Abstract
Disrupted-in-Schizophrenia-1 (DISC-1) is a gene whose mutant truncation is associated with major psychiatric illness with a predominance of schizophrenic symptomatology. We have cloned and characterized rodent DISC-1. DISC-1 expression displays pronounced developmental regulation with the highest levels in late embryonic life when the cerebral cortex develops. In yeast two-hybrid analyses, DISC-1 interacts with a variety of cytoskeletal proteins. One of these, NudE-like (NUDEL), is associated with cortical development and is linked to LIS-1, the disease gene for a form of lissencephaly, a disorder of cortical development. The disease mutant form of DISC-1 fails to bind NUDEL. Expression of mutant, but not wild-type, DISC-1 in PC12 cells reduces neurite extension. As schizophrenia is thought to reflect defects in cortical development that are determined by cytoskeletal protein activities, the cellular disturbances we observe with mutant DISC-1 may be relevant to psychopathologic mechanisms.
Multiple lines of evidence favor a genetic predisposition to schizophrenia (1). Several kinds of studies, including brain imaging and neuropathology, suggest abnormalities in schizophrenic cerebral cortical development that might reflect cytoskeletal disturbances (2–5).
Genetic studies have linked schizophrenia to multiple loci, suggesting the involvement of certain candidate genes as susceptibility factors (6, 7). In a Scottish family, a chromosome (1;11)(q42.1;q14.3) translocation associates with major psychiatric illnesses with a logarithm of odds score of 7.1 (8, 9). This translocation interrupts the coding sequence of Disrupted-in-Schizophrenia-1 (DISC-1), leading to loss of the C-terminal 257 amino acids for DISC-1 protein. Linkage and association studies have also identified the DISC-1 locus as a susceptibility factor for schizophrenia in Finnish families (10).
We have carried out a series of studies to characterize the cellular functions of DISC-1 and ascertain whether these are altered in the mutant form of the protein. We have cloned rodent DISC-1 and generated antibodies to DISC-1 protein. We demonstrate the expression of DISC-1 protein in the brain with marked developmental regulation. By using yeast two-hybrid analysis, we observe interactions of DISC-1 with a variety of structural proteins, one of which is NudE-like (NUDEL), a protein involved in neuronal development that interacts with LIS-1, the disease gene for a form of lissencephaly (11–13). The mutant form of DISC-1 fails to bind to NudE-like (NUDEL). Moreover, expression of mutant, but not wild-type, DISC-1 alters neurite outgrowth in PC12 cells.
Materials and Methods
General Methods and Materials.
Molecular biology reagents were obtained from New England Biolabs, and all other reagents were from Sigma, except as indicated. Protein concentrations were determined by Bradford assay.
Cell Maintenance.
Cell biology reagents were obtained from Invitrogen. PC12 cells were maintained in DMEM with 10% (vol/vol) FBS and 5% horse serum (HS). Differentiation was initiated by the addition of 50 ng/ml nerve growth factor (NGF) with culture medium changed to DMEM with 1% HS and 1% FBS. NGF was supplied every day after differentiation.
Human embryonic kidney (HEK) 293 cells, COS cells, C6 glial cells, and HeLa cells were maintained in DMEM with 10% (vol/vol) FBS. Lymphoblasts from control subjects and patients with Huntington's disease were maintained in RPMI medium 1640 with 10% (vol/vol) FBS (14). Chinese hamster ovary (CHO) cells were maintained in Ham's F12 medium with 10% (vol/vol) FBS.
Cloning of Mouse and Rat DISC-1.
The human DISC-1 full-length cDNA sequence was used as a query to search homologous sequences in the Celera mouse genome database (www.celera.com). Mouse homologous sequences that correspond to exons 4–6 as well as exon 12 and a portion of exon 13 were initially identified within bacteria artificial chromosome clones mCG52715 and mCG57221, respectively. Based on the mouse sequence, PCR primers corresponding to exons 4, 6, and 13 were designed and used to amplify mouse and rat DISC-1 (exons 4–6 and 6–13) from reverse transcription (RT) products prepared from murine P6 cerebellar granule cells (C57BL/6J) and rat embryonic day 14 (E14) embryos, respectively. RT-PCR products of an expected size were subcloned and sequenced. A subsequent database search against the National Center for Biotechnology Information (NCBI) mouse genome database (www.ncbi.nlm.nih.gov) identified a mouse genomic sequence (NW_000349) that contains a putative mouse DISC-1 starting methionine within exon 1. PCR was then performed to amplify sequences from the starting ATG within exons 1–4 of both mouse and rat DISC-1, and the identity of the RT-PCR products was verified by sequencing. The mouse and rat DISC-1 coding sequences, exons 1–4, 4–6, and 6–13, were assembled and used to search the Celera mouse genome database again to identify one long contig (mouse genome assembly ID no. GA_x6k02T2NUPS) spanning 500 kb. All of the sequences of mouse DISC-1 except exons 2, 3, and 7 were available in this contig. Sequences corresponding to exons 2, 3, and 7 were included in other genome assemblies, GA_x6K02T2M244, GA_x6K02T2N3VK, and GA_x6K02T07GAA, respectively.
The sequences of primers used for RT-PCR were as follows: exon 1 sense, 5′-AGAGTCGACCATGCAGGGCGGGGGTCCCCG-3′; exon 4 antisense, 5′-TTCCAACCTCTGTCTCAATGTCTC-3′; exon 4 sense, 5′-GAGACATTGAGACAGAGGTTGG-3′; exon 6 antisense, 5′-CTCCTGAGGGTCTCAGGTGG-3′; exon 6 sense, 5′-GAGCCACCTGAGACCCTCAGG-3′; and exon 13 antisense, 5′-TCTGCGGCCGCTCAGGCCTCGGTTTCCTGAGC-3′.
The GenBank accession numbers for mouse and rat DISC-1 are AY177673 and AY177674, respectively.
Primary Antibodies.
Rabbit polyclonal antibodies were raised against GST with different portions of DISC-1 (amino acids 347–600, 601–854, and 212–537). The antisera were affinity-purified as described.
Commercial primary antibodies used in this study include anti-hemagglutinin (HA) tag antibodies, mAb from BabCO (Richmond, CA), a polyclonal antibody from Medical and Biological Laboratories (Boston), and a polyclonal antibody against myc tag from Oncogene Science.
Tissue Preparation and Western Blotting.
Tissues from adult and embryonic rats were homogenized in ice-cold lysis buffer A [50 mM Hepes, pH 7.3/150 mM NaCl/5 mM MgCl2/5 mM DTT/1 mM PMSF/1 mM EDTA/protease inhibitor mixture (Roche Molecular Biochemicals)]. Cells extracted were mixed with an SDS/PAGE loading buffer containing 50 mM DTT immediately after measurement of protein concentrations.
A secondary antibody for Western blotting was against rabbit IgG conjugated with horseradish peroxidase from Amersham Pharmacia. Enhanced chemiluminescence reagents were obtained from Amersham Pharmacia.
Subcellular Fractionation.
Cells were homogenized with 0.32 M sucrose-containing buffer by using a glass–glass homogenizer, as described (15).
Immunofluorescent Staining.
Cells were fixed with 3.7% (wt/vol) paraformaldehyde followed by permeabilization with 0.1% Triton X-100. After blocking with 1% BSA and 1% normal goat serum in PBS, primary antibodies were incubated for 1 h. A Rhodamine Red-X conjugated secondary antibody (Jackson ImmunoResearch) was incubated at 1:500 dilution for 1 h. Hoechst 33258 (Molecular Probes) was used at 1:500 dilution for 2 min. Confocal microscopy used a Zeiss LSM 410 to obtain phase contrast images. A Zeiss Axiovert 135 microscope, together with a charge-coupled device (CCD) camera (Cool- Snap HQ cooled 12 bit, Roper Scientific, Trenton, NJ) was used to analyze DISC-1 immunofluorescent staining in PC12 cells.
Yeast Two-Hybrid Screen.
Two portions of human DISC-1 (amino acids 347–600 for C1 bait and amino acids 601–854 for C2 bait) were cloned into yeast-expression vector pDBleu, containing the GAL4 DNA-binding domain. These were used to screen a human whole brain cDNA library cloned into pPC 86, containing the GAL4 transactivation domain. Over one million independent clones were screened, and positively interacting proteins were identified by selecting for Leu+, Trip+, His+ growth phenotype. Positive clones were further evaluated for β-galactosidase expression.
Confirmation of Protein Binding.
Immunoprecipitation was performed as described (16). Briefly, constructs with DISC-1, NUDEL, and GAPDH were transfected into HEK 293 cells. Cells were homogenized in a RIPA buffer [50 mM Hepes, pH 7.3/150 mM NaCl/1 mM DTT/1 mM PMSF/protease inhibitor mixture (Roche Molecular Biochemicals)] and spun at 10,000 × g for 20 min to remove insoluble materials. The resulting lysate was mixed with the monoclonal primary antibody against myc tag (as described above), followed by the addition of protein A agarose (Oncogene Science) for the precipitation of protein complex. The agarose pellet was washed five times with PBS.In vitro binding experiments were carried out by using a GST-fusion DISC-1 protein fragment (amino acids 601–854) together with (His)6 fusion proteins of full-length NUDEL or GAPDH protein.
Neurite Outgrowth Assay.
Cells were maintained on chamber slides (Nalge) coated with 0.2% gelatin crosslinked with 0.5% glutaraldehyde. Constructs with full-length DISC-1, mutant form (the C-terminal truncated form) of DISC-1, or GAPDH together with GFP were cotransfected by using Lipofectamine 2000 (Invitrogen). The molar ratio of DISC-1 or GAPDH to GFP construct was 2:1. One day after transfection, the medium was replaced for cell differentiation as described above. Six days after differentiation, cells were fixed with 3.7% (wt/vol) paraformaldehyde and permeabilized with 0.1% Triton X-100, followed by nuclear counterstaining with Hoechst 33258. GFP epifluorescence was used as a marker for cell morphology. Cells with processes longer than three cell body diameters were scored as neurite-harboring neurons. The length of the longest process of each neurite-harboring cell was measured. Cell images were captured with a Roper Scientific CoolSnap HQ cooled 12-bit CCD camera attached to a Zeiss Axiovert 135 microscope. Cell morphology was analyzed in a blind manner.
Results
Cloning of Rodent DISC-1.
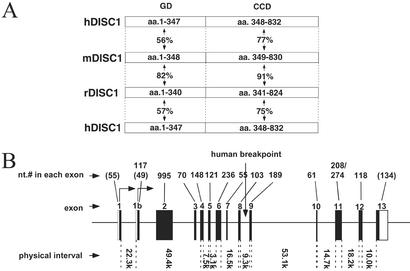
By using PCR techniques and the known human sequence of DISC-1, as well as mouse sequences homologous to portions of the human DISC-1, we have cloned and sequenced cDNAs for mouse and rat DISC-1 (Fig. 1). The amino acid sequence is highly conserved between rat and mouse, but there is much less conservation between rodent and human. The N-terminal globular domain displays only ≈50–60% amino acid identity between rodent and human, whereas the C-terminal coiled-coil domain shows ≈75% identity between rodent and human. The intron/exon structure is essentially identical in the mouse and human genes (17), although we detect an additional exon in the mouse sequence, 1b, which is not evident in human cDNAs.
Figure 1.
Cloning of rodent DISC-1. (A) Schematic diagram of the structure of DISC-1 proteins from the three species and the homology among them. GD denotes the putative globular domain, and CCD denotes the coiled-coil motif-containing domain. (B) Genomic organization of mouse DISC-1, mapped on mouse chromosome 8. The drawing was made by comparing the full-length coding sequence of mouse DISC-1 with a mouse genomic sequence, GA_x6k02T2NUPS, derived from the Celera mouse genome database (www.celera.com). Filled boxes correspond to the coding region of each exon, and open boxes are untranslated regions. Nucleotide numbers in each exon, exon numbers, and physical interval between neighboring exons are indicated. Numbers in parentheses are those that correspond only to coding nucleotides within exons 1 and 13. The entire gene spans >200 kb.
Developmental and Intracellular Expression of DISC-1.
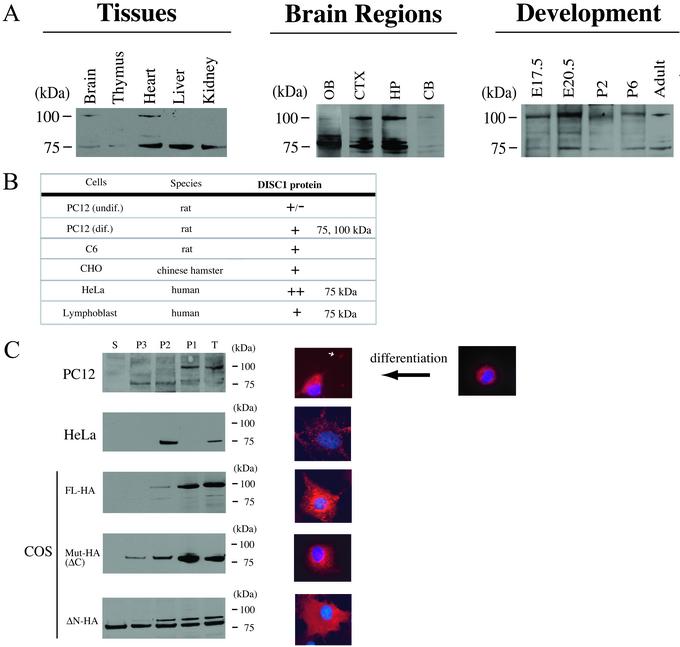
We developed three distinct antibodies to DISC-1, and we examined DISC-1 expression by Western blot analysis. DISC-1 protein is expressed as two distinct bands of 100 and 75 kDa, evident in brain, heart, liver, kidney, and thymus at a lower level in adult rats (Fig. 2A). Marked regional variations occur within the adult brains with higher densities in the olfactory bulb, the cerebral cortex, and hippocampus, and lower expression in the cerebellum.
Figure 2.
DISC-1 protein expression in tissues and cells. Major forms of DISC-1 (100 and 75 kDa) were detected by antibodies against both the N-terminal side (amino acids 347–600 and 212–537) and C-terminal side (amino acids 601–854) of the breakpoint. The specificities were also confirmed by preabsorption experiments. (A) High expression of DISC-1 protein in brain, heart, liver, and kidney, and weak expression in thymus. DISC-1 of 100 kDa is predominant in brain and heart. In adult brains, DISC-1 is enriched in the forebrain. DISC-1 expression is developmentally regulated with an enhanced expression of the 100-kDa band at E20.5. (B) DISC-1 protein expression in cell lines. DISC-1 is expressed in both neuronal and glial cells, as well as some peripheral cells. In PC12 cells, differentiation is associated with augmented expression. (C) DISC-1 distribution is analyzed by subcellular fractionation and immunofluorescent staining. Endogenous DISC-1 protein is enriched in P1 and P2 fractions of differentiated PC12 cells and in P2 of HeLa cells. Exogenously expressed full-length (FL) DISC-1 tagged with HA also occurs in P1 and P2 fractions, whereas mutant DISC-1 with the C-terminal deletion (Mut-HA, ΔC) distributes in P1, P2, and P3 fractions. Another mutant of DISC-1 lacking 290 amino acids of the N-terminal region (ΔN) distributes in all fractions, including the supernatant. Immunofluorescent staining of DISC-1 was analyzed under a confocal microscope. Endogenous and exogenous DISC-1 was stained in red; the nucleus was stained in blue with Hoechst 33258. Dotted DISC-1 staining is observed in the cytoplasm, especially in the perinuclear regions of undifferentiated PC12 cells and HeLa cells. In differentiated PC12 cells, DISC-1 occurs not only in cytoplasm, but also in neurites with higher density at the terminal regions (white arrow indicates DISC-1 in the terminus). Exogenously expressed DISC-1 tagged with HA in COS cells was stained with an anti-HA Ab (red). The dotted staining in perinuclear regions similar to centrosomes reflects full-length DISC-1 (DISC-1 FL). Mutant DISC-1 with the C-terminal deletion occurs in the cytoplasm with smaller dotted structures than DISC-1 FL. The N-terminal truncated form of DISC-1 (DISC-1 ΔN) distributes diffusely in the cytoplasm of COS cells.
DISC-1 expression is developmentally regulated with highest levels at E20.5, somewhat higher than at E17.5. Lower levels at postnatal days 2 and 6 resemble adult values.
DISC-1 occurs in various cell lines (Fig. 2B) with highest levels in HeLa cells. In PC12 cells, differentiation is associated with the augmented expression of DISC-1.
We examined the intracellular distribution of endogenous and exogenous DISC-1 by subcellular fractionation and by immunofluorescent staining (Fig. 2C). In differentiated PC12 cells, the highest levels of DISC-1 occur in the crude nuclear P1 fraction, with lower levels in the P2 (crude mitochondrial) and P3 (crude microsomal) fractions, and negligible levels in the S (soluble) fraction.
By immunohistochemistry, DISC-1 in PC12 cells occurs in punctate structures localized in the cell body when the cells are undifferentiated and proliferating. Once differentiated, DISC-1 staining is observed not only in the cell body but also in neurite-like processes, with higher density in terminal regions. We observed little nuclear staining, suggesting that the P1 localization of DISC-1 mainly reflects nonnuclear membrane and cytoskeletal structures.
The intracellular distribution of endogenous DISC-1 in HeLa cells differs from PC12 cells, as highest densities occur in the P2 fraction. Also, in HeLa cells, DISC-1 is expressed predominantly as a 75-kDa band, whereas in PC12 cells, both the 100- and 75-kDa bands are present. DISC-1 staining in HeLa cells is confined to punctate structures that costain with a mitochondrial marker, MitoTracker (Molecular Probes), fitting with subcellular fractionation experiments localizing the protein to the P2 fraction (Y.O. and A.S., unpublished observations).
To evaluate the intracellular localization of mutant DISC-1, we transfected COS cells with wild type and the disease form with C-terminal 257 amino acid truncation, as well as a form of DISC-1 in which the N-terminal 290 amino acids were deleted (Fig. 2C). Wild-type DISC-1, evident predominantly as a 100-kDa band, is concentrated in the P1 fraction, whereas the disease form of DISC-1, expressed exclusively as the 70-kDa band, is evident in P2 and P3 fractions as well as in P1. The N-terminal truncated protein distributes similarly in P1, P2, P3, and S fractions. Immunohistochemistry reveals a concentration of wild-type DISC-1 in large granular structures in the perinuclear region. The disease form of DISC-1 protein occurs in somewhat smaller punctate perinuclear structures, whereas the N-terminal truncated DISC-1 is expressed in a more diffuse pattern throughout the cell.
DISC-1 Binds Structural Proteins; Mutant DISC-1 Fails to Bind NUDEL.
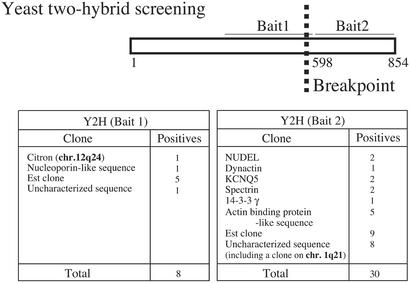
To identify proteins that interact with DISC-1, we conducted yeast two-hybrid analysis with a bait from the N-terminal portion before the breakpoint (Bait 1) and a bait from the C-terminal area (Bait 2; Fig. 3). Bait 2 is composed of amino acids 598–854, which are deleted in individuals carrying the DISC-1 mutation.
Figure 3.
Possible DISC-1 interactors identified by yeast two-hybrid screening. Interactors include cytoskeletal proteins, such as NUDEL and Citron, that are associated with brain development.
A variety of DISC-1 interactors are associated with the cytoskeleton. Bait 1 binds Citron, a protein enriched in glutamatergic synapses, where it binds PSD-95, a major protein of postsynaptic densities, as well as a small G protein, Rho (18, 19).
Bait 2 binds NUDEL, dynactin, spectrin, 14-3-3γ, and the potassium channel KCNQ5, as well as an actin-binding protein-like sequence. Both NUDEL and dynactin interact with dynein and are associated with microtubules. Spectrin is a cytoskeletal protein that binds actin.
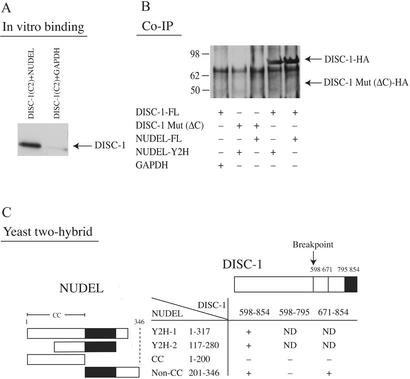
NUDEL occurs in a complex with LIS1, a protein which is mutated in a form of lissencephaly (12, 13, 15, 20). NUDEL plays an essential role in nucleokinesis and neuronal migration (21, 22). As schizophrenia and lissencephaly are associated with cerebral cortical developmental abnormalities, we chose to explore NUDEL–DISC-1 interactions in some detail. We directly demonstrate in vitro binding of DISC-1 and NUDEL (Fig. 4A). We evaluated coimmunoprecipitation of DISC-1 and NUDEL in HEK 293 cells transfected with normal and mutant DISC-1 and NUDEL (Fig. 4B). We observed robust binding of normal DISC-1 to NUDEL, whereas mutant DISC-1 fails to bind.
Figure 4.
Confirmation and characterization of DISC-1–NUDEL interaction. (A) DISC-1–NUDEL interaction by in vitro binding. The interaction is specific, as no interaction of DISC-1 and GAPDH is observed. Representative data from one of three assays are presented. (B) Coimmunoprecipitation of DISC-1 and NUDEL in HEK293 cells. Mutant DISC-1 with the C-terminal deletion fails to bind to NUDEL. Representative data from one of three assays are presented. (C) Mutational analyses of DISC-1–NUDEL interaction. Binding critically depends on the C-terminal end of DISC-1 (amino acids 796–854) and the middle portion of NUDEL (amino acids 201–280) outside of its coiled-coil region.
Both NUDEL and DISC-1 possess coiled-coil domains that, in theory, might interact in a nonspecific fashion. To examine this possibility, we conducted yeast two-hybrid analysis with different portions of NUDEL and DISC-1 (Fig. 4C). The critical domain of NUDEL for binding to DISC-1 consists of amino acids 201–280, which are outside of the coiled-coil domain of NUDEL. The portion of DISC-1 crucial for binding to NUDEL consists of amino acids 795–854, the extreme C terminus, a region predicted to be deleted in individuals with the DISC-1 translocation.
Mutant DISC-1 Alters Neurite Outgrowth.
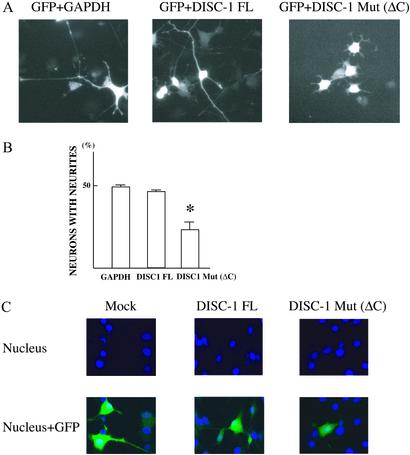
Neurite outgrowth and neuronal migration involve microtubule dynamics as well as remodeling of actin cytoskeleton (23). Accordingly, we examined the influence of DISC-1 transfection on neurite outgrowth of PC12 cells (Fig. 5). Mutant DISC-1 expression does not produce any major morphologic abnormalities. However, it is associated with shorter neurites and with a decreased percentage of neurons bearing neurites (Fig. 5 A and B). Because degenerative processes associated with cell death would alter neurite outgrowth, we examined nuclei of transfected cells. We fail to observe any evidence of apoptosis selectively associated with mutant DISC-1 (Fig. 5C).
Figure 5.
Truncated DISC-1 inhibits neurite outgrowth of PC12 cells. (A) Representative morphology of a PC12 cell transfected with constructs for GAPDH together with GFP (Left). Representative morphology of PC12 cells transfected with constructs of the full-length DISC-1 and GFP (Center). Representative morphology of PC12 cells with the C-terminal truncated mutant DISC-1 with GFP (Right). (B) Mean percentages ± SE of PC12 cells harboring neurites (longer than three cell body diameters) 5 days after transfection of full-length DISC-1, mutant DISC-1, or GAPDH. Mutant, but not full-length, DISC-1 alters neurite outgrowth (P < 0.01). (C) No obvious signs of apoptotic cell death (nuclear condensation and fragmentation) in cells transfected with the mutant or the full-length DISC-1.
Discussion
In the present study, we have characterized the disposition of DISC-1. The protein is expressed in brain with a marked concentration in the forebrain, including the cerebral cortex. Highest levels occur during late embryogenesis when cerebral cortical development takes place. Within cells, DISC-1 is predominantly associated with cytoskeletal elements that mediate neuronal migration and axonal outgrowth in brain development. The localization of DISC-1 differs in HeLa cells, suggesting several roles of DISC-1, depending on biological contexts.
Yeast two-hybrid analysis indicates an interaction of DISC-1 with several cytoskeletal proteins, of which we have explored NUDEL in greatest depth. Preliminary reports by others also detect DISC-1 expression in brain with regional variations and interactions with cytoskeletal proteins, including NUDEL.‡‡,†† NUDEL interacts selectively with the portion of DISC-1 that is deleted in patients. Indeed, NUDEL fails to interact with mutant DISC-1. The DISC-1 mutant truncation has functional consequences, as the expression of mutant, but not normal, DISC-1 alters neuronal outgrowth.
NUDEL is associated with at least two diseases of cerebral cortical development (23). NUDEL binds to LIS-1, which is mutated in a form of lissencephaly (12, 13, 15, 20). Also, NUDEL is downstream of a signaling cascade derived from reelin (23). Mutations in the reelin gene are also associated with a type of lissencephaly with cerebellar hypoplasia (24). The reeler mice, in which reelin gene is disrupted, display abnormal cortical development (25). Recently, several groups reported decreases in reelin expression in schizophrenic brains (26, 27). The relationship of these DISC-1-linked proteins to cerebral cortical abnormalities of several diseases including schizophrenia fits with the notion that the schizophrenia-like disturbances in patients with DISC-1 mutation may reflect cerebral cortical abnormalities.
Further evidence for an association of the DISC-1 translocation with cortical abnormalities comes from our finding that mutant DISC-1 alters neurite outgrowth. Independently of our study, another group also observed an influence of DISC-1 on neurite outgrowth via a DISC-1 interactor (K. Miyoshi, A. Honda, K. Baba, M. Taniguchi, K. Oono, T. Fujita, S. Kuroda, T. Katayama, and M. Tohyama, personal communication). Abundant neuropathologic studies support a disturbance in neuronal migration and process formation during development in patients with schizophrenia (4).
Patients with the DISC-1 mutation predominantly manifest schizophrenic symptomatology. However, some of them display affective disorder. A limited number of individuals with the mutation have, thus far, not developed psychiatric disorders, possibly reflecting incomplete penetrance (9). In relating DISC-1 to the schizophrenic disease process, it is important to bear in mind that the DISC-1 translocation is very rare. Nonetheless, the DISC-1 disturbance may provide valuable insights into the pathogenesis of more common forms of schizophrenia. By analogy, very rare forms of Parkinson's disease are caused by mutations in α-synuclein, whereas other rare cases stem from mutations in parkin (28). Although the α-synuclein disease is rare, α-synuclein protein is concentrated in Lewy bodies that are characteristic of almost all of the patients with Parkinson's disease (29). Protein–protein interaction studies reveal that α-synuclein and parkin are linked together by interactions with the recently discovered protein synphilin (30, 31). Similarly, the DISC-1–NUDEL complex may interact with other proteins to participate in the pathophysiology of more common forms of schizophrenia.
Acknowledgments
We thank Ms. Y. Lema for preparation of the figures; Ms. B. Ziegler for typing the manuscript; Drs. R. Margolis, H. Arai, H. Koizumi, S. Narumiya, and H. Bito for useful comments; Drs. T. Katayama and M. Tohyama for discussion; and Mr. I. Litvinov, Ms. U. Park, Ms. X. Luo, Ms. P. Dulloor, Mr. B. Bae, and M. R. Hara for technical assistance. This work was supported by Stanley Medical Research Institute grants, the National Alliance for Research on Schizophrenia and Depression, the S-R Foundation (Washington, DC) (to A.S.), U.S. Public Health Service Grant NS-34172 and Project 4 of the Johns Hopkins University Stanley Research Center (to C.A.R.), and U.S. Public Health Service Grant MH-18501 and Research Scientist Award DA-00074 (to S.H.S.). Y.O. is a Fellow of the Japan Foundation for Aging and Health.
Abbreviations
- DISC-1
Disrupted-in-Schizophrenia-1
- NUDEL
NudE-like
Footnotes
Kandpal, G., Ma, L., Acton, P., Austin, C. P. & Morris, J. A. (2002) Soc. Neurosci. Abstr. 28.
Millar, J. K., James, R., Christie, S., Taylor, M. S., Devon, R. S., Hogg, G., Muir, W. J., Blackwood, D. H. R. & Porteous, D. J. (2002) Abstr. World Cong. Psychiatr. Genet. 114, 748.
References
- 1.Gottesman I I. Schizophrenia Genetics. New York: Freeman; 1991. [Google Scholar]
- 2.Weinberger D R. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 3.Sawa A, Snyder S H. Science. 2002;296:692–695. doi: 10.1126/science.1070532. [DOI] [PubMed] [Google Scholar]
- 4.Harrison P J. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- 5.Ross C A, Pearlson G D. Trends Neurosci. 1996;19:171–176. doi: 10.1016/s0166-2236(96)10022-9. [DOI] [PubMed] [Google Scholar]
- 6.Berrettini W H. Biol Psychiatry. 2000;48:531–538. doi: 10.1016/s0006-3223(00)00883-0. [DOI] [PubMed] [Google Scholar]
- 7.Karayiorgou M, Gogos J A. Neuron. 1997;19:967–979. doi: 10.1016/s0896-6273(00)80390-6. [DOI] [PubMed] [Google Scholar]
- 8.Millar J K, Wilson-Annan J C, Anderson S, Christie S, Taylor M S, Semple C A, Devon R S, Clair D M, Muir W J, Blackwood D H, Porteous D J. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 9.Blackwood D H, Fordyce A, Walker M T, St. Clair D M, Porteous D J, Muir W J. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, Suhonen J, Ellonen P, Chan G, Sinsheimer J S, et al. Hum Mol Genet. 2001;10:1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa M, Umezu M, Aoki J, Koizumi H, Arai H, Inoue K. FEBS Lett. 2000;479:57–62. doi: 10.1016/s0014-5793(00)01856-1. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki S, Shionoya A, Ishida M, Gambello M J, Yingling J, Wynshaw-Boris A, Hirotsune S. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 13.Niethammer M, Smith D S, Ayala R, Peng J, Ko J, Lee M S, Morabito M, Tsai L H. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 14.Sawa A, Wiegand G W, Cooper J, Margolis R L, Sharp A H, Lawler J F, Jr, Greenamyre J T, Snyder S H, Ross C A. Nat Med. 1999;5:1194–1198. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- 15.Sawa A, Khan A A, Hester L D, Snyder S H. Proc Natl Acad Sci USA. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang M, Jaffrey S R, Sawa A, Ye K, Luo X, Snyder S H. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 17.Millar J K, Christie S, Anderson S, Lawson D, Hsiao-Wei Loh D, Devon R S, Arveiler B, Muir W J, Blackwood D H, Porteous D J. Mol Psychiatry. 2001;6:173–178. doi: 10.1038/sj.mp.4000784. [DOI] [PubMed] [Google Scholar]
- 18.Furuyashiki T, Fujisawa K, Fujita A, Madaule P, Uchino S, Mishina M, Bito H, Narumiya S. J Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Vazquez L, Apperson M, Kennedy M B. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattori M, Adachi H, Tsujimoto M, Arai H, Inoue K. Nature. 1994;370:216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 21.Lambert de Rouvroit C, Goffinet A M. Mech Dev. 2001;105:47–56. doi: 10.1016/s0925-4773(01)00396-3. [DOI] [PubMed] [Google Scholar]
- 22.Hatten M E. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- 23.Ross M E, Walsh C A. Annu Rev Neurosci. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- 24.Hong S E, Shugart Y Y, Huang D T, Shahwan S A, Grant P E, Hourihane J O, Martin N D, Walsh C A. Nat Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 25.Rice D S, Curran T. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- 26.Fatemi S H. Mol Psychiatry. 2001;6:129–133. doi: 10.1038/sj.mp.4000129. [DOI] [PubMed] [Google Scholar]
- 27.Guidotti A, Auta J, Davis J M, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson D R, Impagnatiello F, Pandey G, Pesold C, Sharma R, et al. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Dawson V L, Dawson T M. Neurobiol Dis. 2000;7:240–250. doi: 10.1006/nbdi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 29.Goedert M. Nat Rev Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- 30.Chung K K, Zhang Y, Lim K L, Tanaka Y, Huang H, Gao J, Ross C A, Dawson V L, Dawson T M. Nat Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 31.Engelender S, Kaminsky Z, Guo X, Sharp A H, Amaravi R K, Kleiderlein J J, Margolis R L, Troncoso J C, Lanahan A A, Worley P F, et al. Nat Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]