV38. All the assays were performed in duplicate. IC, internal control (36-bp standard), which is provided in the TRAPeze kit (Intergen Company, Oxford, United Kingdom). (F) Determination of p16INK4a levels in human keratinocytes expressing the viral proteins. One hundred micrograms of protein extracts of the cell cultures indicated in the figure was applied to a 15% polyacrylamide-sodium dodecyl sulfate gel, transferred to a polyvinylidene difluoride membrane, and incubated with an anti-human p16INK4a or β-tubulin (TUB2.1; Sigma) antibody.
The morphology of the keratinocyte cultures at the times indicated is shown in Fig. 5B. RT-PCR analysis revealed that the E6 and E7 genes were actively transcribed in each culture (Fig. 5C). To confirm the epithelial nature of the cells in culture, we checked for the presence of keratins. Western blot and immunofluorescence analyses revealed that all cultures expressed these specific epithelial markers (Fig. 5D and data not shown). Since keratinocytes do not normally express vimentin, an antivimentin antibody was used as a control. As expected, only fibroblasts of the feeder layers were strongly positive for this stain (Fig. 5D). Weak vimentin staining was also observed in keratinocytes, in accord with previously reported findings (51).
In summary, similar to E6 and E7 from the oncogenic HPV16, HPV38 E6 and E7 increase the life span of human primary keratinocytes, an important requirement for the establishment of a tumor phenotype. Since we have shown that the HPV16 and HPV38 E7 proteins have similar properties, it is likely that the differences that we observed in the initial growth pattern of HPV16 E6/E7 and HPV38 E6/E7 cells are due to intrinsic biological properties of HPV38 E6 and HPV16 E6. Indeed, we have observed that in contrast to HPV16 E6, HPV38 E6 is unable to promote p53 degradation (S. Caldeira and M. Tommasino, unpublished data).
Reflecting their proliferative state, HPV38 E6/E7 cells have detectable telomerase activity, although less than HPV16 E6/E7 cells (Fig. 5E). Again, this difference may be dependent on the different properties of the two E6 proteins.
Previous studies have shown that neutralization of the pRb pathway by HPV16 E7 leads to accumulation of high levels of p16INK4a (26). Surprisingly, HPV38 E6/E7 cells do not express p16INK4a (Fig. 5F), in contrast to HPV16 E6/E7 cells. Most likely, loss of p16INK4a expression occurred during the postretroviral infection crisis of HPV38 E6/E7 keratinocytes. However, this event may marginally contribute to the increase in life span of the keratinocytes, since our data described above (Fig. 3) show that HPV38 E7 is competent to overcome the inhibitory function of p16INK4a.
HPV38 DNA is present in nonmelanoma skin cancers.
Since our findings show that HPV38 has in vitro transforming activity, we assessed the relevance of HPV38 infection in skin cancer development. We determined the presence of HPV38 DNA in the two epidermal tumor types, squamous cell carcinoma and basal cell carcinoma, from immunocompetent individuals. In addition, healthy breast skin, which is not normally exposed to the sun, and samples of actinic keratosis lesions, which are heavily exposed to the sun and are potential precursor lesions of squamous cell carcinoma, were included in the analysis. The presence of HPV38 DNA was determined by PCR with specific HPV38 E6 internal primers and confirmed by Southern blotting. The results are summarized in Table 1. HPV38 DNA was more frequently detected in basal cell carcinomas (55%, P < 0.0001), squamous cell carcinomas (46%, P = 0.0011), and in actinic keratosis lesions (32%, P = 0.02) than in healthy skin (10%).
TABLE 1.
PCR analysis of human skin specimens for the detection of HPV38 DNAa
| Skin sample (no. of samples) | No. positive | % Positive (95% C.I.) |
|---|---|---|
| Basal cell carcinoma (n = 69) | 38 | 55 (42.6-67.1) |
| Squamous cell carcinoma (n = 26) | 12 | 46 (26.6-66.6) |
| Actinic keratosis (n = 46) | 15 | 32 (19.5-48.0) |
| Healthy skin (n = 41) | 4 | 10 (2.7-23.13) |
DNA preparation from skin specimens and PCR were performed as described in the text. The confidence interval (C.I.) is given in parentheses. The statistical analysis was performed with Fisher's exact test for comparing two proportions (healthy skin versus actinic keratosis, basal cell carcinoma, or squamous cell carcinoma).
We also examined the presence of HPV10 DNA in a subset of skin cancers (basal cell carcinoma [n = 28] and squamous cell carcinoma [n = 17]) with E6-specific primers. We detected HPV10 only twice (one basal cell carcinoma and one squamous cell carcinoma). In both cases, HPV38 was also present. Thus, in line with the functional data, HPV10 appears to not be associated with malignant skin lesions.
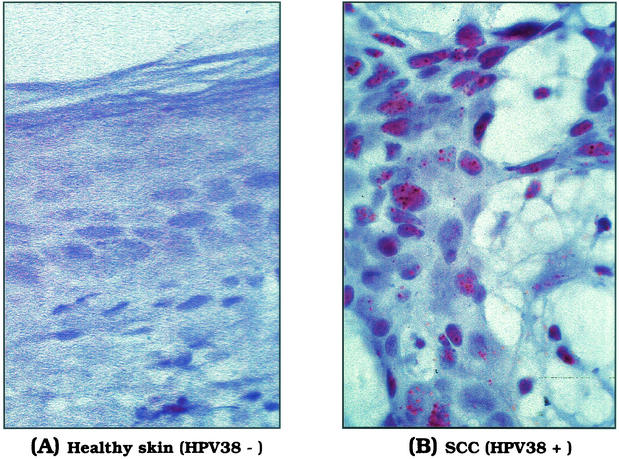
To further demonstrate the presence of HPV38 in the skin lesions, we performed in situ hybridization in a subgroup of the specimens with an HPV38 probe (five HPV38-negative cases and five HPV38-positive cases in PCR analysis). Figure 6 shows representative cases of negative (Fig. 6A, healthy skin) and positive (Fig. 6B, squamous cell carcinoma) HPV38 skin specimens. In the cases analyzed, HPV38 DNA was detected exclusively in the nucleus of cancer cells, while no nuclear staining was observed in healthy tissue (Fig. 6).
FIG. 6.
Detection of HPV38 DNA in skin lesions by in situ hybridization. In situ hybridization was performed with a genomic DNA probe of HPV38. The micrographs show an HPV38-negative healthy skin specimen (A) and a squamous cell carcinoma containing HPV38 DNA (B). Positive signals are seen as punctate, brown precipitates in almost all nuclei of the tumor tissue. Original magnification, ×60.
DISCUSSION
Cancer is considered a cell cycle disease. In this study, we have shown that HPV38 E6 and E7 can deregulate cell cycle control and induce cellular transformation. Similar to the E7 from HPV16, which is the etiological agent of cervical cancer, HPV38 E7 can efficiently inactivate pRb and prevail over the G1/S transition controls. In addition, we have demonstrated for the first time that the early proteins E6 and E7 of a cutaneous HPV type are sufficient to substantially increase the life span of primary human keratinocytes. In fact, the HPV38 E6/E7 keratinocytes are still proliferating. However, the individual contribution of each viral protein to this process remains to be fully characterized in future studies.
In agreement with the in vitro transforming properties of HPV38, we detected HPV38 DNA in approximately 50% of nonmelanoma skin cancers by PCR and Southern blot analysis. In addition, we detected the presence of viral DNA in the nucleus of cancer cells by in situ hybridization. Together, these data suggest a role for HPV38 infection in carcinogenesis. A previous study has also detected HPV38 in nonmelanoma skin cancers, although at lower frequency (14). This difference could be explained by the fact that in our study a specific HPV38 set of primers was used rather than consensus and/or degenerate primers. Indeed, it has been shown that different PCR assays impact on the frequency and spectrum of cutaneous HPV types detected in skin lesions (33).
Skin carcinogenesis is a complex and multifactorial process in which sun exposure is known to play a major role (2, 4). It has been reported that p53 is often mutated in skin cancers (3), while this event is extremely rare in HPV16-induced cervical tumors (48). Our initial data show that HPV38 E6 is not able to induce p53 degradation (Caldeira and Tommasino, unpublished data). Most likely, the UV-mediated DNA damage during sun exposure is responsible for inactivation of p53, rendering E6-mediated p53 degradation unnecessary in skin tumorigenesis. Furthermore, it is possible that HPV38 E6 may prevent apoptosis in a p53-independent manner. In fact, it has been reported that cutaneous as well as mucosal E6s have the ability to inactivate the proapoptotic protein Bak, promoting its degradation (23). We are further investigating whether HPV38 oncoproteins and UV-induced mutations in cellular genes (such as p53 and/or ras) act synergistically in the transformation of the host cell. Our initial data obtained by coexpressing activated ras and HPV38 genes favor this possibility.
It is our belief that the immense variety and ubiquity of HPVs will hamper epidemiological studies, which aim to determine a possible association of specific HPV types with skin cancers in non-epidermodysplasia verruciformis patients. Here, we propose that the establishment of such an association can be facilitated by the analysis of the biological properties of the viral oncoproteins E6 and E7. This approach represents a valid tool to evaluate the carcinogenic potential of different HPV types in future studies. Although our data suggest a role for HPV38 in skin cancer development, they do not exclude the possibility that other cutaneous HPV types are also involved in this event. For instance, further studies are required to evaluate the oncogenicity of other cutaneous HPV types, such as HPV22 and -23, whose E7 proteins are almost 90% similar to HPV38 E7 and are therefore likely to have the same biological activities.
In summary, the data presented will impact on epidemiological, immunological, and clinical studies aiming to establish a causal relationship between the presence of specific HPV types and skin carcinogenesis and to develop therapeutic and preventive strategies for nonmelanoma skin cancers.
Acknowledgments
We are grateful to Harald zur Hausen and Lutz Gissmann for continuous support and interest in our work. We thank all the members of our laboratory and Dirk Breitkreutz for their cooperation, Lutz Edler for the statistical analysis, Bruno Amati, Gordon Peters, and Pascal Tomakidi for providing reagents, and Leonie Ringrose for critical reading of the manuscript.
S.C. was supported by a Praxis XXI doctoral fellowship (Sub Programa Ciencia e Tecnologia do 2° Quadro Comunitario de Apoio). P.B. is supported by Sander Stiftung.
REFERENCES
- 1.Alunni-Fabbroni, M., T. Littlewood, L. Deleu, S. Caldeira, M. Giarre, M. Dell' Orco, and M. Tommasino. 2000. Induction of S phase and apoptosis by the human papillomavirus type 16 E7 protein are separable events in immortalized rodent fibroblasts. Oncogene 19:2277-2285. [DOI] [PubMed] [Google Scholar]
- 2.Ananthaswamy, H. N., S. M. Loughlin, P. Cox, R. L. Evans, S. E. Ullrich, and M. L. Kripke. 1997. Sunlight and skin cancer: inhibition of p53 mutations in UV-irradiated mouse skin by sunscreens. Nat. Med. 3:510-514. [DOI] [PubMed] [Google Scholar]
- 3.Ananthaswamy, H. N., and W. E. Pierceall. 1992. Molecular alterations in human skin tumors. Prog. Clin. Biol. Res. 376:61-84. [PubMed] [Google Scholar]
- 4.Armstrong, B. K., and A. Kricker. 2001. The epidemiology of UV induced skin cancer. J. Photochem. Photobiol. B 63:8-18. [DOI] [PubMed] [Google Scholar]
- 5.Astori, G., D. Lavergne, C. Benton, B. Hockmayr, K. Egawa, C. Garbe, and E. M. de Villiers. 1998. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J. Investig. Dermatol. 110:752-755. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout, R. J., J. N. Bouwes Bavinck, and J. ter Schegget. 2000. Persistence of human papillomavirus DNA in benign and (pre)malignant skin lesions from renal transplant recipients. J. Clin. Microbiol. 38:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout, R. J., L. M. Tieben, H. L. Smits, J. N. Bavinck, B. J. Vermeer, and J. ter Schegget. 1995. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J. Clin. Microbiol. 33:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bickenbach, J. R., V. Vormwald-Dogan, C. Bachor, K. Bleuel, G. Schnapp, and P. Boukamp. 1998. Telomerase is not an epidermal stem cell marker and is downregulated by calcium. J. Investig. Dermatol. 111:1045-1052. [DOI] [PubMed] [Google Scholar]
- 9.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boxman, I. L., R. J. Berkhout, L. H. Mulder, M. C. Wolkers, J. N. Bouwes Bavinck, B. J. Vermeer, and J. ter Schegget. 1997. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J. Investig. Dermatol. 108:712-715. [DOI] [PubMed] [Google Scholar]
- 11.Bringold, F., and M. Serrano. 2000. Tumor suppressors and oncogenes in cellular senescence. Exp. Gerontol. 35:317-329. [DOI] [PubMed] [Google Scholar]
- 12.Cairns, P., T. J. Polascik, Y. Eby, K. Tokino, J. Califano, A. Merlo, L. Mao, J. Herath, R. Jenkins, W. Westra, et al. 1995. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat. Genet. 11:210-212. [DOI] [PubMed] [Google Scholar]
- 13.Ciccolini, F., G. Di Pasquale, F. Carlotti, L. Crawford, and M. Tommasino. 1994. Functional studies of E7 proteins from different HPV types. Oncogene 9:2342-2348. [PubMed] [Google Scholar]
- 14.de Villiers, E. M., D. Lavergne, K. McLaren, and E. C. Benton. 1997. Prevailing papillomavirus types in nonmelanoma carcinomas of the skin in renal allograft recipients. Int. J. Cancer 73:356-361. [DOI] [PubMed] [Google Scholar]
- 15.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, W. L., S. Caldeira, P. Sehr, M. Pawlita, and M. Tommasino. 2001. Determination of the binding affinity of different human papillomavirus E7 proteins for the tumour suppressor pRb by a plate-binding assay. J. Virol. Methods 98:91-98. [DOI] [PubMed] [Google Scholar]
- 17.Frisch, M., and M. Melbye. 1995. New primary cancers after squamous cell skin cancer. Am. J. Epidemiol. 141:916-922. [DOI] [PubMed] [Google Scholar]
- 18.Giarrè, M., S. Caldeira, I. Malanchi, F. Ciccolini, M. J. Leão, and M. Tommasino. 2001. Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a-imposed G1 cell cycle arrest. J. Virol. 75:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartevelt, M. M., J. N. Bavinck, A. M. Kootte, B. J. Vermeer, and J. P. Vandenbroucke. 1990. Incidence of skin cancer after renal transplantation in the Netherlands. Transplantation 49:506-509. [DOI] [PubMed] [Google Scholar]
- 22.Jablonska, S., J. Dabrowski, and K. Jakubowicz. 1972. Epidermodysplasia verruciformis as a model in studies on the role of papovaviruses in oncogenesis. Cancer Res. 32:583-589. [PubMed] [Google Scholar]
- 23.Jackson, S., C. Harwood, M. Thomas, L. Banks, and A. Storey. 2000. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahn, H. S., L. M. Tatham, A. V. Patel, M. J. Thun, and C. W. Heath, Jr. 1998. Increased cancer mortality following a history of nonmelanoma skin cancer. JAMA 280:910-912. [DOI] [PubMed] [Google Scholar]
- 25.Kamb, A., N. Gruis, J. Weaver-Feldhaus, Q. Liu, K. Harshman, S. Tavtigian, E. Stockert, R. Day, III, B. Johnson, and M. H. Skolnick. 1994. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264:436-440. [DOI] [PubMed] [Google Scholar]
- 26.Khleif, S. N., J. DeGregori, C. L. Yee, G. A. Otterson, F. J. Kaye, J. R. Nevins, and P. M. Howley. 1996. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc. Natl. Acad. Sci. USA 93:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremsdorf, D., S. Jablonska, M. Favre, and G. Orth. 1983. Human papillomaviruses associated with epidermodysplasia verruciformis. II. Molecular cloning and biochemical characterization of human papillomavirus 3a, 8, 10, and 12 genomes. J. Virol. 48:340-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levi, F., C. La Vecchia, V. C. Te, L. Randimbison, and G. Erler. 1998. Incidence of invasive cancers following basal cell skin cancer. Am. J. Epidemiol. 147:722-726. [DOI] [PubMed] [Google Scholar]
- 29.Levi, F., L. Randimbison, C. La Vecchia, G. Erler, and V. C. Te. 1997. Incidence of invasive cancers following squamous cell skin cancer. Am. J. Epidemiol. 146:734-739. [DOI] [PubMed] [Google Scholar]
- 30.Majewski, S., S. Jablonska, and G. Orth. 1997. Epidermodysplasia verruciformis. Immunological and nonimmunological surveillance mechanisms: role in tumor progression. Clin. Dermatol. 15:321-334. [DOI] [PubMed] [Google Scholar]
- 31.Mann, D. J., and N. C. Jones. 1996. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr. Biol. 6:474-483. [DOI] [PubMed] [Google Scholar]
- 32.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, T., R. Arndt, E. Christophers, and E. Stockfleth. 2000. Frequency and spectrum of HPV types detected in cutaneous squamous-cell carcinomas depend on the HPV detection system: a comparison of four PCR assays. Dermatology 201:204-211. [DOI] [PubMed] [Google Scholar]
- 34.Miller, D. L., and M. A. Weinstock. 1994. Nonmelanoma skin cancer in the United States: incidence. J. Am. Acad. Dermatol. 30:774-778. [DOI] [PubMed] [Google Scholar]
- 35.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 37.Orth, G., S. Jablonska, M. Jarzabek-Chorzelska, S. Obalek, G. Rzesa, M. Favre, and O. Croissant. 1979. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 39:1074-1082. [PubMed] [Google Scholar]
- 38.Parker, S. L., T. Tong, S. Bolden, and P. A. Wingo. 1996. Cancer statistics, 1996 CA. Cancer J. Clin. 46:5-27. [DOI] [PubMed] [Google Scholar]
- 39.Patrick, D. R., A. Oliff, and D. C. Heimbrook. 1994. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J. Biol. Chem. 269:6842-6850. [PubMed] [Google Scholar]
- 40.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt, A., J. B. Harry, B. Rapp, F. O. Wettstein, and T. Iftner. 1994. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J. Virol. 68:7051-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 43.Shamanin, V., H. zur Hausen, D. Lavergne, C. M. Proby, I. M. Leigh, C. Neumann, H. Hamm, M. Goos, U. F. Haustein, E. G. Jung, et al. 1996. Human papillomavirus infections in nonmelanoma skin cancers from renal transplant recipients and nonimmunosuppressed patients. J. Natl. Cancer Inst. 88:802-811. [DOI] [PubMed] [Google Scholar]
- 44.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 45.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 46.Skehan, P., R. Storeng, D. Scudiero, A. Monks, J. McMahon, D. Vistica, J. T. Warren, H. Bokesch, S. Kenney, and M. R. Boyd. 1990. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82:1107-1112. [DOI] [PubMed] [Google Scholar]
- 47.Stern, R. S. 1999. The mysteries of geographic variability in nonmelanoma skin cancer incidence. Arch. Dermatol. 135:843-844. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, M., D. Pim, and L. Banks. 1999. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 18:7690-7700. [DOI] [PubMed] [Google Scholar]
- 49.Tomakidi, P., D. Breitkreutz, N. E. Fusenig, J. Zoller, A. Kohl, and G. Komposch. 1998. Establishment of oral mucosa phenotype in vitro in correlation to epithelial anchorage. Cell Tissue Res. 292:355-366. [DOI] [PubMed] [Google Scholar]
- 50.Tomakidi, P., N. E. Fusenig, A. Kohl, and G. Komposch. 1997. Histomorphological and biochemical differentiation capacity in organotypic cocultures of primary gingival cells. J. Periodontal Res. 32:388-400. [DOI] [PubMed] [Google Scholar]
- 51.Van Muijen, G. N., S. O. Warnaar, and M. Ponec. 1987. Differentiation-related changes of cytokeratin expression in cultured keratinocytes and in fetal, newborn, and adult epidermis. Exp. Cell Res. 171:331-345. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe, S., T. Kanda, and K. Yoshiike. 1989. Human papillomavirus type 16 transformation of primary human embryonic fibroblasts requires expression of open reading frames E6 and E7. J. Virol. 63:965-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zehbe, I., A. Ratsch, M. Alunni-Fabbroni, A. Burzlaff, E. Bakos, M. Durst, E. Wilander, and M. Tommasino. 1999. Overriding of cyclin-dependent kinase inhibitors by high and low risk human papillomavirus types: evidence for an in vivo role in cervical lesions. Oncogene 18:2201-2211. [DOI] [PubMed] [Google Scholar]
- 54.Zehbe, I., E. Wilander, H. Delius, and M. Tommasino. 1998. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 58:829-833. [PubMed] [Google Scholar]
- 55.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]