Abstract
The VP24 protein of Ebola virus is believed to be a secondary matrix protein and minor component of virions. In contrast, the VP40 protein of Ebola virus is the primary matrix protein and the most abundant virion component. The structure and function of VP40 have been well characterized; however, virtually nothing is known regarding the structure and function of VP24. Wild-type and mutant forms of VP24 were expressed in mammalian cells to gain a better understanding of the biochemical and functional nature of this viral protein. Results from these experiments demonstrated that (i) VP24 localizes to the plasma membrane and perinuclear region in both transfected and Ebola virus-infected cells, (ii) VP24 associates strongly with lipid membranes, (iii) VP24 does not contain N-linked sugars when expressed alone in mammalian cells, (iv) VP24 can oligomerize when expressed alone in mammalian cells, (v) progressive deletions at the N terminus of VP24 resulted in a decrease in oligomer formation and a concomitant increase in the formation of high-molecular-weight aggregates, and (vi) VP24 was present in trypsin-resistant virus like particles released into the media covering VP24-transfected cells. These data indicate that VP24 possesses structural features commonly associated with viral matrix proteins and that VP24 may have a role in virus assembly and budding.
The Ebola and Marburg viruses are members of the Filoviridae family of negative-sense RNA viruses (for reviews, see references 5 and 25). Ebola virus infects both primates and humans and often leads to severe hemorrhagic fever, with mortality rates as high as 90%. There are currently no approved vaccines or antiviral drugs available to prevent and/or treat Ebola virus infections (3). The absence of Ebola virus-specific vaccines and antivirals is of particular concern recently since the filoviruses represent potential agents of bioterrorism. A better understanding of the molecular aspects of Ebola virus replication and pathogenesis will be necessary for the development of effective vaccines and/or antivirals.
Ebola virus encodes seven polypeptides from its RNA genome of ca. 19.0 kb, including the glycoprotein (GP), nucleoprotein (NP), RNA-dependent RNA polymerase (L), VP35, VP30, VP40, and VP24. The functions for many of these viral proteins have been investigated and/or elucidated, whereas the functions for other Ebola virus proteins remain unclear. For example, the surface GP and the VP40 matrix protein of Ebola virus have been well characterized and shown to function in virus attachment and entry and in assembly and budding, respectively (4, 7, 9, 12-15, 17, 20, 22-24, 26, 28, 30). In contrast, virtually nothing is known regarding the structure and function of the VP24 matrix protein of Ebola virus.
Viral matrix proteins in general are often capable of associating with lipid bilayers and self-interacting to form homo-oligomers. These properties are crucial for viral matrix proteins to carry out their function in virion assembly and budding. For example, the VP40 protein is the most abundant virion protein that plays a major role in assembly and budding of Ebola virus (12, 14-17, 28). Recently, the C-terminal region of VP40 was shown to be important for associating with lipid bilayers, while the N-terminal region appears to be important for oligomerization and subsequent assembly of progeny virions (6, 7, 20, 23). In addition, the VP40 protein is capable of budding on its own from mammalian cells in the form of filamentous virus-like particles (VLPs) (12, 14-17, 28). The efficient release of VP40 into VLPs not only is dependent on membrane binding and self-interacting domains but also is dependent on the presence of a late budding domain (L-domain). L-domains have been identified within the N terminus of VP40 and shown to function by mediating interactions with cellular proteins to promote budding (12, 16).
VP24 is considered to be a secondary matrix protein of Ebola virus and a minor component of mature virions (8). Although the function of VP24 remains unknown, as a structural protein VP24 is likely to contribute in some manner to virion assembly. Thus, VP24 would be predicted to possess structural and functional characteristics commonly present in viral matrix proteins. In this study, we sought to elucidate the physical, biochemical, and functional properties of VP24 in mammalian cells. Our findings suggest that VP24 possesses several characteristics often present in viral matrix proteins. For example, VP24 was found to associate strongly with lipid bilayers and oligomerize to preferentially form tetramers under physiological conditions. The structural features of VP24 described here are consistent with a possible role for VP24 in virion assembly. Indeed, VP24 was detected reproducibly in VLPs released from VP24-expressing mammalian cells. The presence of VP24 in VLPs is supported by the observation that VP24 in the supernatant was resistant to digestion with trypsin alone but was sensitive to trypsin in the presence of Triton X-100. Insights provided by these structural and functional analyses will allow for a more complete understanding of the replication cycle of Ebola virus and may ultimately aid in the development of novel anti-Ebola virus therapeutics.
MATERIALS AND METHODS
Cells, plasmids, and virus.
Human 293T or monkey COS-1 cells were maintained in Dulbecco minimal essential medium (Invitrogen/Life Technologies) supplemented with 10% fetal calf serum (HyClone) and 1× penicillin-streptomycin (Invitrogen/Life Technologies). Vero E6 cells (American Type Culture Collection) were cultured in Eagle minimal essential medium medium supplemented with 10% fetal calf serum. Ebola virus Zaire 1976 (EBOZ) was used in the present study. EBOZ samples and products were handled in biocontainment level 4 facilities at the U.S. Army Medical Research Institute of Infectious Disease (USAMRIID; Fort Detrick, Md.).
The VP24 gene from Ebola virus (Zaire) was kindly provided by H.-D. Klenk (Marburg, Germany). The VP24 open reading frame was amplified by standard PCR and inserted into vector pCAGGS digested with ClaI and XhoI by a standard cloning protocol. During the PCR amplification step, a hemagglutinin (HA) epitope tag (YPYDVPDYA) was joined in frame at the C terminus of VP24 to generate pVP24-WT (Fig. 1B). N-terminal deletion mutants of 15, 40, and 85 amino acids were generated by PCR and inserted into pCAGGS to generate pVP24-N15, pVP24-N40, and pVP24-N85, respectively (Fig. 1B). All deletion mutants contain the HA epitope tag at their C termini. The VP40 gene from Ebola virus (Zaire) was inserted into the pCAGGS vector (EcoRI/XhoI) to generate VP40-WT. A c-myc epitope tag was joined to the C terminus of VP40 by using standard PCR and cloning techniques. A eukaryotic expression vector encoding herpes simplex virus type 1 (HSV-1) glycoprotein D (gD) and an anti-gD monoclonal antibody (MAb) were kindly provided by R. Eisenberg (University of Pennsylvania).
FIG. 1.
Ebola virus VP24 amino acid sequence and expression plasmids. (A) Single-letter amino acid sequence of the VP24 protein of Ebola virus (Zaire). Three potential sites for N-linked glycosylation exist at positions 84, 185, and 246, and a region rich in hydrophobic residues is underlined (amino acids 90 to 130). (B) Diagram of eukaryotic expression plasmids encoding VP24-WT, VP24-N15 (N-terminal 15 amino acids deleted), VP24-N40 (N-terminal 40 amino acids deleted), and VP24-N85 (N-terminal 85 amino acids deleted). All VP24 proteins have an epitope tag from the influenza A virus HA protein joined to their C termini. (C) Immunoprecipitation with anti-HA MAb of protein extracts from 293T cells transfected with pCAGGS vector alone (lane 1), VP24-WT (lane 2; 28 kDa), VP24-N15 (lane 3; 26 kDa), VP24-N40 (lane 4; 23 kDa), and VP24-N85 (lane 5; 17 kDa). An asterisk is shown to the right of the various VP24 proteins.
Immunoprecipitation.
Human 293T cells were transfected with pVP24-WT or VP24 deletion mutants by using Lipofectamine and the protocol of the supplier (Invitrogen/Life Technologies). At approximately 36 h posttransfection, cells were radiolabeled with 150 μCi of [35S]Met-Cys (NEN/Perkin-Elmer)/ml for 4 to 6 h. Total cell extracts were prepared in 1× radioimmunoprecipitation assay (RIPA) buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1.0 mM phenylmethylsulfonyl fluoride, and 10 μg each of aprotinin and leupeptin (Roche Biochemicals)/ml. Extracts were clarified for 2 min at 13,000 rpm and then incubated with an anti-HA MAb (Roche Biochemicals) overnight at 4°C. Immune complexes were collected by using protein A-agarose beads (Roche Biochemicals), washed three times with RIPA buffer, and suspended in 1× Laemmli sample buffer. Proteins were analyzed by reducing SDS-10% polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography.
Indirect immunofluorescence.
293T or COS-1 cells in 60-mm dishes (50 to 70% confluent) were transfected with the appropriate plasmid as described above. At 20 to 24 h posttransfection, the cells were fixed with methanol-acetone (1:1) on ice for 20 min. Fixed cells were incubated with anti-HA MAb (Santa Cruz Biotechnology) at 37°C for 30 min, then washed three times in 1× phosphate-buffered saline (PBS), and then washed with fluorescein isothiocyanate-conjugated secondary antibody (Roche Biochemicals) at 37°C for 30 min. Positive cells were visualized by using a Nikon Eclipse E600 fluorescence microscope.
EBOZ-infected cells in eight-well chamber slides were fixed with 10.0% formalin for 24 h. Fixed cells were washed extensively with 1× PBS, treated with proteinase K for 10 min at room temperature, and washed again with 1× PBS. Cells were incubated with a mouse MAb directed against EBOZ VP24 protein (diluted 1:2,000; kindly provided by M. Hart, USAMRIID). Washed cells were then incubated with an anti-mouse immunoglobulin G (IgG; diluted 1:400) secondary antibody (Alexa Fluor 594; Molecular Probes). All antibodies were diluted with antibody diluent containing background reducing components (Dako). Coverslips were affixed to glass slides by using mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole; Vector). Ebola virus infections were conducted within the Biological Safety Level 4 (BSL-4) Laboratory at USAMRIID. Personnel wore positive-pressure protective suits (ILC; Dover, Frederica, Del.) equipped with high-efficiency particulate air (HEPA) filters and supplied with umbilical-fed air.
Cleavage of N-linked oligosaccharides with PNGase F.
Human 293T cells were transfected with pVP24-WT and VP24-WT protein was immunoprecipitated as described above. Immunoprecipitated proteins were separated from protein A-agarose beads by boiling and then incubated in the absence or presence of PNGase F (New England Biolabs) in 0.05 M NaPO4 (pH 7.5) and 1.0% NP-40 at 37°C for 16 h. Samples were suspended in 1× Laemmli sample buffer and then analyzed by SDS-PAGE and autoradiography.
Phase separation with Triton X-114.
Aqueous- versus detergent-phase separation by using Triton X-114 (Sigma) was performed as described previously (2). Briefly, 293T cells were transfected with the appropriate plasmid and radiolabeled as described above. Cell extracts were lysed (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.0% NP-40) on ice for 30 min. Extracts were centrifuged at 13,000 rpm for 1 min to remove cell debris. Clarified extracts were then mixed with an equal volume of 5.0% Triton X-114 in 1× PBS plus protease inhibitors. Phase separation was induced by incubating the samples on ice for 5 min, followed by a 5-min incubation at 37°C. Phase separation was completed by centrifugation of the samples at 13,000 rpm for 5 min. The aqueous and detergent phases were carefully placed into new tubes, and proteins were immunoprecipitated as described above.
Gel filtration and Western blotting.
Human 293T cells were transfected with the appropriate plasmid as described above. At 36 h posttransfection, cells were scraped from the dish and lysed in 1× PBS containing protease inhibitors by three cycles of freeze-thawing at −80°C (5 min) and 37°C (5 min). Cell extracts were clarified twice at 13,000 rpm for 2 min and then filtered to remove cell debris. Total protein was separated by size on a Superdex-200 10/30 high-resolution fast-performance liquid chromatography column (Amersham Biosciences) by using an AKTA 10 purifier system (Amersham Biosciences). The resultant chromatograms depicting absorbance at 280 nm versus the elution volume were obtained by using Unicorn software. Eluted proteins were collected in 0.5-ml fractions, quantitated, and analyzed by reducing SDS-PAGE and Western blotting. VP24-WT and mutants were detected with anti-HA MAb (Roche Biochemicals) and enhanced chemiluminescence (Amersham Biosciences) according to the recommendations of the manufacturer. The molecular mass standards (Bio-Rad) for the gel filtration experiments were of 670, 158, 44, and 17 kDa.
Budding assay for VLPs.
Human 293T cells in 100-mm dishes were transfected with equivalent amounts of the indicated plasmids by using the Lipofectamine reagent (Invitrogen) and the protocol of the supplier. Proteins were radiolabeled with 150 μCi of [35S]Met-Cys (Perkin-Elmer) from 36 to 48 h posttransfection. Culture media was centrifuged at 2,500 rpm for 10 min to remove cellular debris, layered onto a 20% sucrose cushion in STE buffer (0.01 M Tris-HCl [pH 7.5], 0.01 M NaCl, 0.001 M EDTA [pH 8.0]), and centrifuged at 36,000 rpm for 4 h at 4°C. The resulting pellet containing VLPs was suspended in 400 μl of STE buffer overnight at 4°C. After the addition of RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% SDS), the VLPs were immunoprecipitated with the appropriate antiserum (anti-HA MAb for VP24 and anti-c-myc MAb for VP40) and analyzed by SDS-PAGE as described above. Trypsin treatment of VLPs was performed essentially as described elsewhere (14). Total cell extracts from transfected cells were harvested in RIPA buffer, immunoprecipitated with the appropriate antiserum, and analyzed by SDS-PAGE as described previously.
RESULTS
Expression of VP24 in mammalian cells.
Although VP24 is known to be a minor matrix protein within Ebola virions, virtually nothing is known about its physical and biochemical structure and biological function. The amino acid sequence of VP24 reveals an abundance of hydrophobic residues throughout the protein (Fig. 1A). A region spanning amino acids 90 to 130 is particularly rich in hydrophobic residues. In addition to the hydrophobic nature of VP24, the amino acid sequence also reveals three potential sites for N-linked glycosylation (amino acids 84, 185, and 246) and a single cysteine residue at position 27 from the N terminus (Fig. 1A).
To begin to address the physical and structural features of VP24 that may regulate its function, wild-type (WT) and N-terminal deletion mutants of VP24 that contained C-terminal HA epitope tags were expressed in mammalian cells (Fig. 1B). All VP24 WT and mutant proteins were detected readily by immunoprecipitation, and the various truncated forms of VP24 migrated at their predicted molecular weights (Fig. 1C).
Localization of VP24 by immunofluorescence.
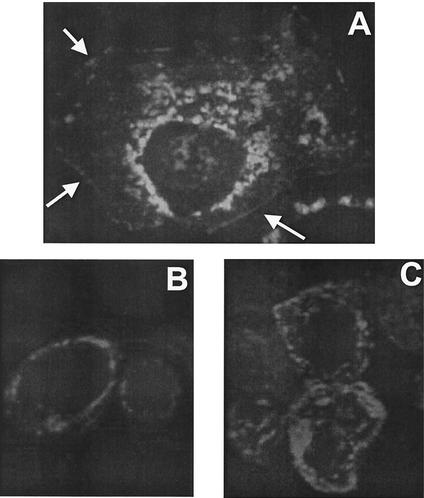
If VP24 functions as a matrix protein in virion assembly and budding, then it is likely to localize in part to the plasma membrane, since Ebola virus replicates in the cytoplasm and buds from the plasma membrane. Indirect immunofluorescence was employed to localize VP24 in transfected 293T or COS cells. As shown in Fig. 2, the pattern of fluorescence indicated that VP24 was present in the cytoplasm but was not detected in the nucleus (Fig. 2A). In repeated experiments, areas of localized fluorescence were observed at the cell periphery, likely representing plasma membrane staining (Fig. 2A, arrows). In addition to localization at the plasma membrane, areas of more intense vesicular fluorescence were observed in the perinuclear region (Fig. 2A). Mock-transfected cells used as a negative control exhibited no fluorescence (data not shown).
FIG. 2.
Intracellular localization of VP24-WT by by indirect immunofluorescence. COS-1 cells were transfected with VP24-WT for 24 h and then fixed for fluorescent staining. (A) An anti-HA MAb served as the primary antibody and fluorescein isothiocyanate-conjugated anti-rat IgG served as the secondary antibody. Arrows indicate localized areas of potential plasma-membrane staining. (B and C) Vero E6 cells infected with Ebola virus (Zaire) were incubated with an anti-VP24 MAb followed by anti-mouse IgG (Alexa Fluor 594).
To determine whether the pattern of localization of VP24 we observed is consistent with its localization in Ebola virus-infected cells, indirect immunofluorescence was performed on EBOZ-infected Vero E6 cells. Similar to transfected cells, VP24 localized predominantly to the perinuclear region in EBOZ-infected cells (Fig. 2B and C). Although present abundantly at the perinuclear region, VP24 was also observed in part at the plasma membrane (Fig. 2C; J. Paragas, unpublished data). Thus, the distribution of VP24 in transfected and infected cells is similar.
VP24 partitions with Triton X-114.
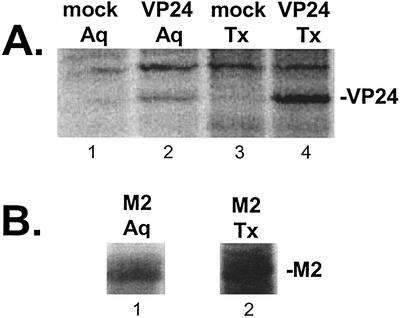
Since VP24 localized, in part, to the plasma membrane, it was of interest to characterize further the nature of the VP24-lipid bilayer interaction. Extracts from human 293T cells expressing either VP24-WT or influenza A virus M2 were lysed and subjected to phase partitioning with Triton X-114 (Fig. 3) to distinguish between peripheral and integral membrane proteins (2). Peripheral membrane proteins will partition predominantly in the aqueous phase, whereas integral membrane proteins will partition predominantly in the detergent phase. Mock-transfected cells served as a negative control. In repeated experiments, the majority of VP24 protein (∼75 to 80%) was detected in the Triton X-114 detergent phase (Fig. 3A, lane 4), whereas a smaller amount (∼20 to 25%) partitioned in the aqueous phase (lane 2). As expected, VP24 was not observed in either the aqueous or detergent phases of mock-transfected cells (Fig. 3A, lanes 1 and 3). The M2 protein of influenza A virus has been shown previously to partition predominantly to the Triton X-114 phase, and M2 served as a control in this experiment (31) (Fig. 3B). These results suggest that VP24 is strongly associated with lipid membranes and may be embedded in the lipid bilayer.
FIG. 3.
Phase partitioning with Triton X-114 detergent. 293T cells were transfected with VP24-WT, transfected with influenza A virus M2, or mock transfected, and cell extracts were subjected to phase partitioning with Triton X-114. (A) Aqueous (Aq) and Triton X-114 (Tx) phases from mock-transfected cells (lanes 1 and 3, respectively) and from VP24-WT transfected cells (lanes 2 and 4, respectively) were separated, immunoprecipitated with anti-HA MAb, and analyzed by SDS-PAGE. The position of VP24-WT in lanes 2 and 4 is indicated. (B) Aqueous (Aq; lane 1) and Triton X-114 (Tx; lane 2) phases from M2-transfected cells were included as a positive control.
VP24 is not modified by N-linked oligosaccharides.
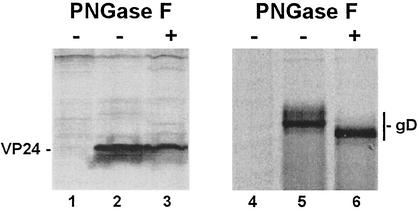
Posttranslational modifications of viral proteins have significant effects on both their structure and function. Because VP24 has three potential sites for N-linked sugars at amino acid positions 84, 185, and 246 (Fig. 1A), we determined whether VP24 contained N-linked oligosaccharides. To assess the glycosylation state of VP24 in mammalian cells, VP24-WT was immunoprecipitated from transfected cell extracts and incubated in the absence or presence of PNGase F (Fig. 4), which removes all N-linked sugars from susceptible proteins. HSV-1 gD (27) was used as a positive control in these experiments (Fig. 4). As demonstrated previously, gD was deglycosylated by PNGase F and migrated faster than the glycosylated (untreated) forms of gD (Fig. 4, compare lanes 5 and 6). In contrast, no change was observed in the migration pattern for VP24 in the absence or presence of PNGase F (Fig. 4, compare lanes 2 and 3). Extracts from mock-transfected cells served as negative controls (Fig. 4, lanes 1 and 4). These results indicate that none of the three potential N-linked glycosylation sites are modified by the addition of oligosaccharides in VP24-transfected 293T cells. It should be noted that the presence of O-linked oligosaccharides on VP24 was not investigated and thus cannot be ruled out.
FIG. 4.
PNGase F treatment of VP24 and gD. 293T cells were mock transfected, transfected with VP24-WT, or transfected with gD of HSV-1. Radiolabeled cell extracts were incubated in the absence (−) or presence (+) of PNGase F, immunoprecipitated with anti-HA MAb (lanes 1 to 3) or anti-gD MAb (lanes 4 to 6), and analyzed by SDS-PAGE. The positions of VP24 (lanes 2 and 3) and gD (lanes 5 and 6) are indicated.
Oligomerization state of VP24.
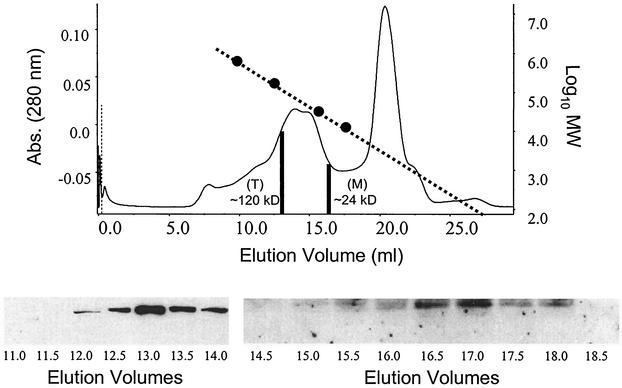
Since the function of numerous viral matrix proteins in assembly and budding is dependent, in part, upon their ability to oligomerize, we determined whether VP24 could self-assemble to form homo-oligomers in mammalian cells. Human 293T cells were either mock transfected or transfected with VP24-WT for 36 h and analyzed by gel filtration and Western blot analyses. As shown, a chromatogram of eluted proteins from VP24-WT-expressing cells contains two distinct peaks (Fig. 5, two vertical black lines). The first peak of VP24 protein was detected in the 13.0-ml elution volume, and a Western blot of elution volumes of 11.0 to 14.0 ml is shown (Fig. 5). The 13.0-ml elution volume corresponds to proteins with molecular masses of ca. 115 to 120 kDa as determined by the elution profile and a plot of the molecular mass standards (Fig. 5, dotted line). Elution of VP24 within these fractions is consistent with the possible formation of tetramers (∼112 kDa) in mammalian cells.
FIG. 5.
Gel filtration and Western blot analyses of VP24-WT. 293T cells were either mock transfected or transfected with VP24-WT, and cell extracts were harvested at approximately 36 h posttransfection. Clarified and filtered protein samples were separated by size on a Superdex-200 10/30 high-resolution fast-performance liquid chromatography column. The elution chromatogram is shown for VP24-WT as a plot of absorbance at 280 nm versus elution volume (in milliliters). Molecular mass standards (670, 158, 44, and 17 kDa) were also separated on the same column, and a plot of their log values versus the elution volume is shown as a dotted line. The two black vertical lines represent the elution volumes where peak activities of VP24 were detected by Western blot analysis. The Western blots representing samples from elution volumes of 11.0 to 14.0 ml and 14.5 to 18.5 ml are shown. A peak of VP24 eluted within the 13.0-ml volume. Elution of VP24 within the 13.0-ml volume corresponds to a protein of ca. 120 kDa (T; tetramer). The second peak of VP24 occurred in an elution volume of 16.5 ml and corresponds to a protein of ca. 28 kDa (M; monomer).
A second peak of VP24 protein was detected in the 16.5-ml elution volume by Western blotting (Fig. 5) Elution of VP24 from the column at this elution volume is consistent with monomers of VP24 (∼28 kDa). As expected, VP24 was not detected by Western blotting in any of the fractions collected from mock-transfected cells (data not shown). These results provide strong evidence that VP24 can oligomerize in mammalian cells and that VP24 may preferentially form tetramers. It should be noted that although the peak of VP24 elution occurred at a position indicative of tetramers, the presence of additional oligomeric forms of VP24 (e.g., dimers, trimers, and pentamers) cannot be ruled out. The ability of VP24 to oligomerize or self-assemble is again consistent with a potential role in virion assembly.
The N-terminal region of VP24 is important for oligomer formation.
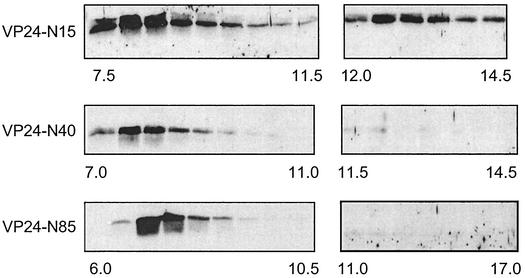
To begin to identify regions of VP24 that are important for oligomerization, we constructed several VP24 mutants with progressively larger N-terminal deletions (Fig. 1B). These N-terminal deletion mutants were analyzed by gel filtration and Western blotting to determine whether their oligomerization properties were similar to those of VP24-WT. Interestingly, the VP24 mutant having the smallest N-terminal deletion (VP24-N15) eluted in a peak corresponding in size to tetramers (Fig. 6, elution volumes 12.5 to 13.5 ml); however, a significant amount of this mutant protein also eluted in a peak corresponding to elution volumes (void volume) of 7.5 to 8.5 ml (Fig. 6). Proteins eluting from the column between 7.5 and 8.5 ml correspond in size to high-molecular-mass multimers or aggregates greater than 670 kDa. In repeated experiments, VP24-WT was not detected by Western blot analysis in elution volumes (7.0 to 9.0 ml) that would correspond to high-molecular-mass aggregates (data not shown). VP24-N40 and VP24-N85 were barely detectable by Western blot analysis in elution volumes that would correspond in size to tetramers (Fig. 6, VP24-N40 [11.5 to 14.5 ml] and VP24N-85 [11.0 to 17.0 ml]). Rather, the majority of these two mutants peaked in elution volumes corresponding in size to high-molecular-mass multimers (Fig. 6; VP24-N40 [7.5 to 8.5 ml] and VP24-N85 [7.0 to 8.0 ml]). It should be noted that monomers of all three N-terminal deletion mutants were detected by Western blot analyis in the appropriate fractions, albeit at levels lower than that observed for VP24-WT (data not shown). Thus, the tendency of VP24 to aggregate appears to increase with the size of the N-terminal deletion.
FIG. 6.
Gel filtration and Western blot analyses of N-terminal deletion mutants of VP24. VP24-N15, VP24-N40, and VP24-N85 were subjected to gel filtration and Western blotting as described in the legend to Fig. 5. VP24 mutants were detected by using anti-HA MAb, and Western blots of elution volumes ranging from 7.5 to 14.5 ml (VP24-N15), 7.0 to 14.5 ml (VP24-N40), and 6.0 to 17.0 ml (VP24-N85) are shown. All lanes represent successive 0.5-ml volumes except for volumes of 11.0 to 17.0 ml for VP24-N85, which represents successive 1.0-ml volumes. The molecular mass markers used in Fig. 5 were also employed in this experiment.
VP24 is present in budding VLPs.
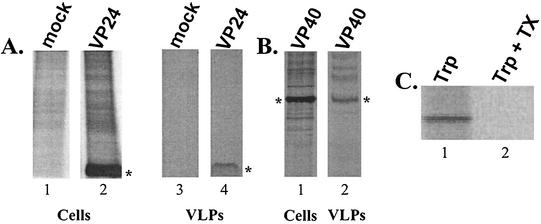
Using a functional budding assay, we were the first to demonstrate that the VP40 protein can bud from cells in the form of VLPs (12, 14, 17, 28). If VP24 plays a role in assembly or budding, we reasoned that perhaps VP24 could bud from mammalian cells in the form of VLPs as well. To address this possibility, cell extracts and VLPs present in the culture media of transfected 293T cells were harvested, immunoprecipitated, and analyzed by SDS-PAGE (Fig. 7). As expected, VP24-WT was readily detected in cell extracts (Fig. 7A, lane 2) and, interestingly, VP24 was also detected, albeit at low levels, in VLPs within the media (Fig. 7A, lane 4). VP24 was not observed in cell and VLP samples from mock-transfected cells (Fig. 7A, lanes 1 and 3). VP40 was used as a positive control for this budding assay and was present in both cell extracts (Fig. 7B, lane 5) and VLPs within the media (Fig. 7B, lane 6). To demonstrate that VP24 was released into the media in a lipid-bound form and not simply secreted as free protein, trypsin digestion of supernatant samples was performed (Fig. 7C). VP24 in supernatant samples was resistant to digestion with trypsin alone (Fig. 7C, lane 1); however, VP24 was sensitive to trypsin only in the presence of Triton X-100 (lane 2). These results strongly suggest that VP24 is released from mammalian cells in lipid-bound particles and thus VP24 may contribute to the processes of assembly and budding of Ebola virus.
FIG. 7.
Budding of VP24 in the form of VLPs. Human 293T cells were transfected with either VP24-WT or VP40-WT expression vectors. (A) Radiolabeled cell extracts from mock-transfected (lane 1) or VP24-transfected (lane 2) cells were immunoprecipitated with anti-HA MAb. Medium samples from mock-transfected (lane 3) or VP24-transfected (lane 4) cells were treated as described in Materials and Methods, and VLPs were immunoprecipitated by using anti-HA antiserum. The asterisks indicate the position of VP24 (lanes 2 and 4). (B) As a positive control for budding and VLP formation, 293T cells were transfected with a VP40 expression plasmid. Cells and media were treated as described above, and radiolabeled proteins were immunoprecipitated with anti-cmyc MAb to detect VP40. Asterisks represent the position of VP40 in the cell extract (lane 5) and in VLPs (lane 6). (C) Supernatant samples from VP24-transfected cells were incubated in the presence of trypsin alone (Trp; lane 1), or trypsin plus Triton X-100 (Trp + Tx; lane 2) before immunoprecipitation.
DISCUSSION
Of all the Ebola virus proteins, VP24 is the least well-characterized in terms of structure and function, although this viral matrix protein of Ebola virus likely has a significant role in viral pathogenesis. In this report, we have elucidated several biochemical, structural, and functional properties of VP24 that provide important insights into the role of VP24 during virus replication. For example, the ability of VP24 to oligomerize under physiological conditions is an important finding. Results from gel filtration and Western blot analysis suggest that VP24 may preferentially form tetramers; however, the formation of additional oligomeric forms of VP24 cannot be ruled out. The ability to self-assemble into a higher-ordered structure is a common feature of viral matrix proteins that function to assemble or build the virus particle. Indeed, the VP40 matrix protein of Ebola virus preferentially forms hexameric building blocks that are believed to be instrumental in promoting virion assembly (7, 20, 23). In addition, VP40, like VP24 (in the present study), aggregates into high-molecular-mass multimers, which presumably require intermolecular interactions. Progressive deletions at the N terminus of VP24 resulted in a progressive increase in aggregate formation, suggesting that the N-terminal amino acids of VP24 are important for efficient tetramer formation. Interestingly, the N-terminal amino acids of VP40 are also important for oligomer formation (6, 7, 23). Alternatively, deletion of these N-terminal amino acids may expose more hydrophobic regions of VP24 that are responsible for increased levels of aggregation. Indeed, amino acids 90 to 130 of VP24 comprise the most hydrophobic regions of VP24 (Fig. 1A), and this region is most highly exposed in the VP24-N85 mutant. Lastly, both divalent cations and pH changes appear to effect oligomerization and stability of VP24 (Z. Han et al., data not shown). The role of these two parameters in protein stability and oligomerization is well documented in the literature (10, 11, 18, 21).
The ability of VP24 to self-assemble and interact strongly with lipid bilayers (Fig. 2 and 3) is consistent with a potential role in virus assembly. Recent findings indicate that Ebola virus buds from lipid raft domains on the plasma membrane (1). Interestingly, preliminary studies involving the extraction of VP24-expressing cells with cold, nonionic detergent suggest that VP24 localizes in part to detergent-resistant lipid membranes, presumably lipid rafts (M. Simpson-Holley and R. N. Harty, unpublished data). Furthermore, the presence of VP24 in budding VLPs (Fig. 7A) strongly supports the possibility of VP24 being important for virus assembly and budding. Together, these results indicate that VP24 is released from mammalian cells in the form of lipid-containing VLPs. Ongoing mutagenesis studies on VP24 will allow us to identify more precisely the regions and motifs that are important for the efficient release of VP24-containing VLPs. It will be of interest to determine whether VP24 buds from cells through a pathway similar to or distinct from that used by VP40. For example, efficient budding of VP40 is dependent on a membrane-binding domain, an oligomerization domain, and a late budding domain (L-domain) (12, 14, 16, 17). In addition, interactions between the VP40 L-domain(s) and cellular proteins such as tsg101 and ubiquitin ligases are believed to play a key role in budding (12, 16). Although we have demonstrated that VP24 is able to interact with lipid membranes and oligomerize, a functional L-domain within VP24 has yet to be identified. However, a motif with the consensus sequence of a functional L-domain (YxxL [19]) is present at amino acids 172 to 175 of VP24.
Molecular aspects of Ebola virus replication and pathogenesis are of increased interest due to the potential use of filoviruses as agents of bioterrorism. A better understanding of viral protein structure and function will ultimately lead to the development of novel treatment and prevention strategies. Findings presented in the present study describe structural, biochemical, and functional properties of the VP24 protein of Ebola virus. All of the data presented here are consistent with a possible role for VP24 in assembly and budding of Ebola virus, and thus VP24 may serve as a target for the development of novel antivirals. It should be noted that VP24 may possess functions other than those involved directly in assembly and budding. For example, VP24 has been implicated recently in playing a role,, along with VP35 and NP, in nucleocapsid assembly (12a). In addition, preliminary data from our laboratory suggest that VP24 may possess ion channel activity in mammalian cells (Han et al., unpublished). Ongoing studies, including the use of reverse genetics (29), will be instrumental in elucidating the importance of VP24 in Ebola virus pathogenesis.
Acknowledgments
We thank H.-D. Klenk, A. Garcia-Sastre, and R. Eisenberg for generously providing reagents. We also thank members of the Harty lab and B. D. Freedman for general comments and discussions.
R.N.H. is supported in part by NIH grant AI46499 and a research grant from the University of Pennsylvania Research Foundation. J.P. is the recipient of a National Research Council fellowship.
REFERENCES
- 1.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 3.Bray, M., and J. Paragas. 2002. Experimental therapy of filovirus infections. Antiviral Res. 54:1-17. [DOI] [PubMed] [Google Scholar]
- 4.Chan, S. Y., R. F. Speck, M. C. Ma, and M. A. Goldsmith. 2000. Distinct mechanisms of entry by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J. Virol. 74:4933-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colebunders, R., and M. Borchert. 2000. Ebola haemorrhagic fever: a review. J. Infect. 40:16-20. [DOI] [PubMed] [Google Scholar]
- 6.Dessen, A., E. Forest, V. Volchkov, O. Dolnik, H. D. Klenk, and W. Weissenhorn. 2000. Crystallization and preliminary X-ray analysis of the matrix protein from Ebola virus. Acta Crystallogr. D Biol. Crystallogr. 56:758-760. [DOI] [PubMed] [Google Scholar]
- 7.Dessen, A., V. Volchkov, O. Dolnik, H. D. Klenk, and W. Weissenhorn. 2000. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 19:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, L. H., M. P. Kiley, and J. B. McCormick. 1985. Descriptive analysis of Ebola virus proteins. Virology 147:169-176. [DOI] [PubMed] [Google Scholar]
- 9.Feldmann, H., V. E. Volchkov, V. A. Volchkova, and H. D. Klenk. 1999. The glycoproteins of Marburg and Ebola virus and their potential roles in pathogenesis. Arch. Virol. Suppl. 15:159-169. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda, M., and K. Mikoshiba. 2000. Distinct self-oligomerization activities of synaptotagmin family: unique calcium-dependent oligomerization properties of synaptotagmin VII. J. Biol. Chem. 275:28180-28185. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda, M., and K. Mikoshiba. 2001. Mechanism of the calcium-dependent multimerization of synaptotagmin VII mediated by its first and second C2 domains. J. Biol. Chem. 276:27670-27676. [DOI] [PubMed] [Google Scholar]
- 12.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 13.Ito, H., S. Watanabe, A. Takada, and Y. Kawaoka. 2001. Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 75:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolesnikova, L., H. Bugany, H. D. Klenk, and S. Becker. 2002. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 76:1825-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 17.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto, L. H., L. J. Holsinger, and R. A. Lamb. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517-528. [DOI] [PubMed] [Google Scholar]
- 19.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruigrok, R. W., G. Schoehn, A. Dessen, E. Forest, V. Volchkov, O. Dolnik, H. D. Klenk, and W. Weissenhorn. 2000. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300:103-112. [DOI] [PubMed] [Google Scholar]
- 21.Salom, D., B. R. Hill, J. D. Lear, and W. F. DeGrado. 2000. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry 39:14160-14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez, A., Z. Y. Yang, L. Xu, G. J. Nabel, T. Crews, and C. J. Peters. 1998. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 72:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scianimanico, S., G. Schoehn, J. Timmins, R. H. Ruigrok, H. D. Klenk, and W. Weissenhorn. 2000. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takada, A., and Y. Kawaoka. 2001. The pathogenesis of Ebola hemorrhagic fever. Trends Microbiol. 9:506-511. [DOI] [PubMed] [Google Scholar]
- 26.Takada, A., S. Watanabe, H. Ito, K. Okazaki, H. Kida, and Y. Kawaoka. 2000. Downregulation of β1 integrins by Ebola virus glycoprotein: implication for virus entry. Virology 278:20-26. [DOI] [PubMed] [Google Scholar]
- 27.Tal-Singer, R., R. J. Eisenberg, T. Valyi-Nagy, N. W. Fraser, and G. H. Cohen. 1994. N-linked oligosaccharides on herpes simplex virus glycoprotein gD are not essential for establishment of viral latency or reactivation in the mouse eye model. Virology 202:1050-1053. [DOI] [PubMed] [Google Scholar]
- 28.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of Ebola virus matrix protein VP40. Virology 283:1-6. [DOI] [PubMed] [Google Scholar]
- 29.Volchkov, V. E., V. A. Volchkova, E. Muhlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H. D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 30.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J., and R. A. Lamb. 1996. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 225:255-266. [DOI] [PubMed] [Google Scholar]