Abstract
Crystal structures of human immunodeficiency virus type 1 (HIV-1) capsid protein (CA) reveal that the last 11 C-terminal amino acids are disordered. This disordered region contains a glycine-rich sequence 353-GVGGP-357 (numbering refers to the initiation methionine of Gag) that is highly conserved within the Gag proteins of HIV-1, HIV-2, and simian immunodeficiency virus, which suggests the importance of this sequence in virus replication. In the present study, we demonstrate that changing any individual residue within this short region in the context of the full-length HIV-1 genome virtually abolishes production of extracellular virus particles, in either the presence or absence of viral protease activity. This severe defect in virus particle production results from impaired Gag multimerization, as well as from decreased Gag association with the cellular membranes, as demonstrated by the results of gradient sedimentation and membrane flotation centrifugation assays. These findings are further supported by the diffuse distribution pattern of the mutant Gag within the cytoplasm, as opposed to the punctate distribution of the wild-type Gag on the plasma membrane. On the basis of these results, we propose that the disordered feature of amino acid stretch 353-GVGGP-357 in the CA crystal forms may have allowed Gag to adopt multiple conformations and that such structural flexibility is needed by Gag in order to construct geometrically complex particles.
Human immunodeficiency virus type 1 (HIV-1) Gag proteins are able to undergo self-assembly and to drive the production of virus-like particles (20). Upon activation of viral protease during or shortly after viral budding, Gag is cleaved at distinct sites; this event leads to the generation of mature proteins that include matrix (MA), capsid (CA), nucleocapsid (NC), and p6, as well as two spacer peptides, p2 and p1 (26). These mature proteins are involved in the construction of subviral structures. MA is located at the inner leaflet of the virus membrane, CA molecules constitute the shell of the conical core, and NC is tightly associated with viral genomic RNA (for a review, see reference 15). The order of protease cleavage at the distinct sites is temporally regulated such that the mature proteins are released in a sequential manner (13, 33, 35, 49, 58). Temporally ordered cleavage of Gag has significant implications for virus morphogenesis. For example, release of p2 from the C terminus of CA is the latest step during Gag processing, and this event triggers morphological transition of the capsid core from a spherical to a conical form (25, 64).
Gag proteins alone are able to generate virus-like particles (20). This Gag function involves three distinct protein domains: the membrane-binding (M) domain, the Gag-Gag interaction (I) domain, and the late (L) domain. The M domain is located at the N terminus of MA and consists of a covalently attached myristate acid, as well as a number of basic residues (20, 23, 56, 67). The myristate moiety anchors Gag into the lipid bilayer, and the basic residues further stabilize Gag-membrane association through electrostatic interactions with the negatively charged phosphate groups. The I domain comprises multiple basic residues within the NC sequences (5-7, 9, 10, 24, 53). This domain promotes Gag-Gag interaction largely by virtue of its high affinity for RNA. The L domain has been mapped to the tetrapeptide motif P(T/S)AP at the N terminus of p6. It assists virus detachment from host cells by recruiting cellular factors such as TSG101 (19, 22, 28, 41, 60; for a review, see reference 16).
In addition to these three functional domains, the C-terminal domain (CTD) of CA (CA amino acids 150 to 231) is also indispensable for virus particle formation (11, 31, 52, 61, 63, 66). Within this domain exists a major homology region (MHR) that is highly conserved among retroviral Gag proteins. The importance of MHR in virus assembly has been demonstrated by a number of mutagenesis studies (4, 11, 12, 40, 42, 51, 52, 61) with the exception of one that described a minor effect of the MHR deletion on virus particle production (57). This latter observation is likely due to the use of an overexpression system to produce Gag proteins. MHR may play essential roles in the multimerization, as well as the membrane targeting, of Gag proteins. The crystal structure analysis of CA reveals a dimer interface that is constituted by a pack of helix 2 between CA molecules (17, 43, 65). Presumably, disruption of this interface by mutations will affect CA-CA interactions, and consequently, inhibit virus assembly.
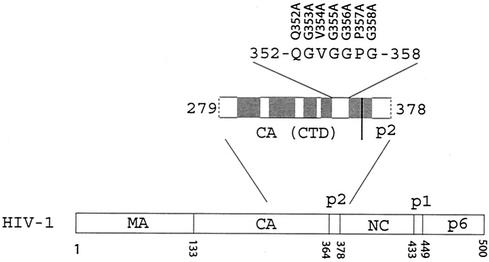
Interestingly, the last 11 to 13 amino acids at the C terminus of CA, i.e., 352-QGVGGPGHKARVL-364 (numbering refers to the initiation methionine of Gag), are disordered in the crystal forms of CA146-231 or CA146-231-p2 (17, 65). Amino acids 359-HKARVL-364 constitute the N-terminal portion of a putative α-helix that extends into the adjacent p2 region (1). This helical structure has been implicated in regulation of virus assembly (1). Immediately upstream of these six amino acids is a glycine-rich motif 353-GVGGPG-358 that is highly conserved in Gag proteins of primate lentiviruses, including HIV-1, HIV-2, and simian immunodeficiency virus (Table 1) (36). This highly conserved feature suggests an important function of these amino acids in virus production. In the present study, we have changed each of six amino acids (353-GVGGPG-358) and measured the effects of these mutations on virus particle formation. Our data demonstrate that altering any of five amino acids (353-GVGGP-357) prevents the formation of large Gag complexes and, consequently, impairs virus particle generation.
TABLE 1.
Conservation of the 352-QGVGGPG-358 amino acids within the primate lentiviral Gag
| Amino acid | % Conservation in HIV-1a | % Conservation in HIV-2 or SIVb |
|---|---|---|
| Q352 | 99 | 100 |
| G353 | 100 | 100 |
| V354 | 100 | 82 |
| G355 | 100 | 96 |
| G356 | 100 | 100 |
| P357 | 100 | 100 |
| G358 | 58 | 96 |
This analysis included sequences from 102 different types of HIV-1, as well as CPZ strains, as summarized previously (36).
A total of 28 strains of HIV-2 or SIV were analyzed.
MATERIALS AND METHODS
Plasmid construction.
An infectious HIV-1 cDNA clone BH10 was employed as starting material for the following mutagenesis experiments. Each of the seven amino acids 352-QGVGGPG-358 was individually changed to alanine in the context of the full-length HIV-1 genome. The mutations thus generated were termed Q352A, G353A, V354A, G355A, G356A, P357A, and G358A (Fig. 1). These mutations were engineered by PCR through use of an antisense primer pNC-A (5′-TTAGCCTGTCTCTCAGTACAATC-3′), together with the following primers: p1840A, 5′-GATGACAGCATGTgcGGGAGTAGGAGGAC-3′; p1843A, 5′-CAGCATGTCAGGcAGTAGGAGGAC-3′; p1846A, 5′-CATGTCAGGGAGcAGGAGGACCCG-3′; p1849A, 5′-GTCAGGGAGTAGcAGGACCCGGCC-3′; p1852A, 5′-GCATGTCAGGGAGTAGGAGcACCCGGCCATAAG-3′; p1855A, 5′-CATGTCAGGGAGTAGGAGGAgCCGGCCATAAGGC-3′; and p1858A, 5′-CAGGGAGTAGGAGGACCCGcCCATAAGGCAAGAG-3′.
FIG. 1.
Illustration of the 352-QGVGGPG-358 sequences located at the C terminus of CA. Each of these amino acids was changed to alanine, and the resultant mutations are shown at the top of the peptide that is illustrated. The domain structure of HIV-1 Pr55Gag is presented at the bottom. On the top is the expanded depiction of the CTD of CA and the adjacent p2 peptide. Locations of the four α-helices within the CTD of CA are depicted as shaded boxes (3, 17, 43, 65). A putative α-helix across the boundary of CA and p2 is also indicated (1). The numbering of amino acids refers to the initiation methionine of Gag.
The mutated nucleotides in each primer are indicated by lowercase letters. The resulting PCR products were used as megaprimers in a second round of PCR, together with the sense primer Sph-S (5′-AGTGCATCCAGTGCATGCAGGGCC-3′). The final PCR products were digested by restriction enzymes SphI and ApaI and inserted into BH10 that had been digested by the same two enzymes.
Cell culture, transfection, and infection.
COS-7 and HeLa cells were grown in Dulbecco modified Eagle medium, and Jurkat cells were grown in RPMI 1640 medium, each supplemented with 10% fetal calf serum. Transfection was performed by using Lipofectamine (Invitrogen, Burlington, Ontario, Canada) in accordance with the manufacturer's instructions. Virus growth in Jurkat cells was monitored by measuring reverse transcriptase (RT) activity in culture fluids at various times.
Measurement of virus production.
At 40 h after transfection of COS-7 or HeLa cells, culture fluids were clarified at 3,000 rpm at 4°C for 30 min in a GS-6R Beckman centrifuge. Virus particles were then pelleted at 35,000 rpm at 4°C for 1 h in a Beckman ultracentrifuge by using an SW41 rotor. The pelleted virus particles were suspended in TNE buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 2 mM EDTA). Amounts of virus particles were determined by either quantitative enzyme-linked immunosorbent assay (ELISA) for HIV-1 p24(CA) (Vironostika HIV-1 Antigen MicroELISA System; Organon Teknika Corp., Durham, N.C.) or RT assays. Virion-associated Gag proteins were assessed by Western blot analyses with monoclonal antibodies (MAbs) against HIV-1 p24(CA) antigen (ID Lab, Inc., London, Ontario, Canada).
Levels of cell-associated Gag proteins were also measured by either quantitative ELISA or Western blot analysis. Briefly, transfected COS-7 or HeLa cells were washed twice with cold phosphate-buffered saline (PBS), scraped from the bottom of the plates, and lysed in Nonidet P-40 [NP-40] lysis buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, 0.02% sodium azide, and a cocktail of protease inhibitors that antagonize the activity of cellular proteases [Roche, Laval, Quebec, Canada]). Cell lysates were first clarified at 3,000 rpm for 30 min at 4°C in a GS-6R Beckman centrifuge and then subjected to quantitative ELISA or Western blot analysis with MAbs against HIV-1 p24(CA) antigen. The efficiency of virus particle production for each construct was represented by the percentage of the virion-associated p24 levels over the total levels of cell- and virion-associated p24.
Fractionation of Gag complexes.
Transfected COS-7 cells were harvested and then suspended in 1 ml of cold TNE buffer containing 50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.1% 2-mercaptoethanol, and protease inhibitor cocktails (Roche). After Dounce homogenization on ice, homogenate was first clarified at 3,000 rpm for 30 min at 4°C in a Beckman GS-6R centrifuge to remove cell debris and nuclei and then placed on the top of 11 ml of a 15 to 50% continuous sucrose gradient (in TNE). These sucrose concentrations, as well as those described in the following, are defined as grams/100 ml. The gradient was centrifuged either at 210,000 × g for 1 h at 4°C or at 100,000 × g for 16 h at 4°C. For each gradient, 12 1-ml fractions were collected. Aliquots of each fraction were analyzed by Western blotting to measure levels of Gag proteins with anti-p24 MAbs. The densities of the gradient fractions were measured through use of the refractometer (Carl Zeiss).
Metabolic labeling of viral Gag proteins with 35S-labeled methionine and cysteine and immunoprecipitation.
At 40 h after transfection, COS-7 cells were starved in Dulbecco modified Eagle medium (lacking methionine and cysteine) at 37°C for 2 h, followed by metabolic labeling with 35S-labeled methionine and cysteine (Tran35S-label; 100 μCi/ml; ICN, Irvine, Calif.) at 37°C for 7 min. Cells were immediately washed twice with ice-cold PBS and then harvested. After Dounce homogenization and clarification as described above, the denucleated homogenate was placed on the top of a 15 to 50% continuous sucrose gradient and centrifuged at 100,000 × g for 16 h at 4°C. Twelve 1-ml fractions were collected for each gradient, and aliquots of each fraction were analyzed either by anti-p24 Western blotting to detect total Gag proteins or by immunoprecipitation to assess 35S-labeled Gag.
Immunoprecipitation was performed as follows. For each fraction, 300 μl of samples were mixed with an equal volume of NET-gel buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% NP-40, 1 mM EDTA [pH 8.0], 0.25% gelatin, and 0.02% sodium azide) containing 1 μg of anti-p24 MAbs (ID Lab, Inc.). After incubation at 4°C for 1 h, 5 μl of protein A-linked Sepharose CL-4B (Amersham Pharmacia Biotech, Inc., Baie d'Urfe, Quebec, Canada) was added for a further 30 min of incubation. The anti-p24 MAb:Gag complexes, bound to protein A, were pelleted and washed twice with NET-gel buffer and once with a buffer containing 10 mM Tris-Cl [pH 7.5] and 0.1% NP-40. The recovered pellets were suspended in 1× sodium dodecyl sulfate-gel loading buffer, boiled for 5 min, and separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. 35S-labeled Gag proteins were visualized by exposure to X-ray films.
Membrane flotation sedimentation.
The flotation assays were performed as described previously (47). Briefly, transfected COS-7 cells were harvested and Dounce homogenized in TNE buffer containing 10% sucrose. A volume of 250 μl of denucleated homogenate was mixed with 1.25 ml of 85.5% sucrose and placed at the bottom of a 5-ml ultracentrifuge tube. On the top of the cell homogenate (containing 73% sucrose) were layered 2.5 ml of 65% sucrose and 1 ml of 10% sucrose (in TNE). The discontinuous gradient was centrifuged at 100,000 × g for 18 h at 4°C. Eight 625-μl fractions were collected for each gradient, and aliquots of each fraction were assessed by anti-p24 Western blotting.
Immunofluorescence staining and confocal microscopy.
Transfected COS-7 cells were first fixed with 4% paraformaldehyde (in PBS) at room temperature for 20 min and then were permeabilized with 0.2% Triton X-100 (in PBS) at room temperature for 10 min. After being blocked with 5% milk (in PBS) for 20 min, cells were first incubated with primary anti-p24 MAbs at 37°C for 1 h, followed by a 1-h incubation with fluorescein isothiocyanate-conjugated secondary antibodies at 37°C. After an extensive washing with PBS, immunofluorescently stained Gag proteins were observed by using a Zeiss LSM410 laser scanning microscope.
Electron microscopy (EM).
At 40 h after transfection, COS-7 cells were fixed with 2.5% glutaraldehyde, followed by treatment with 4% osmium tetroxide. Samples were routinely processed and embedded. Thin-sectioned samples were stained with lead citrate and uranyl acetate and then visualized by using a JEOL JEM-2000 FX transmission electron microscope equipped with a Gatan 792 Bioscan 1,024- by 1,024-byte wide-angle multiscan charge-coupled device camera.
RESULTS
Mutation of the CA sequence 353-GVGGP-357 impairs virus production.
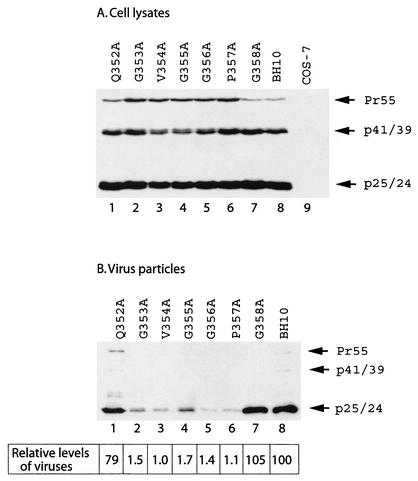
Amino acid sequences 353-GVGGPG-358, located close to the C terminus of CA, are highly conserved within primate lentiviral Gag proteins (Table 1) (36); thus, these amino acids may play important roles in virus production. To pursue this subject, each of these six residues was individually changed to alanine in the context of the HIV-1 genome. The mutations thus generated were termed G353A, V354A, G355A, G356A, P357A, and G358A (Fig. 1). In addition, we altered an upstream amino acid Q352 to alanine to generate Q352A (Fig. 1). Levels of viral Gag proteins were measured either by anti-HIV-1 p24 Western blot analysis or by quantitative anti-p24 ELISA. When the mutant and wild-type DNA constructs were transfected into COS-7 cells, similar levels of intracellular Gag expression were observed for the various DNA constructs (Fig. 2A). Yet the G353A, V354A, G355A, G356A, and P357A mutant Gag proteins, but not Q352A and G358A mutant Gag proteins, displayed defective processing, as shown by the accumulation of Pr55Gag precursor in the case of the former five mutations, but not wild-type BH10 Gag (Fig. 2A). Irrespective of their wild-type expression levels within the cytoplasm, the mutants G353A, V354A, G355A, G356A, and P357A generated levels of extracellular virus particles that were approximately 50- to 100-fold lower than the levels produced by either wild-type BH10 or the mutants Q352A and G358A (Fig. 2B). Similar findings were made by determining the levels of virion-associated RT activity (data not shown). Therefore, amino acids 353-GVGGP-357 are not only required for normal Gag processing but, more importantly, are essential for virus particle production.
FIG. 2.
Virus particle production by mutant and wild-type HIV-1 DNA constructs. COS-7 cells that were transfected with various viral DNA constructs were harvested and then lysed in NP-40 lysis buffer. Virus particles in the culture fluids were pelleted by ultracentrifugation. Western blot analysis with anti-HIV-1 p24 antibodies and quantitative anti-p24 ELISA were performed to determine levels of viral Gag proteins that are either associated with cell lysates (A) or with virus particles (B). Mock-transfected COS-7 cells served as negative controls. The efficiency of virus particle production by each DNA construct was calculated on the basis of the ELISA data, and that of BH10 was arbitrarily set at 100.
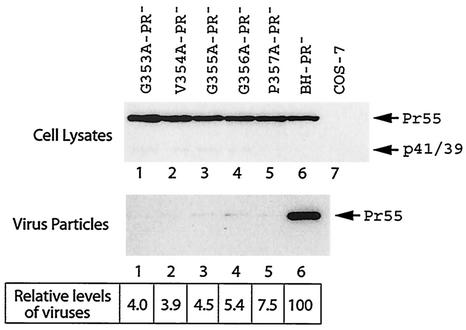
Since mutation of 353-GVGGP-357 affects both Gag processing and virus production, it is possible that the diminution in virus production may result from aberrant Gag processing. To test this possibility, each of the five mutations G353A, V354A, G355A, G356A, and P357A was recombined with the protease-negative mutation D25A, which changes the active site of protease (45). The constructs thus generated were termed G353A-PR−, V354A-PR−, G355A-PR−, G356A-PR−, and P357A-PR−. BH-PR− is identical to BH10, with the exception that the former contains the D25A mutation. Transfection of the former five mutant DNA constructs into COS-7 cells yielded extracellular particles at levels that were approximately 20-fold lower than those generated by BH-PR−. This was in spite of similar levels of Gag protein expression in the cytoplasm among the wild-type and various mutant viruses (Fig. 3). We have also tested these five mutant DNA constructs by transfection of HeLa cells and detected significantly lower quantities of extracellular virus particles in comparison to the wild-type BH-PR− (data not shown). Therefore, the adverse effects of the mutated 353-GVGGP-357 motif on virus particle production are independent of Gag processing.
FIG. 3.
Effects of various Gag mutations on virus particle production in the absence of viral protease activity. Each of the mutant and wild-type DNA constructs, containing a protease-negative mutation D25A (45), was transfected into COS-7 cells. Cell- or virion-associated Gag proteins were analyzed either by anti-p24 Western blotting or quantitative ELISA. Levels of virus particles were calculated on the basis of the ELISA data, and those of BH-PR− were arbitrarily set at 100.
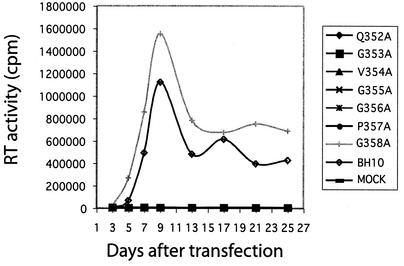
Since mutations G353A, V354A, G355A, G356A, and P357A severely inhibit virus production, they must also have profoundly affected virus replication in CD4+ cells. Indeed, when DNA constructs containing these mutations were transfected into Jurkat cells, infectious viruses could not be detected in culture for up to 25 days, according to the results of RT assays (Fig. 4). In contrast, wild-type BH10 DNA generated high levels of RT activity in culture, suggesting the successful spread of viruses between cells (Fig. 4). As for mutant DNA constructs Q352A and G358A that were able to produce wild-type levels of virus particles in both COS-7 and HeLa cells, Q352A failed to generate infectious viruses in Jurkat cells, whereas G358A yielded viruses as infectious as wild-type BH10 (Fig. 4). The drastic difference in infectiousness between Q352A and G358A correlates with the high conservation of Q352 in contrast to the relatively low conservation of G358 in HIV-1 (Table 1).
FIG. 4.
Replication of mutant and wild-type viruses in Jurkat cells. Two million Jurkat cells were transfected by 1 μg of either mutant or wild-type HIV-1 DNA. Viral growth was monitored by measuring the RT activity in culture fluids at various times. “MOCK” refers to mock-transfected Jurkat cells.
Mutation of the 353-GVGGP-357 sequences restricts Gag multimerization.
Gag assembly involves sequential steps and the formation of a series of intermediate complexes (37, 39, 44, 59). We were interested in understanding whether mutation of the 353-GVGGP-357 motif might have blocked Gag assembly at a particular stage. To pursue this subject, intracellular Gag complexes were analyzed in gradient sedimentation assays. Since mutation of amino acids 353-GVGGP-357 impairs virus production in a manner independent of viral protease activity (see above), we employed viral DNA constructs containing the protease-negative mutation D25A in the following experiments.
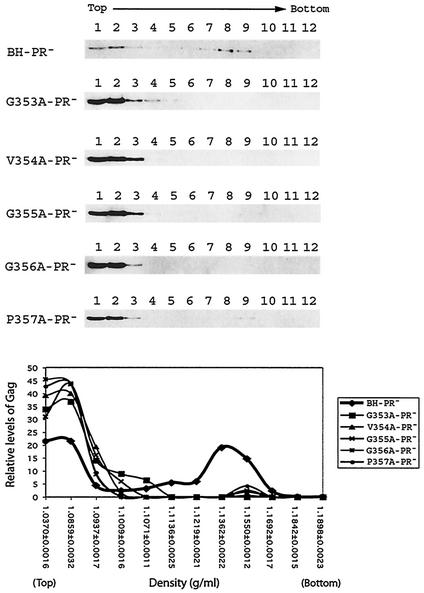
Intracellular Gag complexes were first separated via a continuous sucrose gradient by velocity sedimentation at 210,000 × g for 1 h. Under this condition, particles are separated mainly on the basis of their mass. In the case of wild-type Gag expressed by BH-PR−, they migrated to form two major groups in the gradient (Fig. 5). The group I Gag proteins were located in fractions 1 and 2 and were most likely monomeric; group II were relatively large Gag complexes that were found in the heavier fractions 8 and 9 (Fig. 5). In contrast, the mutant Gag proteins G353A, V354A, G355A, G356A, and P457A were mainly found in the top fractions 1, 2, and 3 (Fig. 5). This demonstrates that the mutant Gag cannot form large complexes as efficiently as wild-type Gag.
FIG. 5.
Fractionation of Gag complexes by velocity sedimentation. HIV-1 DNA clones containing a protease-negative mutation were transfected into COS-7 cells. At 40 h posttransfection, cells were Dounce homogenized, and the denucleated fractions were placed on the top of a continuous 15 to 50% sucrose gradient, followed by centrifugation at 210,000 × g for 1 h at 4°C. Twelve fractions were collected from each gradient, and viral Gag proteins were detected by anti-p24 Western blot analysis. Relative levels of Pr55Gag in each fraction were determined from the intensities of protein bands that were measured by densitometry scanning through the use of the NIH Image program. The relative percentages of Pr55Gag in each fraction versus the total Pr55Gag levels in the gradient were calculated, and the results are plotted against the density of each fraction that was measured by using a refractometer. The density data shown in the graph represent the average for each type of Gag tested.
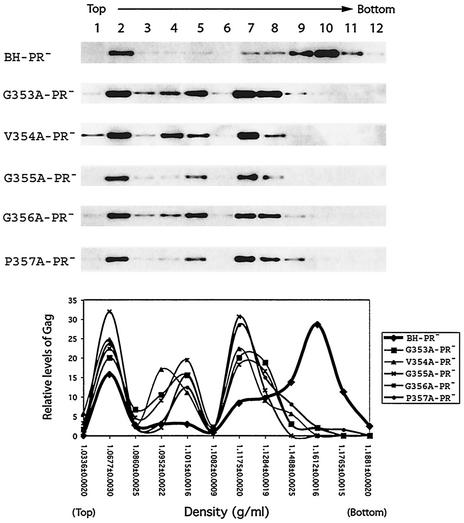
We further characterized intracellular Gag complexes by performing density gradient sedimentation at 100,000 × g for 16 h. In this experiment, particles are separated on the basis of their size and density. The majority of the wild-type Gag population was seen in the relatively high-density fractions, including fractions 7 to 11 (Fig. 6). In particular, fraction 10 contained the highest levels of Gag. These Gag complexes exhibited a density of ca. 1.1612 g/ml and, thus, probably represented nascent virus particles that had been completed but not yet released from the cells. As for mutant Gag proteins, significant quantities of these were recovered in fractions 4 and 5, as well as in fractions 7 and 8, in addition to a major distribution in fraction 2. In contrast to wild-type Gag, fairly low amounts of mutant Gag proteins were seen in fractions 9 and 10 (Fig. 6). These results indicate that assembly of the mutant Gag proteins was blocked at a stage prior to the final formation of virus particles.
FIG. 6.
Fractionation of intracellular Gag proteins by equilibrium density gradient centrifugation. The denucleated COS-7 homogenates were placed on the top of a continuous 15 to 50% sucrose gradient and centrifuged at 100,000 × g for 16 h at 4°C. Measurement of the Pr55Gag levels in each gradient was as described in the legend to Fig. 5.
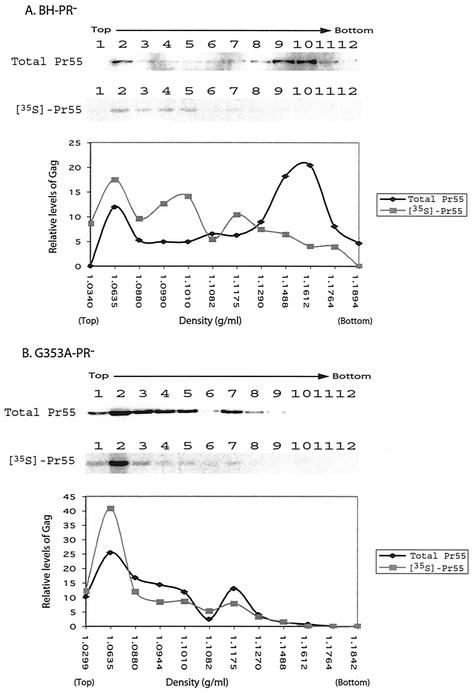
The sedimentation experiments described above had assessed the assembly of Gag proteins at steady state. Next, we followed the migration of newly synthesized Gag in the density gradient after centrifugation at 100,000 × g for 16 h. Accordingly, transfected COS-7 cells were pulse-labeled with 35S-labeled methionine and cysteine for 7 min. Newly synthesized Gag proteins were then assessed by immunoprecipitation with anti-p24 MAbs. In the case of the wild-type Gag, the steady-state and newly synthesized forms displayed strikingly different gradient profiles (Fig. 7A). The majority of the total Gag population had migrated to high-density fractions 9 and 10, as shown by the results of Western blot analyses (Fig. 7A). In contrast, 35S-labeled Gag proteins were mainly found in the light-density fractions from 2 to 5 (Fig. 7A). This indicates that Gag proteins are mainly monomeric or form very small complexes almost immediately after being made, which are converted to larger high-density complexes over time.
FIG. 7.
Fractionation of newly synthesized Gag proteins by equilibrium density gradient sedimentation. Transfected COS-7 cells were pulse-labeled with 35S-labeled methionine and cysteine for 7 min, followed by Dounce homogenization. The denucleated fractions were centrifuged through a continuous 15 to 50% sucrose gradient at 100,000 × g for 16 h at 4°C. Twelve fractions were collected for each gradient, and the samples were assessed for either total Gag population by anti-p24 Western blot analysis (top gel panel) or for newly synthesized Gag by immunoprecipitation (bottom gel panel). Relative levels of either steady-state Gag or newly synthesized Gag in each fraction were calculated through the use of densitometry scanning, and the data are shown in the graphs. The density of each fraction was measured through use of the refractometer and the results are also shown in the graphs. (A) BH-PR−; (B) G353A-PR−.
When the newly synthesized mutant Gag G353A was analyzed by density gradient, this protein displayed a distribution pattern similar to that of the total G353A Gag, except that the newly synthesized molecules were accumulated to a higher extent in fraction 2 (Fig. 7B). Therefore, the assembly of the G353A mutant Gag stops shortly after proteins are made, whereas newly synthesized wild-type Gag molecules are able to proceed to perform high-order multimerization and, eventually, form virus particles.
Mutation of the 353-GVGGPG-357 motif impairs Gag association with cellular membranes.
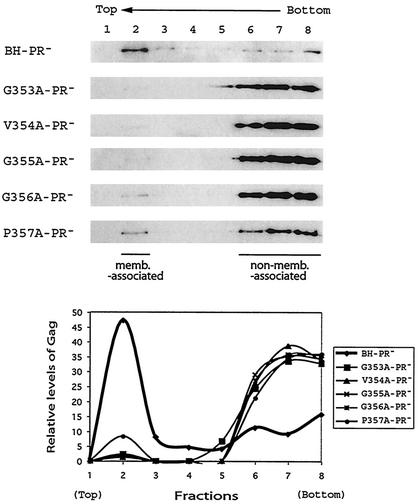
HIV-1 Gag exhibits high affinity for the plasma membrane, a property that is conferred by the membrane-binding signal within the MA sequence. Since the mutant Gag molecules generated in the present study contain an intact MA domain, we decided to study whether they might bind to cellular membranes as efficiently as wild-type Gag. To pursue this subject, denucleated homogenates of transfected COS-7 cells were subjected to membrane flotation sedimentation. In this assay, membrane-bound Gag will migrate from the bottom 73% sucrose layer to the interface between the 10 and 65% sucrose layers (recovered in fraction 2), whereas non-membrane-bound Gag remains within the bottom fractions. Our results show that approximately 40 to 50% of the wild-type Gag was found at the interface between the 10 and 65% sucrose layers, indicating that a major portion of these molecules were associated with cellular membranes (Fig. 8). In contrast, more than 90% of the various mutant Gag proteins remained within the bottom fractions (i.e., fractions 6 to 8), and only low quantities of Gag were detected in fraction 2 (Fig. 8). We conclude that Gag proteins that contain a mutated 353-GVGGP-357 motif are impaired in their ability to bind to cellular membranes and that this defect is probably a major cause of diminished virus production.
FIG. 8.
Effects of the mutated 353-GVGGP-357 motif on Gag membrane binding. Denucleated homogenates of transfected COS-7 cells were subjected to membrane flotation sedimentation as described in Materials and Methods. Membrane-associated Gag (memb.-associated) floats to the interface between the 10 and 65% sucrose layers (fraction 2), whereas non-membrane-associated (non-memb.-associated) Gag remains within the bottom fractions (fractions 6 to 8). Relative amounts of Gag proteins in each fraction were determined by densitometry scanning through use of the NIH Image program, and the results are shown in the graphs.
Mutation of amino acids 353-GVGGP-357 results in diffuse distribution of Gag proteins within the cytoplasm.
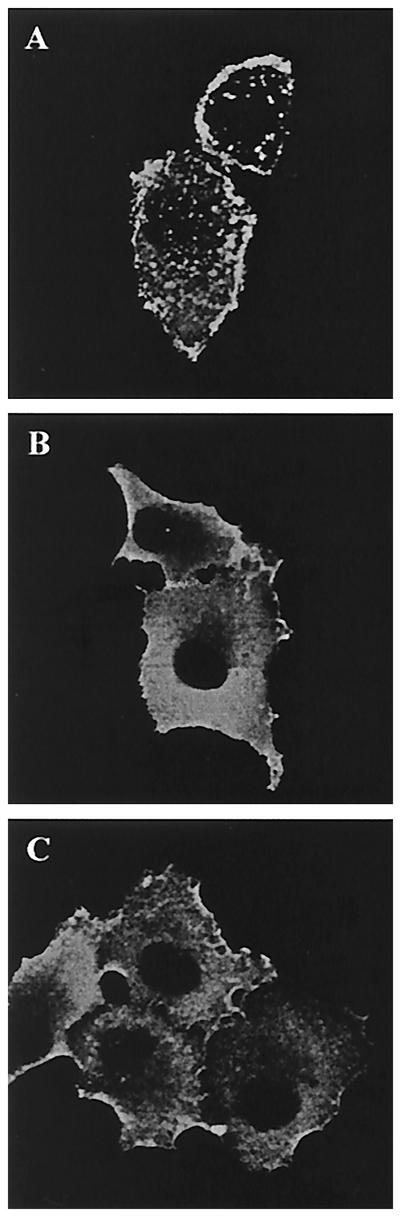
In order to gain further insights into the Gag assembly defects caused by the mutated 353-GVGGP-357 motif, the subcellular distribution of the mutant Gag proteins containing the G356A or P357A substitutions, as well as wild-type BH10 Gag, was studied by immunofluorescence staining by using anti-p24 MAbs as the primary antibodies. The results show that wild-type BH10 Gag was mainly associated with the plasma membrane and displayed a punctate distribution pattern (Fig. 9). In contrast, the mutant Gag proteins G356A and P357A showed a diffuse distribution pattern, and the majority of these molecules were found within the cytoplasm (Fig. 9). Thus, wild-type Gag molecules are able to aggregate and to actively assemble virus particles, whereas the mutant Gag proteins G356A and P357A are less well organized within cells, leading to a low yield of virus production.
FIG. 9.
Immunofluorescence staining of Gag proteins expressed within transfected COS-7 cells. COS-7 cells were transfected with the indicated DNA constructs and fixed with 4% paraformaldehyde at 40 h after transfection. After permeabilization with 0.2% Triton X-100, cells were incubated with anti-p24 MAbs, followed by staining with fluorescein isothiocyanate-labeled secondary antibodies. The immunofluorescence-stained cells were examined by using a Zeiss LSM410 laser scanning microscope. (A) BH10; (B) G356A; (C) P357A.
Mutation of amino acids 353-GVGGP-357 affects both the yield and morphology of HIV-1 particles.
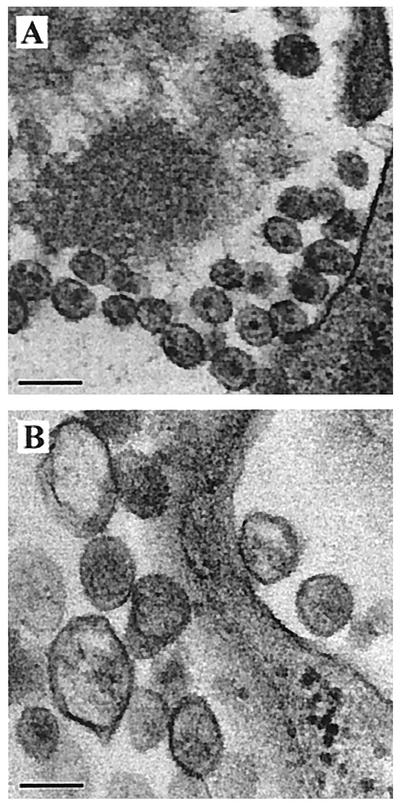
Next, we used EM to investigate the effects of the mutated motif 353-GVGGP-357 on the morphology of HIV-1 particles. The three mutations G353A, V354A, and P357A, as well as wild-type BH10, were studied. Transfection of COS-7 cells with the wild-type BH10 construct led to production of a large number of virus particles, most of which were mature and exhibited condense cores (Fig. 10A). In contrast, transfection with the mutant constructs G353A and V354A did not result in significant viral production (a large number of cells were examined by EM [data not shown]). In addition, these transfected cells rarely showed large electron-dense patches under the plasma membrane, indicating that the mutant Gag proteins had not heavily aggregated beneath the plasma membrane. In the case of P357A, a few viral particle-like structures were detected after extensive searching, but these were heterogeneous in size, displayed irregular morphology and, on average, were larger than wild-type particles (Fig. 10B). A similar phenotype has been observed for HIV-1 mutants containing mutated p2 sequences (1, 34). Therefore, mutation of the 353-GVGGP-357 motif not only results in severely reduced yields of virus particles but also causes aberrant virus morphology.
FIG. 10.
Morphology of virus particles examined by transmission EM. (A) BH10; (B) P357A. Bars, 0.2 μm.
DISCUSSION
In this study, we have identified a peptide motif 353-GVGGP-357 at the C terminus of CA that is essential for virus particle formation. Interestingly, these five amino acids are all needed for efficient virus production. Similar results have been seen with the HIV-1 late domain P(T/S)AP that is highly conserved and positioned at the N terminus of p6. Changing any of the P(T/S)AP amino acids can lead to defects in virus budding (22, 28).
The formation of virus particles involves three major events, including multimerization of Gag, targeting of Gag to the plasma membrane, and the release of virus particles from the host cells. Our data show that mutation of the Gag motif 353-GVGGP-357 adversely affects the first two events and, consequently, leads to severe reductions in virus particle production. Impairment in Gag multimerization is clearly shown by the results of our sedimentation experiments, in which mutant Gag molecules were mainly monomeric or formed fairly small multimers (Fig. 5 and 6). Moreover, these mutant Gag proteins were virtually non-membrane bound, because they could hardly float from the bottom gradient to the interface between the 10 and 65% sucrose layers (Fig. 8). In support of these findings, the results of immunofluorescence staining demonstrated that the mutant Gag proteins were largely diffused within the cytoplasm in contrast to the punctate distribution pattern of wild-type Gag on the plasma membrane (Fig. 9).
Since the mutant Gag proteins generated in the present study contained wild-type MA sequences that possess plasma membrane targeting signals, it is likely that the lack of membrane association was caused, at least in part, by defective multimerization of the mutant Gag molecules. Impaired membrane binding has also been observed for assembly-defective Gag proteins that contain mutated NC sequences (50, 54). Although the mutant Gag proteins of our study possess intact MA sequences, the MA-attached myristate moiety and the downstream basic residues may not have been exposed to an extent necessary to allow efficient membrane binding; this may possibly be due to defective Gag multimerization or incorrect Gag conformation. This concept agrees with the myristyl switch model in which a lesser extent of exposure of myristate acid is believed to be the reason for the weak association of MA with the plasma membrane in comparison with the results observed for Pr55Gag (27, 48, 55, 68). Another possibility is that the binding energy provided by a single membrane-binding domain to the lipid bilayer does not suffice to ensure stable membrane association in the face of defective multimerization (54). Alternatively, our Gag mutations may have caused conformational defects that indirectly lead to disruption of the N-terminal structure of Gag, which subsequently impairs membrane binding. In the meantime, we cannot lose sight of the possibility that mutation of the 353-GVGGP-357 motif may indirectly affect myristylation of Gag.
As opposed to the view that Gag multimerization is a prerequisite for subsequent plasma membrane targeting, a truncated version of Gag, i.e., CA146, that cannot multimerize was shown to display wild-type levels of plasma membrane association (46). One possibility is that this particular form of Gag has adopted a specific conformation that allows maximal exposure of MA signals for plasma membrane binding.
Gag multimerization requires two major domains. The first involves the NC region that promotes Gag-Gag association through binding to RNA as a scaffold (5-7, 9, 10, 24, 53). In support of this idea, removal of RNA by RNase digestion disrupts formation of Gag complexes (32). The second domain encompasses the C-terminal domain of CA (11, 31, 52, 61, 63, 66) and the adjacent spacer peptide, p2 (1, 30, 34, 44, 62). In a structural view, p2 represents an extension of a putative α-helix from the C terminus of CA (1). This CA-p2 region can support efficient virus particle generation in conjunction with the L domain, the MA myristylation signal, and heterologous amino acid sequences that, in place of NC, are able to mediate protein-protein interactions (2).
Molecular details regarding the multimerization activity of CA-p2 start to be revealed by structural studies. CA comprises two structurally independent domains, an N-terminal domain (NTD; residues 1 to 145) and a CTD (residues 150 to 231), that are connected by a short linker (3, 17, 21, 43, 65). A dimer interface exists within the CTD of CA that plays an important role in CA dimerization (3, 17, 43, 65). Reconstructed images of HIV-1 CA tubes, on the basis of cryo-EM at a 30-Å resolution, suggest that the NTD of CA constructs the hexameric ring, whereas the CTD of CA connects each ring to its six neighbors and thus results in the generation of high-order complexes (18, 38). However, an accurate positioning of the CTD, as well as analysis of the detailed molecular interactions involved, await a higher-resolution image. Irrespective of these uncertainties, it is possible that mutation of the CTD sequence 353-GVGGP-357 may have adversely impacted the connection between hexameric rings and, as a result, only hexamers might have been formed. In support of this hypothesis, our data show that Gag proteins containing the mutated 353-GVGGP-357 sequences were unable to form large complexes.
Interestingly, the few resolved crystal structures of CA, one of which contains the p2 sequence, all reveal a disordered feature of the last 11 to 13 C-terminal amino acids (17, 43, 65). Thus, the C-terminal 11 amino acids of CA must adopt multiple conformations and, as a consequence, cannot be easily crystallized. The glycine-rich motif 353-GVGGP-357 studied here are among these 11 amino acids. Glycine has an H atom as its side chain and thus leaves sufficient space for adjacent residues to rotate. Due to this property, glycine represents a common turning point in protein structures. We suspect that this glycine-rich motif may be responsible for structural flexibility on the part of these 11 CA amino acids, as well as the adjacent p2 sequences. Accordingly, 353-GVGGP-357 could confer structural flexibility to both CA and full-length Gag.
Structural polymorphism has been shown to be an important property, allowing chemically identical subunits to establish geometrically distinct interactions with neighboring subunits and to eventually construct complex structures, especially sealed geometrical structures such as virus particles (29). According to the quasi-equivalent assembly theory, interchangeable formation between hexamers and pentamers by the same protein molecule is a basic rule for the construction of a virus particle (8). Disturbing the relative stabilities of hexagonal and pentagonal morphological units by mutation of the capsid protein can severely interfere with proper virus assembly (14). The same principle has been applied to understand construction of the HIV-1 core. According to the conical model, HIV-1 cores are composed of hexagonal lattices that are closed by pentagonal defects at the narrow and wide ends (18, 38). In addition to the ability to form hexamers and pentamers, CA can also construct different types of tubes that exhibit a high degree of polymorphism. In order to build these many types of structures, CA must be able to assume a variety of conformations. This high structural flexibility of CA is believed to result, at least in part, from the linker segment (CA residues 146 to 149) that connects the NTD and the CTD (3, 38). Considering the structurally disordered feature of the amino acid stretch 353-GVGGP-357, we propose that this glycine-rich sequence may also assist CA to adopt distinct conformations and to play its role in construction of asymmetric cores.
Acknowledgments
We thank Shan Cen and Rabih Halwani for valuable suggestions in regard to performance of the sedimentation experiments; Véronique Bériault, Laurent Chatel-Chaix, Andrew Mouland, and Daniel Larocque for help in immunofluorescence staining and microscopy; and Mervi Detorio and Maureen Oliveira for measuring p24 levels and RT activity.
This research was supported by grants from the Canadian Institutes of Health Research and by the Fonds de la Recherche en Sante du Quebec (FRSQ). Chen Liang is a Chercheur-Boursier of the FRSQ.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspar, D. L., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1-24. [DOI] [PubMed] [Google Scholar]
- 9.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 11.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebbets-Reed, D., S. Scarlata, and C. A. Carter. 1996. The major homology region of the HIV-1 gag precursor influences membrane affinity. Biochemistry 35:14268-14275. [DOI] [PubMed] [Google Scholar]
- 13.Erickson-Viitanen, S., J. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retrovir. 5:577-591. [DOI] [PubMed] [Google Scholar]
- 14.Flasinski, S., A. Dzianott, J. A. Speir, J. E. Johnson, and J. J. Bujarski. 1997. Structure-based rationale for the rescue of systemic movement of brome mosaic virus by spontaneous second-site mutations in the coat protein gene. J. Virol. 71:2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 18.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 19.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 20.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 21.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 22.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross, I., H. Hohenberg, and H.-G. Kräusslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 25.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H.-G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, L. E., R. C. Sowder, T. D. Copeland, S. Oroszlan, and R. E. Benveniste. 1990. Gag precursors of HIV and SIV are cleaved into six proteins found in the mature virions. J. Med. Primatol. 19:411-419. [PubMed] [Google Scholar]
- 27.Hermida-Matsumoto, L., and M. D. Resh. 1999. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55gag and p17MA. J. Virol. 73:1902-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, J. E., and J. A. Speir. 1997. Quasi-equivalent viruses: a paradigm for protein assemblies. J. Mol. Biol. 269:665-675. [DOI] [PubMed] [Google Scholar]
- 30.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. [DOI] [PubMed] [Google Scholar]
- 31.Kattenbeck, B., A. von Poblotzki, A. Rohrhofer, H. Wolf, and S. Modrow. 1997. Inhibition of human immunodeficiency virus type 1 particle formation by alterations of defined amino acids within the C terminus of the capsid protein. J. Gen. Virol. 78:2489-2496. [DOI] [PubMed] [Google Scholar]
- 32.Khorchid, A., R. Halwani, M. A. Wainberg, and L. Kleiman. 2002. Role of RNA in facilitating Gag/Gag-Pol interaction. J. Virol. 76:4131-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konvalinka, J., A.-M. Heuser, O. Hruskova-Heidingsfeldova, V. M. Vogt, J. Sedlacek, P. Strop, and H.-G. Kräusslich. 1994. Proteolytic processing of particle-associated retroviral polyproteins by homologous and heterologous viral proteinases. Eur. J. Biochem. 228:191-198. [DOI] [PubMed] [Google Scholar]
- 34.Kräusslich, H.-G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kräusslich, H.-G., H. Schneider, G. Zybarth, C. A. Carter, and E. Wimmer. 1988. Processing of in vitro-synthesized Gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J. Virol. 62:4393-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuiken, C., B. Foley, et al. 1999. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 37.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243:78-93. [DOI] [PubMed] [Google Scholar]
- 38.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 39.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multi-step, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 42.McDermott, J., L. Farrel, R. Ross, and E. Barklis. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J. Virol. 70:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 44.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morin, N., E. Cherry, X. Li, and M. A. Wainberg. 1998. Cotransfection of mutated forms of human immunodeficiency virus type 1 Gag-Pol with wild-type constructs can interfere with processing and viral replication. J. Hum. Virol. 1:240-247. [PubMed] [Google Scholar]
- 46.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paillart, J. C., and H. G. Gottlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Platt, E. J., and O. K. Haffar. 1994. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc. Natl. Acad. Sci. USA 91:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Provitera, P., A. Goff, A. Harenberg, F. Bouamr, C. Carter, and S. Scarlata. 2001. Role of the major homology region in assembly of HIV-1 Gag. Biochemistry 40:5565-5572. [DOI] [PubMed] [Google Scholar]
- 52.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasakumar, N., M. L. Hammarskjold, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tritch, R. J., Y.-S. E. Cheng, F. H. Yin, and S. Erickson-Viitanen. 1991. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 65:922-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Poblotzki, A., R. Wagner, M. Niedrig, G. Wanner, H. Wolf, and S. Modrow. 1993. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology 193:981-985. [DOI] [PubMed] [Google Scholar]
- 62.Wang, C.-T., H.-Y. Lai, and J.-J. Li. 1998. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J. Virol. 72:7950-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, C.-T., J. Stegeman-Olsen, Y. Zhang, and E. Barklis. 1994. Assembly of HIV GAG-B-galactosidase fusion proteins into virus particles. Virology 200:524-534. [DOI] [PubMed] [Google Scholar]
- 64.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, H., and H.-G. Kräusslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55:85-92. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, W. H., D. J. Hockley, M. V. Nermut, Y. Morikawa, and I. M. Jones. 1996. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J. Gen. Virol. 77:743-751. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, W., and M. D. Resh. 1996. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70:8540-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]