Abstract
We have reinvestigated the subcellular distribution of the duck hepatitis B virus (DHBV) core protein in infected duck hepatocytes and in transfected cells. By using indirect immunofluorescence, the protein was found to be localized not only in the cytoplasm, as described previously, but also within the cell nucleus, being concentrated in distinct, brightly staining nuclear core bodies (NCBs). In colocalization studies using confocal microscopy, the NCBs were found exclusively in the periphery of nuclear subdomains characterized as splicing factor compartments and distal to other subnuclear domains. Also relevant for their functional significance is that the NCBs formed during the establishment of virus infection, i.e., at very low overall concentrations of newly synthesized core protein, and persisted throughout all stages of infection. Moreover, a subset of NCBs colocalized with foci of pregenomic DHBV RNA present at concentrations detectable by fluorescence in situ hybridization. Taken together, these findings indicate that a minor fraction of the DHBV core protein molecules escapes the major cytoplasmic assembly pathway to accumulate in specific subnuclear domains, and they furthermore suggest that these NCBs serve a role in the synthesis and/or maturation of the DHBV RNA pregenome.
The hepadnaviruses (hepatitis B viruses [HBVs]) are a family of small, enveloped DNA viruses comprising the medically important human HBV and other members infecting rodents and birds (9). The HBVs are characterized by containing a small, circular, noncovalently closed DNA genome and having a compact genome organization and also a unique replication strategy that involves reverse transcription of an RNA pregenome. Early in infection, the incoming DNA genome is delivered to the host cell nucleus, where it is converted from an open circle to a covalently closed circular form, which in turn serves as the transcriptional template for pregenomic and subgenomic RNA synthesis (9, 31). Out of a small set of gene products, the polymerase and the core protein assemble with the pregenomic RNA to form an immature, RNA-containing nucleocapsid. Genome maturation progresses in a multistep process involving reverse transcription and finally leading to mature nucleocapsids containing the open circular DNA genome as present in the virion (35). Early in infection, these nucleocapsids deliver the genome back to the nucleus, amplifying the pool of intranuclear DNA templates (36). At later stages with abundant envelope protein expression, mature nucleocapsids attach to intracellular membranes containing budding envelope structures, initiating virion formation and secretion (22).
According to the basic replication cycle outlined above, the core protein serves primarily as the structural, nucleic acid binding component of the nucleocapsid and is additionally a cofactor in viral DNA synthesis, including reverse transcription. These functions require, as essential elements, several clusters of arginine residues which are located in the carboxy-terminal segment of the amino acid sequence following the capsid assembly domain (13, 24, 28). These sequence elements are additionally part of a nuclear localization signal, which is involved in targeting the nucleocapsid to the nuclear pore for genome import and which is multipartite in mammalian hepadnaviruses (6, 17, 39) and monopartite in the avian duck HBV (DHBV) (21). The nuclear localization signal function seems to be counterbalanced, probably by phosphorylation (17, 19; A. K. Weigand, A. Knaust, and H. Schaller, unpublished data), thus precluding nuclear accumulation of core protein and supporting the cytoplasmic localization and capsid assembly observed for all hepadnaviruses in productively infected or transfected cells. However, a predominant nuclear localization of HBV core protein has been reported to occur in various experimental systems, such as in the livers of chronic HBV carriers (3) or of HBV-transgenic mice (11, 12), as well as in a particular HBV-transfected cell line (40). Electron microscopy of liver sections and further biochemical analysis revealed that this phenotype correlated with the presence of capsids free of viral nucleic acid, a result interpreted as arising from unbalanced import of core protein subunits (11; H. W. Zentgraf, C. Kuhn, and H. Schaller, unpublished results). No functional significance has so far been attributed to these observations, and presently the assumption prevails that intranuclear core protein assembly may be a functionally irrelevant by-product of disregulated intracellular core protein traffic.
Further support for this notion come from studies with DHBV, an animal model that has been extensively employed to elucidate the molecular biology of hepadnavirus replication in vivo and in vitro. In contrast to the results with mammalian hepadnaviruses, for DHBV only cytoplasmic and no intranuclear core protein accumulation has been reported (see e.g., references 8 and 41), except for a recent report describing unidirectional nuclear import and retention of assembly-incompetent, green fluorescent protein (GFP)-fused DHBV core polypeptides (21). In a follow-up to these observations, we have now used the DHBV system to readdress the question of the general nature of nuclear core protein import and of its potential functional significance. Modifying previous reports, we demonstrate that a minor fraction of the assembly-competent, full-length protein escapes the major assembly pathway to accumulate in the cell nuclei of DHBV-infected duck hepatocytes. There it forms distinct nuclear core bodies (NCBs) that map very close to two subnuclear splicing factor compartments and that accumulate pregenomic DHBV RNA to concentrations occasionally detectable by fluorescence in situ hybridization (FISH). Taken together, these findings suggest that the hepadnavirus core protein has a so-far-unrecognized, nonstructural function in the posttranscriptional viral RNA metabolism.
MATERIALS AND METHODS
Virus, plasmids, and antibodies.
DHBV subtype 16 was used throughout the study. pCD16, a plasmid starting at position 2520 under control of the cytomegalovirus immediate-early promoter-enhancer, has been described previously (26), as have pCDcore, a plasmid expressing the DHBV core protein as the only gene product (21), and pHBV-P7, a plasmid containing the full-size DHBV genome bearing a TTG→TAG mutation at DHBV nucleotide 183 that introduces a stop codon at amino acid 5 of the polymerase open reading frame (29). The core protein was detected by immunofluorescence with a polyclonal anticore antiserum (D087) as described previously (21). We performed colocalization studies of the NCBs with several focus-forming nuclear factors by using the following antibodies: a mouse anti-P80-coilin monoclonal antibody (immunoglobulin G2a) (provided by A. Lamond, University of Dundee, Dundee, Scotland United Kingdom); an anti-splicing cofactor SC35 monoclonal antibody (immunoglobulin G1) (Sigma); a guinea pig antinucleolin antibody (30) (provided by M. Schmidt-Zachmann, DKFZ, Heidelberg, Germany); and a mouse anti-Sm protein (systemic lupus erythematosus marker) monoclonal antibody (clone Y12; Neomarkers). Anti-nuclear pore complex monoclonal antibody 414 was provided by D. Görlich, ZMBH, Heidelberg, Germany.
Cell culture, infection, and transfection.
DHBV-positive livers were obtained from 4- to 6-week-old ducks infected congenitally or experimentally. Primary duck hepatocytes (PDHs) were prepared and cultivated as described previously (14). For infection of negative PDHs, DHBV-containing duck serum was applied to one well of a 12-well culture dish containing 8 × 105 cells at a multiplicity of infection of 100. After 14 h of incubation, the cells were washed and further cultivated. After 3 days, the cells were fixed and immunostained for DHBV core protein as described below. The human hepatoma cell line HuH7 was cultivated on coverglass chamber slides (Nunc) and transfected with the plasmids by using a standard calcium phosphate protocol. For immunofluorescence analysis, the cells were fixed 3 to 4 days after transfection as described below.
Immunofluorescence and Western blotting.
At 3 days postinfection or posttransfection, the cells, PDHs, or transfected HuH7 cells were washed with phosphate-buffered saline (PBS) and fixed with 3% formalin for 30 min. The fixed cells were permeabilized with 0.25% Triton X-100 in PBS and immunostained with a rabbit polyclonal antibody against the DHBV core protein (D087) as a primary antibody and an fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin as a secondary antibody. Cell nuclei were isolated by using a standard protocol based on the method of Widnell and Tata (38). Isolated nuclei were washed twice in 1% Triton X-100 in 10 mM Tris-5 mM MgCl2-0.25 M sucrose. Chromatin was extracted by digestion with 400 U of DNase I per ml in cytoskeletal buffer {10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.8], 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 20 mM vanadyl riboside complex, 1 mM 4-(2-aminoethyl)benzene sulfonyl fluoride} for 30 min at 37°C followed by washing twice with 1 M NaCl buffer (cytoskeltal buffer adjusted to 1 M NaCl) (25).
For costaining, the samples were incubated at the same time with D087 and one of the different primary mouse antibodies. FITC-conjugated anti-rabbit or tetramethyl rhodamine isocyanate-conjugated anti-mouse immunoglobulin was used as a secondary antibody. Fluorescence was analyzed either with a conventional microscope or with a Leica TCS NT confocal laser scanning microscope (63×/1.23 objective). For monitoring core expression biochemically (see Fig. 2), cells were lysed in sample buffer and equivalents of 8 × 104 cells per time sample were loaded on a sodium dodecyl sulfate gel. Western blotting analysis was performed as described previously (22).
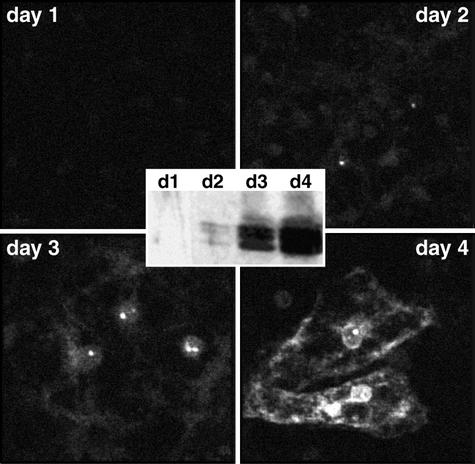
FIG. 2.
Kinetics of NCB formation after DHBV infection. PDHs were infected in vitro with a multiplicity of 100 DNA-containing DHBV particles per cell. After 1, 2, 3, or 4 days, cells were fixed, immunostained for DHBV core protein, and analyzed with a confocal microscope (main panels). In parallel, cell lysates were analyzed by Western blotting for DHBV core protein content (inset).
In situ hybridization.
To generate an antisense probe specifically detecting the pregenomic RNA (from restriction site AvrII to EcoRI, nucleotide [nt] 403 to 3018 of DHBV16), we used the plasmid pCD16, displaying a T7 promoter after the NheI restriction site in antisense direction. After deleting the AvrII-NheI fragment from pCD16 (nt 911 to 3335) and linearizing the obtained plasmid with EcoRI, we generated a digoxigenin-labeled RNA probe by in vitro transcription with T7 polymerase as detailed by the manufacturer (Boehringer Mannheim). To generate a sense probe detecting genomic DHBV DNA (from nt 3018 to 391 of DHBV16), we subcloned the EcoRI-BamHI fragment of pCD16 (nt 504 to 2166) downstream of an SP6 promoter sequence of plasmid pSPT19 (Boehringer Mannheim). The resulting plasmid was cut with BglII, serving as starting product for in vitro transcription.
To examine colocalization of DHBV RNA relative to DHBV core protein, we used the protocol used by Seguin et al. (32). PDHs prepared from a noninfected duck liver were cultivated on glass coverslips and infected as described above. At 3 days postinfection, coverslips were washed with PBS and fixed at room temperature for 10 min in 4% paraformaldehyde-PBS buffered to pH 7.4. Following fixation, coverslips were washed twice with PBS and stored in 70% ethanol at 4°C. For prehybridization, each coverslip was inverted onto 100 μl of prehybridization solution (50% deionized formamide, 2× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 5× Denhardt's reagent, 1 mg of herring sperm DNA per ml), and the chamber was sealed with Parafilm and incubated for 1 h at 37C. The appropriate RNA probe was resuspended at 1 μg/μl in fresh hybridization solution and heated at 80°C for 10 min. The coverslips were then placed on 30 μl of fresh hybridization solution, and the chambers were sealed with Parafilm and incubated overnight at 37°C. Unbound probe was removed by four 15-min washes in 50% deionized formamide-2× SSPE at 37°C. Samples were then incubated with 1% blocking reagent (Boehringer Mannheim) containing the rabbit polyclonal anti-DHBV core antibody. Samples were then washed and incubated with both FITC-conjugated sheep anti-digoxigenin antibody (1/40 dilution) and Texas Red-conjugated goat anti-rabbit immunoglobulin antibody (1/80 dilution). Coverslips were mounted in antibleach medium (Boehringer Mannheim), and the fluorescence was analyzed with a Leica TCS NT confocal laser scanning microscope (63×/1.23 objective). To test for colocalization of DHBV DNA, we followed the same protocol except that (i) the fixed cells were heated at 70°C for 2 min in 70% deionized formamide-2× SSPE prior to hybridization with the sense DHBV RNA probe and (ii) the cells were incubated with the sense DHBV RNA probe at 42°C overnight.
RESULTS
DHBV core protein localizes to distinct nuclear foci in DHBV-infected cultured hepatocytes.
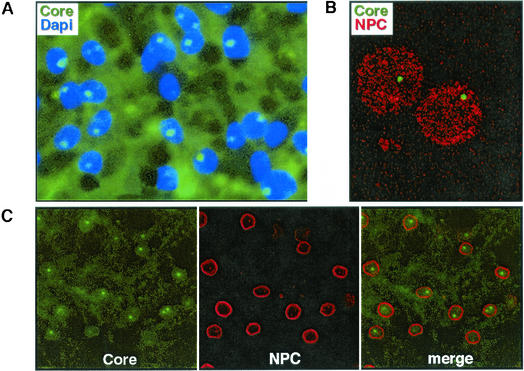
PDHs that had been prepared from a DHBV-infected animal were analyzed, after fixation with paraformaldehyde, for the subcellular distribution of the core protein by using indirect immunofluorescence with a DHBV core-specific antiserum. As shown in Fig. 1A, microscopic analysis of such samples revealed that all DHBV core-positive cells contained, in addition to the diffuse cytoplasmic staining, DHBV core protein in one, or occasionally several, brightly staining, spherical foci which apparently resided within the cell nucleus. No such NCBs were detected in noninfected control cells (not shown here; see Fig. 2, day 1). The NCBs were also detected, although with somewhat reduced intensity and frequency, after fixation with methanol, a result arguing against artifacts due to the fixation procedure. Moreover, their nuclear localization was confirmed by confocal laser scanning microscopy sectioning through fixed PDHs, or cell nuclei that had been prepared from DHBV-infected duck livers, and additionally immunostained for the nuclear pore complex, a marker delineating the nuclear membrane (Fig. 1B and C). Interestingly, the NCB pattern was affected neither by destruction of the nuclear membrane with 1% Triton X-100 nor by subsequent chromatin extraction (Fig. 1B), suggesting a rather stable structure which is not attached to chromatin.
FIG. 1.
DHBV core protein forms nuclear foci in DHBV-infected hepatocytes. Primary duck hepatocytes (A and C) or isolated nuclei (B) were fixed and immunostained for DHBV core protein. After counterstaining for chromatin (4′,6′-diamidino-2-phenylindole [DAPI]) (A) or the nuclear pore complex (NPC) (B and C), the cells or nuclei were analyzed by conventional fluorescence microscopy (A) or confocal laser scanning microscopy (B and C). The NCBs remained stable in the isolated nuclei after removal of the outer nuclear membrane and chromatin extraction (B) (for details, see Materials and Methods).
NCBs arise during the establishment of viral infection.
To characterize the kinetics of NCB formation after experimental DHBV infection of PDHs, cells were fixed at daily intervals, stained with an anti-DHBV core antiserum, and analyzed by confocal microscopy. A parallel set of infected cells was lysed and analyzed by Western blotting for total core protein content. As shown in Fig. 2, nuclear foci of core protein appeared as early as day 2 postinfection, a time point when cytoplasmic core protein was not yet detected by immunostaining and when the protein was barely detectable by Western blotting (Fig. 2, inset). Thereafter, the NCBs remained essentially unchanged in size and numbers and persisted in the infected cells throughout prolonged cell culture up to 4 weeks (data not shown). For the functional relevance of these observations, it is important to note that the accumulation of core protein in NCBs, and in the cell nucleus in general, occurred well before the high steady-state levels of cytoplasmic core protein were reached, thus making an artifactual aggregation in the nuclear compartment unlikely. In contrast, NCBs were formed less frequently in transfected cells producing core protein more rapidly and to high cytoplasmic concentrations (see below).
NBC formation does not require any further viral component.
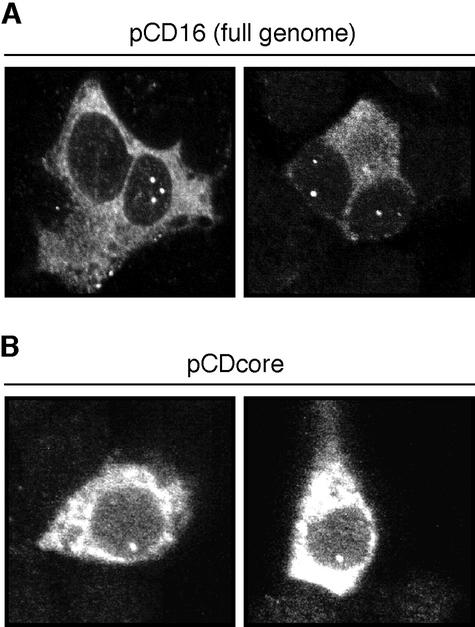
Next, we wanted to assess whether NCB formation would depend on viral gene products other than the core protein or on the presence of the intact DHBV replication machinery. To this end, hepatoma cells of human or avian origin (Huh7 and LMH, respectively) were transfected with a set of plasmids containing different genomic and subgenomic DHBV DNA segments (for details, see Materials and Methods). The human cell line was preferentially chosen for these and further transfection experiments to facilitate subsequent immunofluorescence studies determining colocalization with cellular marker proteins (see below). At 2 days posttransfection, the cells were fixed, immunostained, and analyzed by confocal microscopy for the distribution of the DHBV core protein. Expression from pCD16, a plasmid expressing a replication-competent DHBV RNA pregenome under the control of the cytomegalovirus immediate-early promoter, resulted only in about 5 to 10% of core-positive cells in an NCB pattern similar to the one observed in infected duck hepatocytes (Fig. 3A). A comparable intracellular DHBV core distribution was observed in cells transfected with a related genomic plasmid displaying a stop codon at the beginning of the polymerase open reading frame (data not shown) or with a plasmid in which all DHBV coding sequences following the core gene, as well as the upstream encapsidation signal, had been deleted (pCDcore) (Fig. 3B). These results demonstrate that the accumulation of core protein in nuclear bodies is an intrinsic property of the protein itself, predicting an interaction with specific cellular partners. They further indicate that structures relevant for such interactions are conserved between avian and human cells.
FIG. 3.
Formation of nuclear core protein foci does not depend on additional viral components. HuH7 cells were transfected with pCD16, encoding the full DHBV genome (A), or pCDcore, a plasmid expressing the core protein only (B). At 2 days posttransfection, the cells were fixed, immunostained for DHBV core protein, and analyzed with a confocal microscope. For each transfection, two examples of DHBV core-positive cells with NCBs were selected.
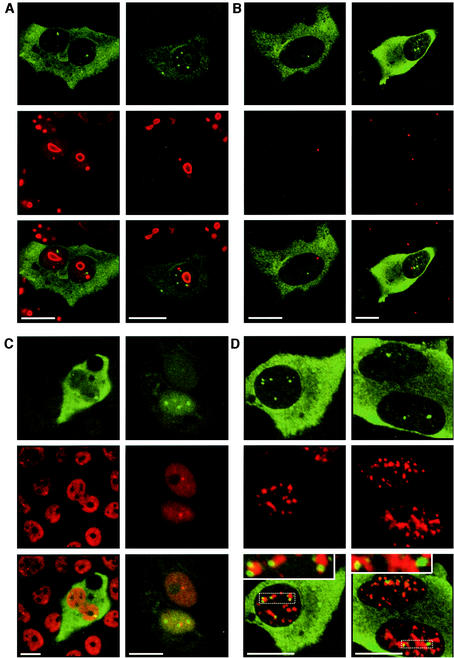
NCBs are located in the immediate vicinity of interchromatin granule speckles containing the spliceosome assembly cofactor SC35.
The cell nucleus contains distinct classes of subnuclear bodies, including nucleoli, splicing speckles, and coiled bodies (Cajal bodies), etc., which are either attached to chromatin or located in the interchromatin space (33, 18). To define whether NCB domains were associated with known subnuclear compartments, we performed colocalization studies by confocal fluorescence microscopy, using antibodies detecting various focus-forming nuclear proteins. Analysis of immunostained NCBs in pCD16-transfected HuH7 cells revealed that these occupied quite characteristic positions relative to reference structures identified by antibodies specific for nucleoli (N038), coiled bodies (p80/coilin), Sm proteins, and pre-mRNA splicing cofactor SC35 (Fig. 4): NCBs localized occasionally at the border of nucleoli (in about 1 out of 10 cases [Fig. 4A]), always distal to coiled bodies (in no case out of more than 20 [Fig. 4B]), and preferentially in the immediate vicinity of Sm speckles (in about 5 out of 10 cases [Fig. 4C]) but without exact overlap between domains stained for the marker protein or the core protein (visualized at a higher magnification in Fig. 4C, second column).
FIG.4.
NCBs map at distinct nuclear subdomains. HuH7 cells were transfected with a plasmid encoding the full DHBV genome (pCD16). At 2 days posttransfection, the cells were fixed and coimmunostained for DHBV core protein (green fluorescence) and for either nucleolus (N038 antibody) (A), coiled bodies (anti-p80/coilin antibody) (B), Sm protein (Y12 antibody) (C), or spliceosome (anti-splicing factor SC35 antibody) (D) (red fluorescence). They were then analyzed with a confocal microscope. Sequential excitation and scanning of the two fluorescent channels (separate excitations at 488 and 568 nm) were used to avoid cross bleeding of the fluorochromes between channels. For each antibody, two representative cells are shown. Bars, 10 μm. The insets in panels D represent threefold enlargements of the area marked by the dotted lines.
Even more significantly, NCBs always mapped at the periphery of SC35-positive substructures. This is exemplified in the two panels of Fig. 4D showing eight strong NCBs (in two focal planes through three nuclei), seven of which are also covered in the threefold-enlarged inserts. Although SC35 speckles represent rather extended intranuclear substructures, NCBs were never found to map within these domains, nor were they detected in the much more extended space in between. Even weak NCBs that were occasionally observed very close to but slightly detached from SC35 speckles, like the one detectable at the higher magnification in the upper right corner of the left inset in Fig. 4D, were found to be directly attached to the nearby SC35-positive substructure when viewed in successive focal planes (data not shown). Taken together, these observations strongly argue that the association of NCB structures with SC35 speckles is specific, suggesting a functional correlation.
Sm proteins have functions in the biogenesis and are components of a subset of the small ribonucleoprotein particles that catalyze pre-mRNA splicing (27). SC35 speckles are assumed to be nuclear compartments for storage or recycling of splicing factors or to be more directly involved in the splicing mechanism (7, 10, 15, 16). Thus, our results indicate that the NCBs are part of distinct nuclear subdomains, which are located in the close vicinity of cellular RNA splicing compartments.
Foci of pregenomic DHBV RNA colocalize with core protein in NCBs.
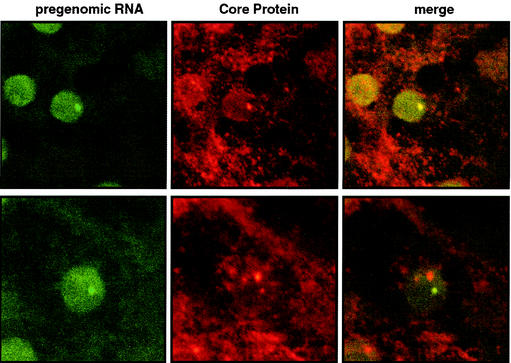
The results described above suggest that NCB formation reflects a specific association of core protein with a nuclear compartment involved in the control of alternative RNA splicing of the DHBV RNA pregenome (26). In a first attempt to assess this possibility, we used FISH to test whether NCB also contained viral nucleic acids. DHBV-infected PDHs were fixed and hybridized under the appropriate annealing conditions (see Materials and Methods) with an RNA probe complementary to a sequence unique for the RNA pregenome and absent from all subgenomic DHBV RNAs (map positions 1 to 415). As a control, we used a probe of opposite polarity assaying for antisense or double-stranded viral nucleotide sequences. Within the sensitivity reached, no signal was observed with the latter probe or with a non-DHBV-specific control RNA (not shown), which is not surprising in view of the approximately 20 copies of DHBV DNA template per cell (4, 36). In contrast to the case for these controls, we saw a distinct spot-like concentration of the genomic RNA probe in about 1% of the infected hepatocytes. As exemplified in Fig. 5 by two representative RNA-positive cells, these pregenomic RNA dots consistently colocated with an NCB. Conversely, most NCBs were without RNA dots, even in cells where other NCBs overlapped with DHBV RNA foci, and there was also no clear correlation of the intensities of core staining and the pregenome signal between individual NCB structures (Fig. 5, lower panel). While this absence of RNA dots in most NCBs is probably due to the insensitivity of our RNA analysis, the strict colocalization of all of the DHBV RNA foci detected with NCBs nevertheless confirms that both structures occupy the same nuclear compartment. Furthermore, this colocalization strongly suggests that these domains play a discrete role in the viral RNA metabolism at the level of hepadnaviral RNA synthesis and/or maturation in the cell nucleus.
FIG. 5.
Foci of DHBV pregenomic RNA colocalize with NCBs in a subset of infected hepatocytes. DHBV-infected PDHs were stained for pregenomic DHBV RNA by FISH (green fluorescence) (for details, see Materials and Methods) and immunostained for DHBV core protein (red fluorescence). Analysis was performed with a confocal microscope (sequential excitation and scanning of the two fluorochromes). The upper and lower panels show two representative examples of RNA-positive cells. Note that in the lower panel, the RNA colocalize with a weak NCB and not with a nearby strong NCB in the same nucleus.
DISCUSSION
While the hepadnavirus core proteins were up to now believed to be destined solely to become a structural and functional component of the cytoplasmic nucleocapsid and the secreted virion, we now demonstrate that a fraction of the assembly-competent DHBV core protein subunits accumulate in specific nuclear subdomains, termed NCBs, for which we propose a role in the biosynthesis of the unspliced DHBV RNA pregenome.
The formation of these NCBs probably depends on the availability of yet-unassembled core protein subunits, which are capable of rapid nucleocytoplasmic shuttling: NCBs formed preferentially at the low overall DHBV core concentrations as present during establishment of DHBV infection, whereas they were only rarely observed in DHBV-transfected cells with high DHBV core concentrations leading to cytoplasmic capsid assembly. In keeping with this interpretation, an NCB-like nuclear retention pattern was also observed in cells transfected with an assembly-incompetent GFP fusion with the C-terminal half of DHBV core (GFP/C125-262) but not in those transfected with a fusion to the assembly-competent full-length DHBV core (21). Together, our data indicate that the DHBV core protein subunit carries the potential for selective, high-affinity retention at yet-undefined subnuclear structures. The structures targeted seem to be limited in size and numbers, as they were already largely saturated during establishment of infection, so that the NCBs represented only a minor fraction of the total cellular core protein once the steady state of productive infection had been reached. Our findings further show that this process did not require the presence of other virus-encoded interaction partners, at least not for formation of the basic NCB structure. However, as demonstrated by the colocalization of pregenomic DHBV RNA, our data do not exclude a nonessential participation of additional viral gene products such as the viral P protein, which is known to bind to the genomic RNA packaging signal as well as to contain interaction domains for core protein sequences (20).
At present, our data allow only speculation as to the potential NCB functions. The first clues come from colocalization studies showing (i) an overlap with foci accumulating the DHBV RNA pregenome and (ii) a preferential location adjacent to nuclear speckles associated with the SR protein SC35, which is prototypic of essential, multifunctional splicing factors required at different steps of spliceosome assembly and modulating constitutive and alternative pre-mRNA splicing (7, 10, 37, 42). Actively transcribed cellular genes have been shown to locate at the periphery of these subcellular domains, supporting the existence of compartments coupling synthesis and maturation of primary transcripts (15, 16, 18, 33, 34). In analogy, the NCBs may be part of factories involved in the synthesis and maturation of nascent DHBV transcripts, acting either by binding to the DHBV minichromosome as reported for HBV by Bock et al. (1) or as a more general nucleic acid interaction partner. Additionally, and not mutually exclusive, the DHBV core protein, being a nucleocytoplasmic shuttling RNA binding protein (21), may also participate in the nuclear export of the unspliced pregenomic DHBV RNA, as exemplified by the role of structural virus proteins in the ribonucleoprotein complex incorporating the unspliced RNA genomes of influenza A virus (2).
In this context it is important to recall that the DHBV RNA pregenome is subject to alternative splicing (26) and is expected to pass through a cellular splicing compartment. Furthermore, the hepadnavirus core proteins, containing an RNA binding motif and an SR-rich protein interaction domain, appear to be related to the SR protein splicing cofactor family, an assumption supported by the recent observations that the HBV core protein is the target of SR-specific protein kinases (5). The formation of DHBV NCBs at the periphery of nuclear splicing speckles thus suggests the existence of a particular speckle subcompartment (23) which is used by DHBV to facilitate the nuclear interaction of its unspliced RNA pregenome with the core protein as a putative viral RNA export adaptor. In further speculation, the DHBV nuclear core protein might also be envisaged to be directly involved in the regulation of alternative splicing of the DHBV RNA pregenome. In initial attempts to prove this concept by RNA analysis of core gene knockout mutants, we have observed enhanced ratios of spliced RNA relative to the pregenome but have not obtained conclusive answers due to overlapping effects from cis-acting elements also influencing pregenome splicing (B. Zachmann-Brand and H. Schaller, unpublished data).
Finally, we may ask whether NCB formation is a feature generally associated with hepadnavirus replication. To our knowledge, an NCB-like pattern has so far not been detected in hepatocytes infected by mammalian hepadnaviruses, and even on close inspection, we have observed nuclear foci of HBV core protein in only a very minor fraction of HBV-transfected cells. This difference from the pronounced NCB formation in DHBV parallels the absence of any significant or functionally relevant RNA splicing in the replication of the mammalian hepadnaviruses and is therefore consistent with the hypothesis that the NCBs play an SR protein-like role in the maturation of the DHBV RNA pregenome.
Acknowledgments
We thank Bärbel Glass for the preparation of PDHs and for providing duck liver tissue and sera. We are also grateful to Angus Lamond, Marion Schmidt-Zachmann, Dirk Görlich, and Christa Kuhn for antibodies and to Karin Coutinho for expert editorial assistance.
This work was supported by a ZMBH fellowship to H.M., a grant (Scha 134/11-1) from the Deutsche Forschungsgemeinschaft to H.S., and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Bock, C. T., S. Schwinn, S. Locarnini, J. Fyfe, M. P. Manns, C. Trautwein, and H. Zentgraf. 2001. Structural organization of the hepatitis B virus minichromosome. J. Mol. Biol. 307:183-196. [DOI] [PubMed] [Google Scholar]
- 2.Bui, M., E. G. Wills, A. Helenius, and G. R. Whittaker. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu, C. M., and Y. F. Liaw. 1987. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology 92:220-225. [DOI] [PubMed] [Google Scholar]
- 4.Civitico, G. M., and S. A. Locarnini. 1994. The half-life of duck hepatitis B virus supercoiled DNA in congenitally infected primary hepatocyte cultures. Virology 203:81-89. [DOI] [PubMed] [Google Scholar]
- 5.Daub, H., S. Blencke, P. Habenberger, A. Kurtenbach, J. Dennenmoser, J. Wissing, A. Ullrich, and M. Cotten. 2002. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J. Virol. 76:8124-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckhardt, S. G., D. R. Milich, and A. McLachlan. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 65:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, X. D., and T. Maniatis. 1992. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science 256:535-538. [DOI] [PubMed] [Google Scholar]
- 8.Galle, P. R., H. J. Schlicht, M. Fischer, and H. Schaller. 1988. Production of infectious duck hepatitis B virus in a human hepatoma cell line. J. Virol. 62:1736-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 10.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., V. Martinez, Y. T. Loh, C. E. Rogler, and F. V. Chisari. 1994. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J. Virol. 68:5469-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatton, T., S. Zhou, and D. N. Standring. 1992. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 66:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hild, M., O. Weber, and H. Schaller. 1998. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J. Virol. 72:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, S., and D. L. Spector. 1996. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 133:719-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez-Garcia, L. F., and D. L. Spector. 1993. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73:47-59. [DOI] [PubMed] [Google Scholar]
- 17.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamond, A. I., and W. C. Earnshaw. 1998. Structure and function in the nucleus. Science 280:547-553. [DOI] [PubMed] [Google Scholar]
- 19.Liao, W., and J. H. Ou. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 69:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lott, L., B. Beames, L. Notvall, and R. E. Lanford. 2000. Interaction between hepatitis B virus core protein and reverse transcriptase. J. Virol. 74:11479-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mabit, H., K. M. Breiner, A. Knaust, B. Zachmann-Brand, and H. Schaller. 2001. Signals for bidirectional nucleocytoplasmic transport in the duck hepatitis B virus capsid protein. J. Virol. 75:1968-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabit, H., and H. Schaller. 2000. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J. Virol. 74:11472-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mintz, P. J., and D. L. Spector. 2000. Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 129:241-251. [DOI] [PubMed] [Google Scholar]
- 24.Nassal, M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickerson, J. A., G. Krockmalnic, K. M. Wan, and S. Penman. 1997. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc. Natl. Acad. Sci. USA 94:4446-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obert, S., B. Zachmann-Brand, E. Deindl, W. Tucker, R. Bartenschlager, and H. Schaller. 1996. A splice hepadnavirus RNA that is essential for virus replication. EMBO J. 15:2565-2574. [PMC free article] [PubMed] [Google Scholar]
- 27.Pannone, B. K., and S. L. Wolin. 2000. Sm-like proteins wRING the neck of mRNA. Curr. Biol. 10:R478-R481. [DOI] [PubMed] [Google Scholar]
- 28.Schlicht, H. J., R. Bartenschlager, and H. Schaller. 1989. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J. Virol. 63:2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlicht, H. J., G. Radziwill, and H. Schaller. 1989. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell 56:85-92. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt-Zachmann, M. S., B. Hugle-Dorr, and W. W. Franke. 1987. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 6:1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seguin, B., A. Staffa, and A. Cochrane. 1998. Control of human immunodeficiency virus type 1 RNA metabolism: role of splice sites and intron sequences in unspliced viral RNA subcellular distribution. J. Virol. 72:9503-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spector, D. L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9:265-315. [DOI] [PubMed] [Google Scholar]
- 34.Spector, D. L., X. D. Fu, and T. Maniatis. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10:3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 36.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 37.Wang, H. Y., X. Xu, J. H. Ding, J. R. Bermingham, Jr., and X. D. Fu. 2001. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7:331-342. [DOI] [PubMed] [Google Scholar]
- 38.Widnell, C. C., and J. R. Tata. 1964. A procedure for the isolation of enzymically active rat-liver nuclei. Biochem. J. 92:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh, C. T., Y. F. Liaw, and J. H. Ou. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 64:6141-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh, C. T., S. W. Wong, Y. K. Fung, and J. H. Ou. 1993. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc. Natl. Acad. Sci. USA 90:6459-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, M., and J. Summers. 1994. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J. Virol. 68:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]