Abstract
The β subunits of voltage-gated Ca2+ channels are known to be regulators of the channels' gating properties. Here we report a striking additional function of a β subunit. Screening of chicken cochlear and brain cDNA libraries identified β4c, a short splice variant of the β4 subunit. Although β4c occurs together with the longer isoforms β4a or β4b in the brain, eye, heart, and lung, the cochlea expresses exclusively β4c. The association of β4c with the Ca2+-channel α1 subunit has slight but significant effects on the kinetics of channel activation and inactivation. Yeast two-hybrid and biochemical assays revealed that β4c interacts directly with the chromo shadow domain of chromobox protein 2/heterochromatin protein 1γ (CHCB2/HP1γ), a nuclear protein involved in gene silencing and transcriptional regulation. Coexpression of this protein specifically recruits β4c to the nuclei of mammalian cells. Furthermore, β4c but not β4a dramatically attenuates the gene-silencing activity of chromobox protein 2/heterochromatin protein 1γ. The β4c subunit is therefore a multifunctional protein that not only constitutes a portion of the Ca2+ channel but also regulates gene transcription.
The proper regulation of gene transcription is essential for cellular differentiation and proliferation as well as for processes such as long-term memory. Precise mechanisms therefore must assure that each cell type expresses only the appropriate subset of genes. Gene silencing is one mechanism for achieving an appropriate gene-expression pattern (1). Heterochromatin proteins 1 (HP1s) were first identified as responsible for position-effect variegation, a type of gene silencing (2). HP1s specifically recognize and directly bind a lysine residue near the carboxyl terminus of histone H3 that is methylated by a unique methyltransferase, SUV39H1 (3–5). These proteins are constituents of a heterochromatin complex in which transcription is inactive. Because of the impact of gene silencing, it is important to understand the factors that can initiate or maintain this process.
Ca2+ signals play an indispensable role in the activity of most cells. Ca2+ influx in response to the depolarization of presynaptic terminals triggers vesicle fusion and mediates neurotransmission (6). Elevation of the intracellular Ca2+ concentration stimulates several signaling cascades (7). Ca2+ signals additionally propagate to the nucleus, where they affect gene expression and influence biological events including neuronal survival, axon outgrowth, and plasticity (7). Precise spatial and temporal control of the intracellular Ca2+ concentration therefore is mandatory. Voltage-gated Ca2+ channels localized in the plasma membrane contribute importantly to Ca2+ homeostasis. Each of these channels comprises a pore-forming α1 subunit as well as accessory β, α2-δ, and sometimes γ subunits (8). The diversity of the channel subunits and variety of their combinations confer the unique properties of the channels in each cell type. Although distinct channel subunits have been implicated in particular physiological responses, their exact functions have not been elucidated fully. The results of the present study suggest that one isoform of β subunit is not only a constituent of Ca2+ channels but also participates with an HP1 in the regulation of gene silencing.
Materials and Methods
Cloning and Tissue Distribution of β4 Splice Variants.
Cloning of cDNAs encoding β4 subunits was performed as described in Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org (9).
RT-PCR analyses were carried out as reported (9). The equivalent of 1 μg of total RNA from various organs of the chicken was used for each PCR (Fig. 1B). In nested PCRs, 3 μl of the first PCR product (30 μl), which was obtained from 150 ng of total RNA with primers 12F and 5R, was amplified again by using primers 20F and 19R (Fig. 4). Fragments of 195 and 254 bp were amplified from β4c and β4a/b, respectively. The sequences of primers specific to chicken β4 subunits were: 12F, 5′-GTGGGAATTCTTCTTCAAG-3′; 20F, 5′-GTCTTGGAGAAATGGTTTCTGG-3′; 5R, 5′-GTGATTGATAGTGTCTGCATCCAGG-3′; and 19R, 5′-CTTAGCAAGAGAAATGTCG-3′.
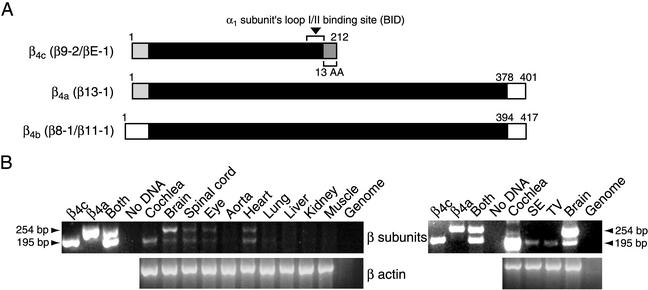
Figure 1.
Cloning and distribution of chicken Ca2+-channel β4 subunits. The smaller amplified fragment (195 bp) corresponds to β4c and the larger (254 bp) to β4a/b. (A) Structure of β4-subunit proteins. The β-interaction domain (BID) for binding of α1 subunits is indicated. The 13-aa sequence unique to β4c occurs in its carboxyl terminus because of the absence of exon d. β11-1 is identical to β8-1 except in the 3′-untranslated region, where the former contains two additional exons. β13-1 displays a unique 5′ sequence derived from an alternative exon encoding an amino-terminal sequence that is nearly identical to that of the β3 subunit. (B Left) Tissue distribution of β4 subunits analyzed by RT-PCR in the chicken. (Right) Results of nested RT-PCRs with two pairs of the primers described in the Fig. 4. β-actin amplification was used for positive control experiments. Muscle, skeletal muscle; SE, sensory epithelium; TV, tegmentum vasculosum.
Electrophysiology.
Xenopus laevis oocytes of stages V and VI were isolated by treatment with collagenase as described (10). After having been injected with 20 nl of a mixture of rat brain α1A and α2-δ in the absence or presence of either chicken β4a or β4c cDNA (1 μg/μl at a 1:2:3 ratio), oocytes were incubated for 3–4 days. Ba2+ currents then were measured as described in the legend to Fig. 5, which is published as supporting information on the PNAS web site.
Yeast Two-Hybrid Assays.
Yeast two-hybrid screening was performed as described (9). A cDNA fragment corresponding to amino acid residues 16–212 of β4c was produced by the PCR, inserted into the pBDGAL4 vector (Stratagene), and used as a bait for screening. For the mapping of β4c and chromobox protein (CHCB)2-binding sites, additional baits of β4c in the pBDGAL4 vector and additional preys of CHCB2 in the pADGAL4 vector were generated by PCRs.
Antibodies.
A rabbit polyclonal antiserum was raised against a (His)6-tagged fusion protein containing amino acid residues 16–212 of chicken β4c and purified against an equivalent GST fusion protein. Mouse monoclonal anti-HP1γ antibody was purchased from Chemicon. Anti-myc and anti-GFP antibodies were obtained from CLONTECH.
Subcellular Fractionation.
Purification of nuclear proteins from chicken brain was conducted as described (11, 12).
Biochemical Binding Assays.
GST pull-down and coimmunoprecipitation experiments were performed as described (9).
Cellular Labeling.
Full-length β4c and CHCB2 sequences were subcloned into pEGFP-C2 and pCMV-myc vectors (CLONTECH), respectively, and then transfected in tsA201 cells. Proteins were visualized directly by GFP fluorescence or by incubation with anti-myc antibody followed by Texas red-labeled secondary antibodies. Immunohistochemistry was done with 10-day-old chickens as described (9). Cochleae were isolated after fixation by perfusion with 4% formaldehyde in PBS solution. Twelve-micrometer-thick cryosections were incubated with antibodies against β subunits and HP1γ and visualized with fluorescein-conjugated anti-rabbit and Texas red-labeled anti-mouse antisera, respectively. Images were obtained with a laser confocal microscope (MRC-1024, Bio-Rad).
Chloramphenicol Acetyltransferase (CAT) Assays.
Cos-1 cells were grown in 48-well plates and transiently transfected with various plasmids. To normalize transfection efficiency, the cells of each well were cotransfected with 0.1 μg of pCMV-β-galactosidase plasmid. CAT and β-galactosidase activities were measured with CAT-ELISA and β-galactosidase-ELISA kits (Roche). All CAT activities were normalized by dividing the measured value by the corresponding β-galactosidase activity (13). The normalized CAT activity of each combination was divided further by the mean value of control experiments (pM1-mock+G5SV40CAT), resulting in a relative CAT activity.
Results
The dominant voltage-gated Ca2+ channels of cochlear hair cells exhibit unique molecular and functional properties (14–16). The channels are composed of α1D subunits containing three unusual segments and yield an L-type Ca2+ current that has a relatively negative threshold of activation, rapid activation and deactivation kinetics, and little or no Ca2+-dependent inactivation. To identify the regulatory β subunits associated with these channels, we performed RT-PCR on the total RNA from chicken cochlear sensory epithelia with degenerate primers that hybridize with homologous sequences of the four distinct β subunits reported (17, 18). The amplified DNA fragment, which belonged to the β4 subunit family, was used to probe chicken cochlear and brain cDNA libraries. We isolated one clone from the cochlea (βE-1) and four clones from the brain (β8-1, β9-2, β11-1, and β13-1), all of which represented splice variants of the β4 subunit (Fig. 4). Three of these, clones β8-1, β11-1, and β13-1, are highly homologous to the human β4b and β4a isoforms reported previously (18, 19). The cochlear clone βE-1 and the brain clone β9-2, however, represent splice variants of β4 of which we know no previous report. Although the 3′ noncoding region of β9-2 differs from that of βE-1, the two cDNAs encode the same polypeptide (Figs. 1A and 4). The first half of β9-2 and βE-1 is identical to that of β13-1. In β9-2 and βE-1, however, the absence of a 59-bp exon causes a frame shift. The ensuing stop codon results in the formation of a short β4a isoform designated chicken β4c (Figs. 1A and 4).
To examine the tissue distributions of β4c and β4a/b, we conducted semiquantitative RT-PCR analysis of various tissues of the chicken with primers common to all the β4 isoforms. mRNAs encoding both β4c and β4a/b were detected in the brain, spinal cord, eye, and heart (Fig. 1B). Transcripts of the two classes occurred in similar amounts in all organs save the brain, in which β4a/b was expressed more abundantly than β4c. The cochlea alone expressed only β4c. We further tested the expression of β4c in the cochlear sensory epithelium, which includes hair cells and supporting cells, and in the tegmentum vasculosum, the vascularized secretory epithelium of the ear. Nested PCRs with two primer pairs revealed that both tissues express predominantly β4c but not β4a/b (Fig. 1B). These results suggest that the different variants of β4 subunits subserve tissue- or organ-specific functions.
The β-interaction domain, which comprises 30 amino acids near the middle of a β subunit, is essential for the binding of β to α1 subunits (20). The association of the two subunits generally increases the amplitude and modifies the kinetics of Ca2+ currents. Because it contains a canonical interaction domain except for the last two amino acids, the β4c subunit would be expected to bind to an α1 subunit and act as a channel regulator. Consistent with this supposition, a GST fusion protein containing the cytoplasmic loopI/II of the α1D subunit clearly interacted with His-tagged β4c (data not shown).
To investigate the physiological effects of β4c on Ca2+ channels, we compared the Ca2+ currents in Xenopus oocytes expressing α1 and α2-δ subunits in the presence or absence of either β4c or β4a (Fig. 5). Because our experiments revealed that the hair cell's α1D subunit was not functional under our assay conditions, we analyzed the α1A subunit that is reported to occur with β4 subunits in the brain (21). Coexpression of the β4a subunit caused a hyperpolarizing shift of ≈10 mV in the activation and inactivation curves accompanied by a decrease in the voltage sensitivity of activation. In addition, the β4a subunit slowed inactivation by increasing the amplitude of its slow component (Table 1 and Fig. 5). By contrast, coexpression of the β4c subunit barely affected the properties of the channel, because the voltages for activation and inactivation, as well as the two time constants of inactivation, were not significantly modified. The addition of β4c did lower the slope of the activation curve significantly and reduce the amplitude of the slow component of inactivation (Table 1 and Fig. 5). The reactivation rate was not affected by either β subunit. Together with the clear detection of β4c subunit expressed in Xenopus oocytes on Western blots (data not shown) and the protein's capability of interacting with the loopI/II of the α1 subunit, these results indicate that β4c is a functional Ca2+-channel β subunit. They also imply a physiological role of the secondary interaction sites localized in the carboxyl-terminal half of β4a in the regulation of the channel's voltage dependence and inactivation kinetics (22, 23).
Table 1.
The effects of β subunits on the properties of Ca2+ channels in Xenopus oocytes
| Injected β subunit | VIN, mV | kIN, mV−1 | R, % | VACT, mV | kACT, mV−1 | EREV, mV | n |
|---|---|---|---|---|---|---|---|
| None | −13.9 ± 0.7 | 5.6 ± 0.5 | 0.0 ± 2.0 | 4.7 ± 0.6 | −5.7 ± 0.2 | 50.5 ± 2.9 | 6 |
| β4a | −23.4 ± 0.5* | 6.3 ± 0.2 | 3.0 ± 2.0* | −6.0 ± 0.7* | −4.3 ± 0.2* | 54.4 ± 0.7 | 11 |
| β4c | −14.1 ± 1.0 | 5.7 ± 0.4 | 0.0 ± 3.0 | 3.2 ± 0.9 | −4.7 ± 0.2* | 54.6 ± 2.3 | 11 |
All oocytes were injected with rat brain α1A and α2-δ subunits. The Ba2+ currents and kinetics of each combination were analyzed as described in the Fig. 5 legend.
Significantly different from α1A and α2-δ without β (P < 0.05).
Because the effect of β4c on Ca2+ currents is relatively small, we postulated that β4c has other functions. We therefore sought binding partners of β4c by two-hybrid screening of a cDNA library from the chicken's cochlear sensory epithelium. Thirty-one positive clones were isolated, all belonging to CHCBs (24), also termed HP1s (25). Members of the CHCB/HP1 family contain a chromo domain and a chromo shadow domain (CSD; Fig. 2A; ref. 26). The chromo domain is essential for gene-silencing activity (27) through its specific binding to lysine 9 of histone H3 (3, 4). The CSD acts as a scaffold that assembles various nuclear proteins such as transcriptional intermediary factors 1 (11), SP100 (13, 28), Ku70 (29), and SUV39H1 (30).
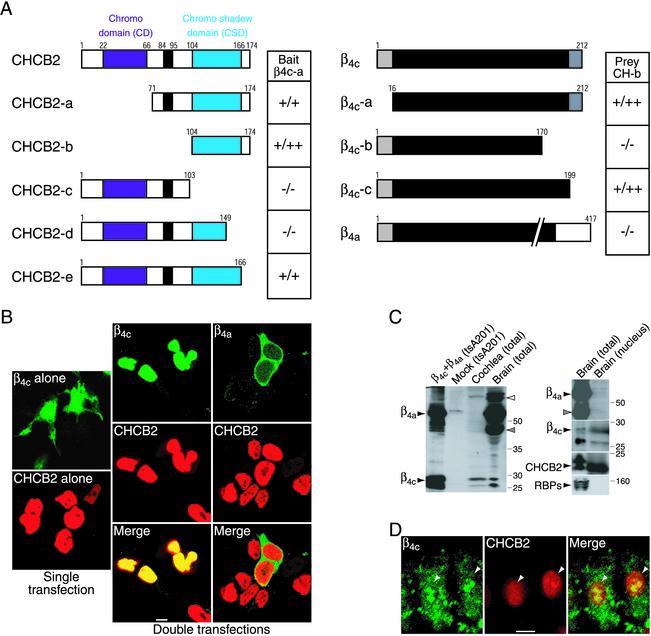
Figure 2.
Interaction of β4c with CHCB2. (A) Mapping of binding sites by the yeast two-hybrid assay. Five fragments of CHCB2 in the prey vectors and three fragments of β4c and full-length β4a in the bait vectors were tested with β4c-a in the bait vector (Left) and CHCB2-b in the prey vector (Right), respectively. The interactions were scored by β-galactosidase activity and His prototrophy. The black bars in the CHCB2 constructs delineate the serine-rich regions. (B) CHCB2 recruits β4c but not β4a to the nuclei of transfected tsA201 cells. Expressed alone, GFP-tagged β4c (fluorescein, green) has a diffuse cytoplasmic and nuclear distribution (Upper Left), and myc-CHCB2 (Texas red, red) is localized only in the nuclei (Lower Left). (Center) When coexpressed with CHCB2, β4c moves into the nuclei, where its distribution completely overlaps that of CHCB2. (Right) Coexpression with CHCB2 does not change the cytoplasmic distribution of β4a. (Scale bar, 3 μm.) (C) Identification of β-subunit proteins in chicken tissues. (Left) Affinity-purified antibody against β4c was used to identify β subunits in lysates of chicken total cochlea (third lane) and brain (forth lane). For Left and Right, each lane was loaded with 80 μg of protein produced by extraction in 0.5% Nonidet P-40. Proteins from tsA201 cells expressing β4a and β4c (first lane) and mock-transfected cells (second lane) were analyzed as positive and negative controls, respectively. (Right) β4c (second segment), but neither β4a (top segment) nor the 40-kDa β subunit (top segment, gray arrowhead), was detected together with CHCB2 (third segment) in the nuclear fraction from the brain. Total protein (first lanes) and nuclear protein (second lanes) from chicken brain were analyzed on Western blots with the various antibodies. An antibody against Rab-interacting molecule-binding protein 2 was used to probe the nuclear fraction as a negative control (bottom segment). (D) Colocalization of β4c with CHCB2 in cochlear hair cells. The chicken hair cells were immunolabeled with anti-β (fluorescein, green) and anti-HP1γ (Texas red, red) antisera. The proteins occur together in nuclei (arrowheads). (Scale bar, 3 μm.)
Because all the clones isolated by two-hybrid screening retained a CSD, this domain seemed likely to be responsible for the interaction with β4c. To identify the precise interaction sites between CHCB2 and β4c, we conducted two-hybrid assays with deletion mutants. The constructs CHCB2-a and CHCB2-b, which contained the CSD but not the chromo domain, were able to bind β4c (Fig. 2A). The CHCB2-c construct that lacked the CSD and the carboxyl terminus of CHCB2 could not interact with β4c. CHCB2-e, which did not contain the carboxyl terminus of CHCB2 but retained the whole CSD, associated with β4c. Deletion of last 17 amino acids of the CSD (CHCB2-d), however, abolished the interaction. The interaction therefore requires the carboxyl terminus of the CSD.
A mutant of β4c lacking 13 residues at the carboxyl terminus, β4c-c, could bind CHCB2 (Fig. 2A), so the interaction does not require a sequence unique to β4c. However, the deletion of 29 residues at the carboxyl terminus of β4c (β4c-b) abolished the interaction (Fig. 2A). Although the long isoform β4a contains the binding site for CHCB2, it was found not to associate (Fig. 2A). A potential CHCB2-binding site therefore may be masked in β4a. Consistent with the yeast two-hybrid assays, GST fusion proteins of the CSD from CHCB2 (CHCB2-b) clearly interacted in tsA201 cells with GFP-tagged β4c but not with β4a. Coimmunoprecipitation yielded a consistent result: anti-myc antibodies precipitated β4c but not β4a in the presence of myc-tagged CHCB2 (Fig. 6, which is published as supporting information on the PNAS web site).
We next examined the effect of coexpression with CHCB2 on the subcellular distribution of β4c. When expressed alone in tsA201 cells, GFP-tagged β4c displayed a principally cytoplasmic distribution with modest labeling of nuclei (Fig. 2B). When coexpressed with myc-tagged CHCB2, however, β4c accumulated in nuclei, where it was perfectly colocalized with CHCB2 (Fig. 2B). This nuclear translocation was specific for β4c, because CHCB2 did not recruit the long isoform β4a to nuclei (Fig. 2B). Furthermore, coexpression with either HP1α or HP1β redistributed β4c into the nuclei (data not shown), so not only HP1γ/CHCB2 but also other types of HP1s can bind β4c.
We developed an affinity-purified antiserum against amino acid residues 16–212 of β4c. On Western blots, the antiserum detected β4a, β4c, and β1b subunits expressed in tsA201 cells (data not shown), indicating that it recognizes all the β subunits. Immunoblotting analysis showed that the antiserum detected five major bands in chicken-brain lysate (Fig. 2C). The protein of 60 kDa is likely to represent β4a and/or β4b, because β4a transfected in tsA201 cells exhibited almost the same size. The protein of 75 kDa (Fig. 2C, open arrowhead) might correspond to β1 or β2, and that of 40 kDa might represent another β subunit (31, 32). The proteins of 26 and 28 kDa found after expression of β4c in tsA201 cells were identical in size to those from brain lysate, indicating that β4c protein occurs in the brain. The smallest protein (26 kDa) in both brain and tsA201 cell lysates could be a degradation product of β4c. Consistent with the result of semiquantitative RT-PCR, the brain expresses long β4 isoforms more abundantly than β4c. On the other hand, chicken cochlea expresses predominantly β4c and small amounts of the 75-kDa β subunit but no other long β-subunit isoform (Fig. 2C). Furthermore, we detected both β4c and CHCB2 but not long β isoforms in the nuclear fraction isolated from the brain (Fig. 2C). The β4c signal in nuclei did not result from cytoplasmic contamination, because Rab-interacting molecule-binding proteins (RBPs), which occur at synaptic terminals (9), were absent from the same preparation (Fig. 2C). In addition, immunohistochemical labeling of the chicken cochlear hair cells revealed that β4c is expressed in nuclei as well as in the cytosol and at the plasma membrane; at least in part, the subunit colocalizes with nuclear CHCB2 (Fig. 2D). β4c therefore occurs together with CHCB2 in the nuclei of native cells.
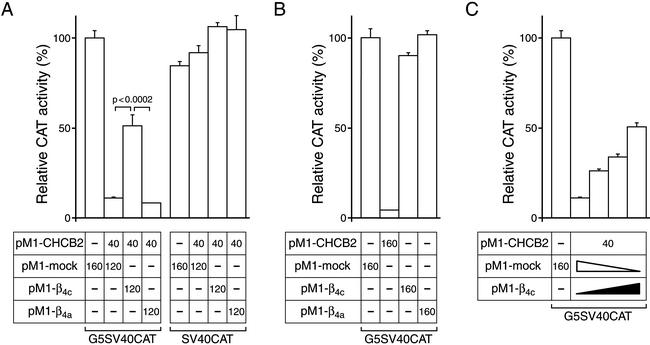
We also investigated the functional impact of the interaction between β4c and CHCB2. Proteins involved in gene silencing are reported to suppress transcription regionally, rather than promoter- or sequence-specifically, through the generation of a DNA–histone complex that is inaccessible to DNA-binding proteins (1). The GAL4-CAT reporter assay therefore provides a useful means of analyzing the gene-silencing activity of CHCBs/HP1s (11, 13). In Cos-1 cells, expression of a fusion protein comprising the GAL4-(1–147) DNA-binding domain and CHCB2 strongly suppressed CAT activity (10.9 ± 0.8% of control, n = 10) by comparison to cells transfected with mock plasmids (100 ± 3.6%, n = 10; Fig. 3A). The gene-silencing activity of CHCB2 was decreased dramatically when β4c was cotransfected (50.9 ± 6.2%, n = 10). On the other hand, the coexpression of β4a had little effect on CHCB2-induced transcriptional repression (7.9 ± 0.4%, n = 10). All the combinations of the transfected constructs conferred essentially the same CAT activity on Cos-1 cells when they were transfected with the SV40CAT reporter-gene plasmid lacking the GAL4 site, indicating that the effect of β4c is specific. β4c prevented CHCB2 from silencing CAT-gene transcription in a concentration-dependent manner (Fig. 3C). Because β4c itself did not influence gene transcription (Fig. 3B), the effect of the subunit depends on its interaction with CHCB2.
Figure 3.
β4c regulates transcriptional repression activity of CHCB2. Cos-1 cells were transiently transfected with the different combinations of plasmids displayed beneath the histograms. (A) β4c but not β4a attenuates the transcriptional repression activity of CHCB2. (B) β subunits themselves have no effect on transcription. The pM1-CHCB2 plasmid was transfected as a control. (C) β4c suppresses the gene-silencing activity of CHCB2 in a concentration-dependent manner. pM1-β4c plasmid (0, 40, 80, or 120 ng) was transfected in conjunction with 120, 80, 40, or 0 ng of pM1-mock plasmid, respectively. CHCB2 (40 ng of pM1-CHCB2) and CAT reporter gene (100 ng of G5SV40CAT) were expressed in all cells except for the control (first result), in which 160 ng of pM1 mock plasmid was transfected instead. The numbers in the boxes indicate in nanograms the amounts of plasmid DNAs used for transfection. In each of the experimental series, the total amount of transfected pM1-vector constructs was identical. The histograms display CAT activities relative to those of the appropriate controls, and error bars show the standard errors (A, n = 10; B and C, n = 5).
Discussion
We have demonstrated that an ion-channel accessory protein, the β4c subunit of the Ca2+ channel, interacts directly with a nuclear protein and regulates gene silencing. The mechanism of the inhibitory effect of β4c on CHCB2-induced transcriptional repression remains unknown. HP1s form hetero- or homodimers through interactions between their CSDs (33, 34). However, our GST pull-down assays revealed that β4c did not inhibit the direct interaction between CHCB2 and HP1α (data not shown). Furthermore, β4c competed with neither SUV39H1 nor transcriptional intermediary factor 1β (data not shown), proteins that associate directly with the CSDs of HP1s and mediate gene silencing (11, 30). These negative results suggest that β4c prevents CHCBs from interacting with unknown molecules involved in CHCBs/HP1s-mediated gene silencing. Alternatively, β4c might alter the conformation of CHCB2 and thereby reduce its transcriptional repression.
The cell-junction proteins CASK and β-catenin can directly enter nuclei, bind specific transcriptional factors, and regulate gene transcription (35, 36). Phosphorylation by GSK3-β destabilizes β-catenin and prevents its translocation to nuclei (37). Although we have not determined what triggers the redistribution of β4c, a modification such as phosphorylation or dephosphorylation might initiate the process. Most intriguingly, the transit of β4c from Ca2+ channels to nuclear HP1s/CHCBs may be induced by changes in electrical activity and thus constitute a feedback system between neuronal responses and gene transcription. Although we could not detect the β4c subunit by RT-PCR from rat brain (data not shown), it remains possible that β4c occurs and acts as a transcriptional regulator in mammals. The lethargic (lh) mouse, which displays ataxia and seizures, has been reported recently to have a 4-nt insertion into a splice donor site of the β4 subunit, resulting in a lack of β4 protein (31, 38). The absence of controlled gene silencing mediated by β4c may be one of the factors that induce the lethargic phenotype.
Supplementary Material
Acknowledgments
Dr. N. Lehming (Max-Delbrück-Laboratorium) and Drs. P. Chambon and R. Losson [Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale (INSERM), Universite Louis Pasteur] kindly provided the CAT-assay plasmids and Flag-tagged HP1s, and Dr. S. Heller (Massachusetts Eye and Ear Infirmary) contributed the basilar-papilla cDNA libraries. Dr. J. Nargeot and members of our research group offered valuable comments on the manuscript. H.H. was supported by Human Frontier Science Program Grant LT0318/1999-B, P.C. was supported by Association pour la Recherche contre le Cancer and by Association Francaise contre les Myopathies, and F.L. was supported by INSERM, the North Atlantic Treaty Organization, and The Philippe Foundation. This research was supported by Grant DC00317 to A.J.H., who is an Investigator of The Howard Hughes Medical Institute.
Abbreviations
- HP1
heterochromatin protein 1
- CHCB
chromobox protein
- CAT
chloramphenicol acetyltransferase
- CSD
chromo shadow domain
References
- 1.Moazed D. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 2.Eissenberg J C, James T C, Foster-Hartnett D M, Hartnett T, Ngan V, Elgin S C. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Zegerman P, Partridge J F, Miska E A, Thomas J O, Allshire R C, Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 5.Rea S, Eisenhaber F, O'Carroll D, Strahl B D, Sun Z W, Schmid M, Opravil S, Mechtler K, Ponting C P, Allis C D, Jenuwein T. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 6.Lin R C, Scheller R H. Annu Rev Cell Dev Biol. 2000;16:19–49. doi: 10.1146/annurev.cellbio.16.1.19. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh A, Greenberg M E. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 8.Catterall W A. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 9.Hibino H, Pironkova R, Onwumere O, Vologodskaia M, Hudspeth A J, Lesage F. Neuron. 2002;34:411–423. doi: 10.1016/s0896-6273(02)00667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cens T, Mangoni M E, Nargeot J, Charnet P. FEBS Lett. 1996;391:232–237. doi: 10.1016/0014-5793(96)00704-1. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorski K, Carneiro M, Schibler U. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 13.Lehming N, Le Saux A, Schuller J, Ptashne M. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis R S, Hudspeth A J. Nature. 1983;304:538–541. doi: 10.1038/304538a0. [DOI] [PubMed] [Google Scholar]
- 15.Kollmar R, Montgomery L G, Fak J, Henry L J, Hudspeth A J. Proc Natl Acad Sci USA. 1997;94:14883–14888. doi: 10.1073/pnas.94.26.14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kollmar R, Fak J, Montgomery L G, Hudspeth A J. Proc Natl Acad Sci USA. 1997;94:14889–14893. doi: 10.1073/pnas.94.26.14889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hullin R, Singer-Lahat D, Freichel M, Biel M, Dascal N, Hofmann F, Flockerzi V. EMBO J. 1992;11:885–890. doi: 10.1002/j.1460-2075.1992.tb05126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellano A, Wei X, Birnbaumer L, Perez-Reyes E. J Biol Chem. 1993;268:12359–12366. [PubMed] [Google Scholar]
- 19.Helton T D, Horne W A. J Neurosci. 2002;22:1573–1582. doi: 10.1523/JNEUROSCI.22-05-01573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Waard M, Pragnell M, Campbell K P. Neuron. 1994;13:495–503. doi: 10.1016/0896-6273(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig A, Flockerzi V, Hofmann F. J Neurosci. 1997;17:1339–1349. doi: 10.1523/JNEUROSCI.17-04-01339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker D, Bichet D, Campbell K P, De Waard M. J Biol Chem. 1998;273:2361–2367. doi: 10.1074/jbc.273.4.2361. [DOI] [PubMed] [Google Scholar]
- 23.Walker D, Bichet D, Geib S, Mori E, Cornet V, Snutch T P, Mori Y, De Waard M. J Biol Chem. 1999;274:12383–12390. doi: 10.1074/jbc.274.18.12383. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi K, Hidema S, Mizuno S. Exp Cell Res. 1998;242:303–314. doi: 10.1006/excr.1997.4082. [DOI] [PubMed] [Google Scholar]
- 25.Jones D O, Cowell I G, Singh P B. BioEssays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Elgin S C. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 27.Platero J S, Hartnett T, Eissenberg J C. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song K, Jung Y, Jung D, Lee I. J Biol Chem. 2001;276:8321–8327. doi: 10.1074/jbc.M008779200. [DOI] [PubMed] [Google Scholar]
- 30.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McEnery M W, Copeland T D, Vance C L. J Biol Chem. 1998;273:21435–21438. doi: 10.1074/jbc.273.34.21435. [DOI] [PubMed] [Google Scholar]
- 32.De Waard M, Witcher D R, Pragnell M, Liu H, Campbell K P. J Biol Chem. 1995;270:12056–12064. doi: 10.1074/jbc.270.20.12056. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen A L, Oulad-Abdelghani M, Ortiz J A, Remboutsika E, Chambon P, Losson R. Mol Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 34.Brasher S V, Smith B O, Fogh R H, Nietlispach D, Thiru A, Nielsen P R, Broadhurst R W, Ball L J, Murzina N V, Laue E D. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsueh Y P, Wang T F, Yang F C, Sheng M. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 36.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 37.Morin P J. BioEssays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 38.Burgess D L, Jones J M, Meisler M H, Noebels J L. Cell. 1997;88:385–392. doi: 10.1016/s0092-8674(00)81877-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.