Abstract
The VP40 protein of Ebola virus can bud from mammalian cells in the form of lipid-bound, virus-like particles (VLPs), and late budding domains (L-domains) are conserved motifs (PTAP, PPxY, or YxxL; where “x” is any amino acid) that facilitate the budding of VP40-containing VLPs. VP40 is unique in that potential overlapping L-domains with the sequences PTAP and PPEY are present at amino acids 7 to 13 of VP40 (PTAPPEY). L-domains are thought to function by interacting with specific cellular proteins, such as the ubiquitin ligase Nedd4, and a component of the vacuolar protein sorting (vps) pathway, tsg101. Mutational analysis of the PTAPPEY sequence of VP40 was performed to understand further the contribution of each individual motif in promoting VP40 budding. In addition, the contribution of tsg101 and a second member of the vps pathway, vps4, in facilitating budding was addressed. Our results indicate that (i) both the PTAP and PPEY motifs contribute to efficient budding of VP40-containing VLPs; (ii) PTAP and PPEY can function as L-domains when separated and moved from the N terminus (amino acid position 7) to the C terminus (amino acid position 316) of full-length VP40; (iii) A VP40-PTAP/tsg101 interaction recruits tsg101 into budding VLPs; (iv) a VP40-PTAP/tsg101 interaction recruits VP40 into lipid raft microdomains; and (v) a dominant-negative mutant of vps4 (E228Q), but not wild-type vps4, significantly inhibited the budding of Ebola virus (Zaire). These results provide important insights into the complex interplay between viral and host proteins during the late stages of Ebola virus budding.
Ebola virus (Zaire; EBOZ) is a member of the Filoviridae family of negative-sense RNA viruses (for a review, see reference 41). Ebola virus infection of nonhuman primates and humans results in severe hemorrhagic fever with high rates of mortality. As a biological safety level 4 pathogen, Ebola virus affords investigators many unique challenges in developing effective vaccines (24). Nevertheless, our understanding of the molecular aspects of Ebola virus replication and pathogenesis has progressed, and further studies will provide important insights for the future development of vaccines and/or antivirals.
Of the seven proteins encoded by Ebola virus, VP40 is the most abundant structural protein and functions in assembly and budding (11). As such, VP40 is the functional homolog of both the matrix (M) protein of rhabdoviruses (e.g., vesicular stomatitis virus [VSV]) and the gag polyprotein of retroviruses (e.g., human immunodeficiency virus type 1 [HIV-1]). Interestingly, VP40, M, and gag proteins can associate with the plasma membrane and bud as virus-like particles (VLPs) from mammalian cells in the absence of additional viral proteins (7, 8, 10, 15, 17-19, 21, 25, 26, 28, 30, 31, 37, 42, 44-46). Efficient budding of these viral matrix proteins is dependent, in part, upon the presence of conserved amino acid motifs termed late budding domains (L-domains) (12). To date, motifs conserved in viral matrix proteins that can function as L-domains include PPxY, PTAP, and YxxL (where “x” is any amino acid). Viral L-domains function during a late stage of virus-cell separation (15, 20, 30, 44); however, the mechanism by which they promote virus-cell separation remains unclear. Increasing evidence suggests that L-domains function as docking sites to recruit host proteins, therefore facilitating the pinching-off stage of virus budding. Although candidate cellular proteins thought to be involved in virus budding have been identified, the precise molecular mechanism by which these virus-host interactions facilitate budding remains unclear (9, 14, 16, 18, 23, 29, 32-34, 36, 38, 39, 43, 47).
The VP40 protein is unique among the viral matrix proteins in that it possesses overlapping L-domain motifs at its amino terminus (7PTAP10 and 10PPEY13) (17). The presence of an intact PTAP motif was shown to enhance the efficiency of release of both Ebola virus VP40 and HIV-1 gag (14, 29). PTAP is thought to mediate its function in budding through an interaction with the cellular protein tsg101, a member of the vacuolar protein sorting (vps) pathway within mammalian cells (6, 9, 14, 29, 36, 43). In addition, the PTAP peptide from VP40 was shown to interact with and colocalize with tsg101 at the plasma membrane of transfected cells (29).
Along with tsg101, the vps4 protein has also been implicated in the process of virus budding (6, 14, 35). vps4 is a component of the vps machinery of the cell and is a member of the AAA family of ATPases (2, 3). vps4 is thought to function downstream of tsg101 during cellular protein sorting and invagination of vesicles into the cellular multivesicular body. Recent evidence suggests that the ATPase activity of vps4 may be important for efficient budding of HIV-1 (6, 14, 35). Indeed, dominant-negative mutants of vps4 that are enzymatically inactive were shown to inhibit release of HIV-1 (14).
A potential role for the PPxY motif during budding of VP40 has also been postulated (17, 19). PPxY motifs have been shown to function as L-domains within the M proteins of rhabdoviruses, the gag protein of type C retroviruses (Rous sarcoma virus), and the VP40 protein of Ebola virus (8, 13, 16-18, 20, 34, 44-46). Like PTAP, PPxY is also known to mediate interactions with specific cellular proteins. For example, the PPxY motifs of VSV M, Rous sarcoma virus gag, and EBOZ VP40 can interact with WW-domains of cellular ubiquitin ligases such as Nedd4/Rsp5 (13, 16-18, 23, 34, 47). Furthermore, physical interactions between the PPxY motif and Nedd4/Rsp5 resulted in the covalent addition of ubiquitin to the viral matrix proteins (16, 17, 23, 33, 39, 47). Of particular interest to the mechanism of virus budding is the fact that mono-ubiquitination of cellular and perhaps viral proteins is thought to serve as a signal for entry of these proteins into the vps pathway (22). The presence of two potential L-domains within VP40 is intriguing, and it is possible that both motifs contribute in a redundant or sequential manner to facilitate virus budding. Thus, a more thorough understanding of the contribution of these overlapping L-domains to virus budding and of host protein interactions with L-domains of VP40 would provide new insights into late stages of Ebola virus replication. The results from such studies may ultimately lead to the development of novel antiviral therapies.
We have generated here mutations within the PTAPPEY region of VP40 to more clearly define the role of these overlapping motifs both in VP40 budding and in mediating functionally significant interactions with cellular proteins. Our results suggest that both the PTAP and the PPEY motifs are important for the efficient release of VP40-containing VLPs. In addition, we demonstrate for the first time that the PTAPP and PPEY motifs are both movable within full-length VP40 and separable in their ability to function as L-domains. Finally, we show that (i) the PTAP motif of VP40-WT recruits tsg101 into budding VLPs, (ii) the tsg101-VP40 interaction recruits VP40 into lipid raft domains, and (iii) a dominant-negative mutant of vps4 significantly inhibits the budding of Ebola virus (Zaire).
MATERIALS AND METHODS
Cells and virus.
Human 293T cells were used for transfection experiments due to their high efficiency of transfection, and Vero E6 cells were used in all other experiments since they are utilized most often for the growth of Ebola virus in tissue culture. Human 293T cells were maintained in Dulbecco minimal essential medium (Life Technologies, Rockville, Md.) supplemented with 10% fetal calf serum (HyClone) and 1× penicillin-streptomycin (Life Technologies). Vero E6 cells (American Type Culture Collection) were maintained in Eagle minimal essential medium containing 10% fetal calf serum. Ebola virus Zaire 1976 (EBOZ) was employed in these studies. All virus experiments were conducted in biocontainment level 4 facilities at the U.S. Army Medical Research Institute of Infectious Disease (USAMRIID; Fort Detrick, Md.).
Plasmids and antibodies.
The VP40 open reading frame of EBOZ was amplified and inserted into the EcoRI/XhoI restriction sites of the pCAGGS/MCS vector to yield pVP40-WT by standard PCR and cloning techniques, respectively. The c-myc epitope tag (EQKLISEEDL) was joined to either the N terminus or the C terminus of all VP40 proteins as indicated in Fig. 1. Site-directed mutagenesis and PCR were utilized to construct all mutants of VP40 (Fig. 1). All plasmids containing introduced mutations and/or deletions were confirmed by automated DNA sequencing. The expression plasmid encoding hemagglutinin (HA)-tagged tsg101 was kindly provided by P. Bates (University of Pennsylvania), and plasmids encoding vps4-WT and the dominant-negative mutant vps4-E228Q were kindly provided by W. Sundquist (University of Utah). A monoclonal antibody (MAb) directed against the HA epitope tag (Roche Biochemicals, Indianapolis, Ind.) was used according to the protocol of the supplier. Monoclonal (9E10; University of Pennsylvania Cell Center) and polyclonal (Abcam, Ltd., Cambridge, United Kingdom) antibodies against the c-myc epitope tag were used according to protocols of the suppliers.
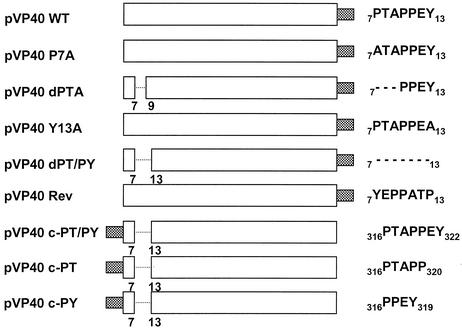
FIG. 1.
Schematic diagram of VP40 expression plasmids. The name of each plasmid is indicated on the left. The amino acid sequence and location of the L-domain(s) within each construct is shown at the right. Amino acid residues are indicated by their single-letter code. Amino acid substitutions are indicated in boldface type. The dashed lines indicate amino acid deletions. The shaded box indicates the position of the in-frame c-myc epitope tag.
VP40 budding assay.
Briefly, 293T cells were transfected with equal amounts of the indicated plasmids by using Lipofectamine (Invitrogen). First, 150 μCi of [35S]Met-Cys (Perkin-Elmer) was added to each 100-mm dish from 36 to 48 h posttransfection. Culture medium was harvested and clarified at 2,500 rpm for 10 min, loaded over a 20% sucrose cushion in STE buffer (0.01 M Tris-HCl [pH 7.5], 0.01 M NaCl, 0.001 M EDTA [pH 8.0]), and centrifuged at 36,000 rpm for 4 h at 4°C. Pelleted VLPs were suspended in 400 μl of STE buffer overnight at 4°C. Radioimmunoprecipitation assay buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) was added, and the VLPs were immunoprecipitated with the appropriate antiserum. Transfected cells were harvested in radioimmunoprecipitation assay buffer, immunoprecipitated with the appropriate antiserum, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Protease protection experiments were performed essentially as described by Jasenosky et al. (19).
Immunofluorescence and confocal microscopy.
Vero E6 cells were transfected with the appropriate plasmids as described above. The cells were washed with 1× phosphate-buffered saline (PBS) at 24 h posttransfection and fixed in 1× PBS-4% formaldehyde for 20 min at room temperature. Cells were then permeabilized as necessary with 1× PBS containing 0.2% Triton X-100 and stained with primary antibodies to the c-myc (Abcam) epitope tag. Secondary antibodies conjugated to the Alexa 488 fluorophore (Molecular Probes, Ltd.) were used. Coverslips were mounted in ProLong Antifade (Molecular Probes, Ltd.). Microscopy was performed with a Bio-Rad Radiance 2000 confocal microscope by using Lasersharp 2000 software (Bio-Rad, Ltd.). To generate images of whole cells, serial optical planes of focus (at ∼0.5-μm intervals) were taken on the z axis through the depth of the cell, and images were merged into one by using the projection algorithm in Lasersharp 2000 (Bio-Rad). z axis images were collected by using the vertical scanning function in this program.
Isolation of detergent-insoluble and -soluble fractions.
Vero E6 cells were transfected with the indicated plasmids (10 μg of total DNA) as described above. Transfected cells were placed on ice at 24 h posttransfection and treated with 1× PBS containing TNE buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 15 mM EDTA) plus 1.0% Triton X-100. After 30 min, cells were harvested by scraping and centrifuged at 14,000 × g at 4°C. Supernatants were then separated from the pellets, and SDS-PAGE and Western blotting analyses were performed. Proteins were detected with antibodies to either the c-myc epitope tag (Abcam) or the HA epitope tag (Roche), followed by the addition of enhanced chemiluminescence reagents (Amersham Biosciences, Ltd.).
vps4 transfection and Ebola virus infection.
293T cells were transfected in suspension with 5.0 μg of the eukaryotic expression plasmids green fluorescent protein (GFP)-vps4-WT or GFP-vps4-E228Q by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 24 h posttransfection, cells were infected with Ebola virus Zaire 1976 at a multiplicity of infection (MOI) of 1.0. Culture media from Ebola virus-infected cells were harvested at 0, 3, 5, and 7 days postinfection. Samples were assayed by plaque titration on Vero E6 cells. All experiments involving Ebola virus-infected samples were performed in biological safety level 4 biocontainment laboratories at the USAMRIID.
RESULTS
Expression and budding of VP40 as VLPs.
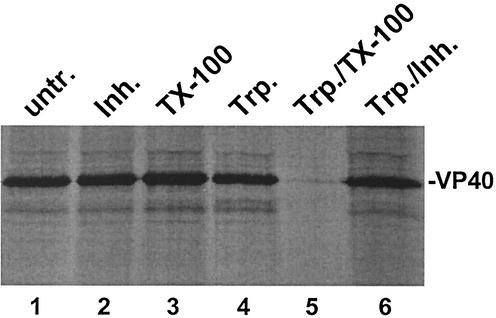
To assess the contribution of overlapping L-domain motifs to VP40 budding, mutations were introduced to disrupt the PTAP motif alone, the PPEY motif alone, or both motifs (Fig. 1). The ability of both wild type and mutants of VP40 to bud as VLPs was assessed by using a functional budding assay. To first demonstrate that VP40-WT is indeed released from mammalian cells in the form of VLPs, a protease protection assay was performed, followed by immunoprecipitation (Fig. 2). Culture medium from VP40-WT-transfected cells was layered onto a 20% sucrose cushion, and lipid-containing particles were pelleted. VLPs in the resultant pellet were divided into six equivalent samples. VP40-containing VLPs were either untreated (Fig. 2, lane 1), treated with trypsin inhibitor alone (lane 2), Triton X-100 alone (lane 3), trypsin alone (lane 4), trypsin plus Triton X-100 (lane 5), or trypsin plus trypsin inhibitor (lane 6). Importantly, VP40 was not degraded in the presence of trypsin alone (Fig. 2, lane 4); however, VP40 was completely degraded by trypsin in the presence of Triton X-100 detergent. These results confirm previous findings (19) and substantiate the reliability of this assay in measuring the budding efficiency of VP40-containing VLPs.
FIG. 2.
Protease protection assay for VP40-containing VLPs. VLPs were harvested from the media of cells transfected with VP40-WT and were either left untreated (lane 1) or treated as described in Materials and Methods with trypsin inhibitor (lane 2), Triton X-100 (lane 3), trypsin (lane 4), trypsin plus Triton X-100 (lane 5), and trypsin plus trypsin inhibitor (lane 6). VP40-WT was detected by immunoprecipitation and SDS-PAGE analysis.
Both PTAP and PPEY motifs of VP40 are important for the efficient budding of VLPs.
The ability of specific mutants of VP40 to bud from mammalian cells as VLPs was assessed by using the functional budding assay described above (Fig. 3). VP40-WT and mutants P7A, dPTA, Y13A, and dPT/PY were detected readily at equivalent levels in cell extracts (Fig. 3A, lanes 2 to 6). Mock-transfected cells served as a negative control (lane 1). As expected, VP40-WT was present in VLPs (Fig. 3B, lane 2). Interestingly, mutants P7A, dPTA, and Y13A were also readily detected in VLPs (Fig. 3B, lanes 3 to 5, respectively). Quantitation by phosphorimager analysis indicated that the levels of P7A and dPTA mutants were similar to that of VP40-WT in VLPs (Fig. 3C). Also, the level of mutant Y13A in VLPs was twofold lower than that of VP40-WT (Fig. 3C). In contrast, although the dPT/PY mutant, which lacks both the PTAP and the PPEY motifs, was readily detected in cell extracts (Fig. 3A, lane 6), this mutant was barely detectable in VLPs (Fig. 3B, lane 6). These results suggest that each motif alone can function as an L-domain. It should be noted that the appearance of VP40 as a doublet in VLPs has been described previously (17, 19) and that the smaller protein species is translated from an internal AUG codon. The internally translated VP40 species was present in VLPs of P7A and dPTA upon longer exposure of the film.
FIG. 3.
Mutational analysis of the PTAP and PPEY motifs of VP40. 293T cells were transfected with pCAGGS vector alone (mock, lane 1), pVP40-WT (lane 2), pVP40-P7A (lane 3), pVP40dPTA (lane 4), pVP40Y13A (lane 5), or pVP40dPT/PY (lane 6). VP40 proteins were detected in cell extracts (A) and in budding VLPs (B) by immunoprecipitation with polyclonal antiserum against the c-myc epitope tag. (C) Levels of VP40 proteins detected in VLPs were quantitated by phosphorimager analysis. The level of pVP40-WT was normalized to 100%, and the relative percentages of VP40 mutants P7A, dPTA, Y13A, and dPT/PY in VLPs are indicated.
Orientation and localization of PTAPPEY within full-length VP40 effects budding efficiency.
Viral L-domains are believed to exert their function by acting as protein-protein interaction modules, conceivably usurping the normal role of proteins within the cell to promote virus release (12). As docking sites for cellular proteins, these viral L-domains would be predicted to function when moved to a new location within the viral protein and, indeed, this phenomenon has been observed (8, 27, 31). To determine whether the L-domains of VP40 retain their function when moved within the full-length protein, plasmids were generated to express VP40 proteins with the PTAPPEY motif deleted from the N terminus, and either PTAPP alone (c-PT), PPEY alone (c-PY), or PTAPPEY (c-PT/PY) inserted downstream of amino acid 316 at the C terminus of VP40 (see Fig. 1). In addition to placing the various motifs at the C terminus of VP40, a fourth construct was generated to determine whether the orientation of the PTAPPEY motif was important for VLP formation. The pVP40-Rev plasmid (Fig. 1) was generated to encode full-length VP40 containing the sequence YEPPATP (PTAPPEY reversed) at amino acids 7 to 13 at the N terminus.
The ability of these four VP40 mutants to bud from cells as VLPs was assessed by using a functional budding assay (Fig. 4). Human 293T cells were transfected with the indicated plasmid, and cells and VLPs were analyzed by immunoprecipitation. Equivalent amounts of VP40-WT and the four mutant proteins were detected in cell extracts (Fig. 4A, lanes 2 to 5 and lane 7). Extracts from mock-transfected cells served as a negative control (Fig. 4A, lanes 1 and 6). As postulated, the VP40-cPT/PY was detected in budding VLPs (Fig. 4B, lane 3). Interestingly, VP40-cPT and VP40-cPY were also detected readily in budding VLPs (Fig. 4B, lanes 4 and 5), indicating that each motif alone was movable and functional as an L-domain. Lastly, VP40-Rev was not detected in budding VLPs (Fig. 4B, lane 7), indicating that the orientation of PTAPPEY was critical for efficient budding of VP40. Interestingly, levels of VP40-cPT/PY, VP40-cPT, and VP40-cPY in VLPs were reproducibly three- to fourfold greater than that of VP40-WT, as determined by phosphorimager analysis (Fig. 4C). In sum, these data indicate that the PTAPP and PPEY motifs alone are moveable motifs within the VP40 protein and that the L-domain function of PTAPPEY is ablated when the amino acid sequence is reversed (YEPPATP).
FIG. 4.
Budding assay of VP40 proteins containing C-terminal L-domains. Human 293T cells were transfected with pCAGGS vector alone (mock, lanes 1 and 6), pVP40-WT (lane 2), pVP40-cPT/PY (lane 3), pVP40-cPT (lane 4), pVP40-cPY (lane 5), or pVP40Rev (lane 7). Radiolabeled cell extracts (A) and pelleted VLPs (B) were immunoprecipitated with anti-c-myc MAb, and immunoprecipitated proteins were analyzed by SDS-PAGE. (C) Levels of VP40 mutants detected in the VLPs were quantitated by phosphorimager analysis. The level of VP40-WT in VLPs was normalized to 100%, and the relative percentages of VP40 mutants c-PT/PY, c-PT, and c-PY in VLPs are indicated.
Intracellular localization of VP40-WT and VP40 mutants.
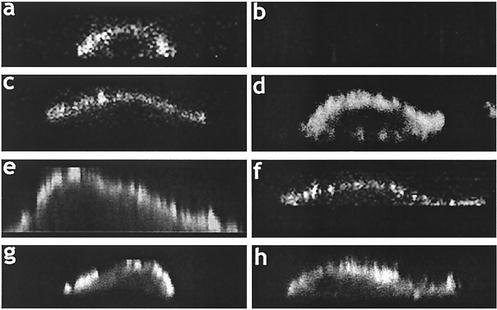
Indirect immunofluorescence and confocal microscopy were performed to confirm that both budding-competent and budding-defective forms of VP40 exhibited similar localization patterns in transfected cells. Indeed, confocal images of VP40-WT (Fig. 5a), P7A (Fig. 5c), dPT/PY (Fig. 5d), dPTA (Fig. 5e), c-PT (Fig. 5f), c-PT/PY (Fig. 5g), and VP40-Rev (Fig. 5h) demonstrated plasma membrane localization. Cells transfected with empty vector alone served as a negative control (Fig. 5b). These results confirm that the cellular localization patterns of the various VP40 proteins were not altered by the introduced mutations, and thus the inability of specific VP40 mutants to bud as VLPs correlates with an impaired L-domain(s) function.
FIG. 5.
Intracellular localization of VP40-WT and indicated mutants by immunofluorescence and confocal microscopy. Confocal images displaying plasma membrane staining are shown for cells transfected with VP40-WT (a), P7A (c), dPT/PY (d), dPTA (e), c-PTAP (f), c-PT/PY (g), and VP40-Rev (h). (b) As a negative control, cells were transfected with empty pCAGGS vector alone.
tsg101 is incorporated into VP40-containing VLPs.
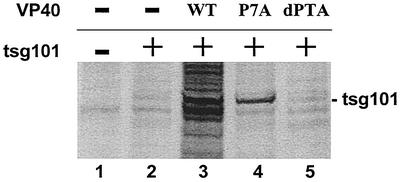
One of the cellular proteins implicated in budding of HIV-1 and Ebola virus is tsg101 (6, 14, 29). tsg101 has been shown to interact with the PTAP L-domain present in HIV-1 gag and Ebola virus VP40 by virtue of its N-terminal ubiquitin enzyme 2 variant domain (14, 29). In addition, VP40 and tsg101 were found to colocalize at the plasma membrane of transfected cells (29). We hypothesized that if the VP40-tsg101 interaction and colocalization are biologically significant, then tsg101 may be present in budding VLPs. To address this question, 293T cells were transfected with tsg101 alone or cotransfected with tsg101 and the indicated VP40 plasmids (Fig. 6). The presence of tsg101 in VLPs was assessed by immunoprecipitation with an anti-HA MAb. VLPs from mock-transfected cells served as a negative control (Fig. 6, lane 1). In the absence of the VP40-WT, tsg101 was barely detected in budding VLPs (Fig. 6, lane 2). In contrast, in the presence of either VP40-WT (lane 3) or the P7A mutant (lane 4), tsg101 was clearly evident in budding VLPs. It has been shown previously that the P7A mutation does not significantly disrupt binding to tsg101 (14). Finally, only background levels of tsg101 were detected in VLPs formed from cells transfected with tsg101 and VP40-dPTA (Fig. 6, lane 5). These results indicate that tsg101 is released into VP40-containing VLPs and that incorporation of tsg101 into VLPs is dependent on its ability to interact with the PTAP motif of VP40. Equivalent levels of tsg101 were detected in all cell extracts, and equivalent levels of VP40-WT, VP40-P7A, and VP40-dPTA were detected in appropriate cell extracts (data not shown).
FIG. 6.
Recruitment of tsg101 into budding VLPs. Human 293T cells were transfected with pCAGGS (mock, lane 1), tsg101-HA alone (lane 2), or cotransfected with tsg101-HA and pVP40-WT (lane 3), pVP40-P7A (lane 4), or pVP40-dPTA (lane 5). VLPs were harvested from the culture medium at 48 h posttransfection. The presence of tsg101-HA in VLPs was detected by immunoprecipitation with anti-HA MAb. The position of tsg101-HA is indicated.
tsg101 recruits VP40 into lipid raft domains.
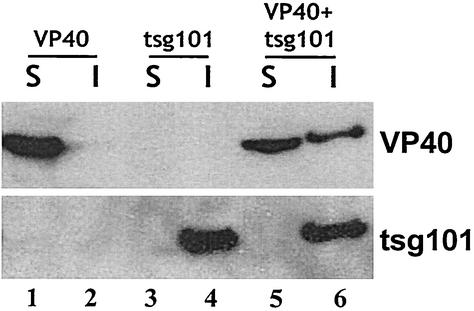
It has been reported recently that Ebola virus buds from lipid raft domains on the plasma membrane (4). Lipid raft domains have been defined by their insolubility in nonionic detergent at 4°C. Therefore, since tsg101 is postulated to be involved in the budding process, we wanted to determine whether tsg101 and/or VP40 are present in lipid raft domains. Vero E6 cells were transfected with VP40-WT alone, tsg101 alone, or VP40-WT plus tsg101, and detergent-soluble fractions were separated from detergent-insoluble fractions in the presence of 1.0% Triton X-100 at 4°C. The presence of VP40-WT and tsg101 in each fraction was determined by Western blotting (Fig. 7). In the absence of tsg101, VP40-WT was present in the soluble fraction but not in the insoluble fraction (Fig. 7, lanes 1 and 2). This finding suggests that association with detergent insoluble lipid fractions is not an intrinsic property of VP40. In contrast, tsg101 alone was present exclusively in the insoluble lipid fraction (Fig. 7, compare lanes 3 and 4), implying that tsg101 associates with detergent-insoluble membranes in the cell. Since tsg101 is not known to associate with detergent-insoluble matrices such as the cytoskeleton, it is reasonable to infer that tsg101 associates with lipid raft microdomains either directly or indirectly by interacting with another membrane-associated protein. This conclusion is supported further by flotation of detergent-extracted material in a discontinuous sucrose gradient, in which tsg101 was detected in low-density fractions representing lipid rafts (M. Simpson-Holley and R. N. Harty, unpublished data). Interestingly, when VP40-WT and tsg101 were coexpressed, VP40-WT was detected in both the soluble and the insoluble fractions (Fig. 7, lanes 5 and 6), whereas tsg101 remained within the insoluble fraction (lane 6). This finding suggests that tsg101 interacts with and recruits VP40-WT into lipid raft microdomains from which Ebola virus is released.
FIG. 7.
VP40-WT and tsg101 associate and colocalize in detergent-insoluble fractions. Vero cells were transfected with VP40-WT alone, tsg101 alone, or VP40-WT plus tsg101. Cells were lysed in 1.0% Triton X-100 at 4°C, and extracts were separated into detergent-soluble (S) and detergent-insoluble (I) fractions. VP40-WT (top panel) and tsg101 (bottom panel) were detected in soluble and/or insoluble fractions by Western blot analysis.
Dominant-negative mutant of VPS-4 inhibits budding of Ebola virus.
The vps4 ATPase is known to function downstream of tsg101 in the vps pathway within the cell (2, 3). It has been postulated that the enzymatic activity of vps4 may provide the energy necessary to promote the final pinching-off step of virus budding. Indeed, dominant-negative mutants of vps4 (E228Q blocks ATP hydrolysis and K173Q blocks ATP binding) have been shown to inhibit the budding of HIV-1 (14). We sought to determine whether a dominant-negative mutant of vps4 could inhibit budding of infectious Ebola virus.
Human 293T cells were first transfected with either vps4-WT or vps4-E228Q for 24 h and then infected with Ebola virus Zaire 1976 at an MOI of 1.0 (Fig. 8). Titers of infectious virus harvested from cell supernatants at the indicated times were determined by plaque assay. Viral titers from cells expressing vps4-WT reached 7.0 × 101, 4.5 × 104, and 6.0 × 106 PFU/ml on days 3, 5, and 7, respectively (Fig. 8). In comparison, viral titers from cells expressing the E228Q mutant reached 7.5 × 100, 5.0 × 103, and 5.0 × 105 PFU/ml on days 3, 5, and 7, respectively. This 10-fold difference in viral titer was reproducible in two independent experiments containing duplicate samples for each time point. It should also be noted that budding of VP40 alone was inhibited significantly in cells cotransfected with the E228Q mutant of vps4 (data not shown). Taken together, these data suggest that the ATPase activity of vps4 plays a role in facilitating efficient budding of Ebola virus.
FIG. 8.
A dominant-negative mutant of vps4 inhibits budding Ebola virus. Human 293T cells were transfected with vps4-WT (black bars) or vps4-E228Q (striped bars) for 24 h and then infected with Ebola virus Zaire 1976 at an MOI of 1.0. Medium samples were harvested at 3, 5, and 7 days postinfection, and titers of infectious virus were determined by plaque assays on Vero E6 cells. Cells transfected with vps4-WT yielded virus titers of 7.0 × 101, 4.5 × 104, and 6.0 × 106 PFU/ml on days 3, 5, and 7, respectively. Cells transfected with vps4-E228Q yielded virus titers of 7.5 × 100, 5.0 × 103, and 5.0 × 105 PFU/ml on days 3, 5, and 7, respectively. The virus titers shown represent the average of two independent experiments containing duplicate samples for each time point. Mock-transfected and Ebola virus-infected cells yielded virus titers of 2.0 × 107 PFU/ml on day 7 postinfection (data not shown).
DISCUSSION
The VP40 protein of Ebola virus is unique in that it contains two overlapping L-domain motifs at its N terminus; however, the relative contribution of each of these motifs in promoting budding was unclear. We present here evidence that the PTAP and PPEY motifs are functionally redundant and can act independently as L-domains. VP40-dPT/PY, which lacks the entire PTAPPEY sequence (Fig. 1), was barely detectable in budding VLPs. In contrast, VP40 proteins with mutations in either the PTAP or PPEY motifs were released in VLPs at levels virtually identical (≤2-fold) to that of VP40-WT, as determined by immunoprecipitation (Fig. 3) and Western blotting (data not shown). Confocal microscopy demonstrated that budding defects exhibited by specific VP40 mutants were not due to their mislocalization in transfected cells (Fig. 5) but rather to a disruption of L-domain activity. These results suggest that the PTAP and PPEY motifs of VP40 are functionally redundant in this budding assay. It has been reported previously that the PTAP motif of VP40 may be dominant over the PPEY motif (29).
To further determine whether the PPEY motif of VP40 could function independently as an L-domain, the PPEY motif was inserted adjacent to amino acid 316 at the C terminus of VP40-dPT/PY to yield VP40-c-PY (see Fig. 1). The PPEY motif within VP40-c-PY was able to provide L-domain function and promote budding of VP40-c-PY to levels greater than or equal to that of VP40-WT (Fig. 4). Thus, the PPEY motif not only can function independently as an L-domain but also is movable within full-length VP40. Similarly, the PTAPP motif can also function independently when placed at the C terminus of VP40 (Fig. 4). Therefore, we conclude that in the context of this functional budding assay, the PTAPP and PPEY motifs are functionally redundant. However, whether one motif is more dominant than the other in Ebola virus-infected cells remains to be determined. Lastly, not all VP40 proteins of Ebola and Marburg viruses contain two overlapping motifs. For example, the VP40 protein of Marburg virus (Popp strain) possesses only a PPxY motif at its N terminus (5), which likely serves as an L-domain.
Although the PTAP and PPEY motifs demonstrate redundancy with regard to budding, they are believed to serve as docking sites for mediating interactions with distinct cellular proteins (6, 12). The movable nature of the PTAPP and PPEY motifs within VP40 is consistent with their predicted role as docking sites for host proteins. Furthermore, the inability of VP40-Rev to bud indicates that the orientation of the amino acids sequences comprising the L-domain motifs is critical for efficient budding (Fig. 4), a finding consistent with their role as docking sites for host proteins.
The PPxY motif is known to interact physically and functionally with WW-domains of cellular proteins, particularly those present in the mammalian ubiquitin ligase, Nedd4 (13, 17, 18, 23, 34, 47). As a result of this virus-host interaction, VP40 and other viral matrix proteins are covalently modified by ubiquitin (16, 17, 33, 39, 40, 47). Increasing evidence suggests that mono-ubiquitination of viral matrix proteins plays a key role in facilitating virus budding (16, 17, 33, 39, 40, 47). On the other hand, the PTAP motif has been shown to interact with tsg101 (9, 14, 29, 43); a cellular protein that functions, in part, as a sorting component of the vps pathway (1). Intriguingly, tsg101 also possesses the ability to bind to mono-ubiquitinated proteins and is thought to direct these modified proteins into the vps pathway of the cell (22). Moreover, recent evidence suggests that tsg101 and Nedd4 may in fact interact in mammalian cells (6), providing a persuasive link between viral L-domains, ubiquitin, and the vps pathway.
In this report, coexpression of VP40 and tsg101 in mammalian cells yielded two important findings. First, VP40 was able to recruit tsg101 into budding VLPs (Fig. 6). A wild-type PTAP (VP40-WT) or mutant ATAP (VP40-P7A) motif, which can still interact with tsg101 (14), was required for recruitment of tsg101 into budding VLPs. These results are consistent with reported findings that a truncated form of tsg101 is recruited into HIV-1 virions via an interaction with the PTAP motif of HIV-1 gag (9). These results are also consistent with previous findings that VP40 colocalizes with tsg101 at the plasma membrane (29).
Second, a VP40-tsg101 interaction results in the redistribution of VP40 from the detergent-soluble fraction alone to both detergent-soluble and -insoluble fractions (Fig. 7). Interestingly, we demonstrate for the first time that tsg101 alone is present predominantly in detergent-insoluble fractions (Fig. 7). These findings are of particular significance, since it has been reported recently that Ebola virus buds and perhaps enters cells via lipid raft domains on the plasma membrane (4). Bavari et al. demonstrated that VP40 alone is not detectable in lipid rafts (4). In addition, their results suggest that GP of Ebola virus not only plays a role in directing VP40 into lipid rafts but also is responsible for efficient formation of filamentous, budding VLPs (4). Our finding that VP40 alone is not readily detectable in lipid rafts is consistent with those of Bavari et al. (4). However, our data and those of others clearly indicate that VP40 alone can bud from cells in the form of VLPs in the absence of GP (17, 19, 29, 30, 42). Although GP undoubtedly plays an important role in virus assembly and budding, GP is not essential for promoting the budding of VP40 in the form of VLPs (17, 19, 29, 30, 42). In addition, our data suggest that cellular proteins such as tsg101 may also contribute to the recruitment of VP40 into lipid raft domains. It is tempting to speculate that appropriate positioning of VP40 in the plasma membrane is dependent on its interaction with both cellular (tsg101) and viral (GP) proteins. Exactly where tsg101 and VP40 interact in the cell and the mechanism by which the tsg101-VP40 complex localizes to the plasma membrane remain to be determined.
Since the vps4 ATPase is one of the final effectors in the vps pathway and functions downstream of tsg101, we postulated that a dominant-negative mutant of vps4 may disrupt budding of VP40-containing VLPs. Dominant-negative mutants of vps4 have been shown to inhibit budding of infectious HIV-1 particles by more than 200-fold in cotransfection experiments, whereas WT vps4 inhibited budding of HIV-1 by only two- to fourfold (14). We demonstrate that overexpression of vps4-E228Q inhibits budding of infectious Ebola virus by 10-fold compared to that of WT vps4 (Fig. 8). One possibility for the observed difference in the levels of inhibition of budding between HIV-1 (200-fold) and Ebola virus (10-fold) is that the experimental conditions (e.g., cotransfection experiment for HIV-1 versus the transfection-infection experiment for Ebola virus) were not similar.
Based on data presented here and those of others (17, 19, 29, 30, 42), it is clear that efficient budding of Ebola virus is dependent on functional L-domains within VP40 and on their ability to interact with and recruit host proteins. Studies are ongoing to determine whether mono-ubiquitination of VP40 mediated by a PPEY-WW-domain interaction occurs in vivo and whether ubiquitin or ubiquitinated forms of VP40 are present in Ebola virions. In addition, experiments to elucidate the importance of amino acids surrounding the L-domains of VP40 in promoting virus budding are under way. An interesting observation from our studies is that VP40 proteins containing a free C terminus were released as VLPs more efficiently than those containing a free N terminus. This may reflect the need for the free C terminus of VP40 to interact effectively with either the plasma membrane or perhaps with itself to promote budding. The composition of amino acids flanking the L-domain motifs may also affect the efficiency of VP40 budding. Lastly, although VP40 is unique in that it contains overlapping L-domains, other viral matrix proteins contain more than one potential L-domain. For example, the M protein of VSV contains a PSAP motif just downstream of the well-characterized PPxY motif (18). It will be of interest to determine whether viral matrix proteins in general possess functionally redundant L-domains.
Acknowledgments
We thank P. Bates and W. Sundquist for kindly providing us with reagents and members of the Bates laboratory for fruitful discussions.
J.M.L. was supported by NIH training grant AI007324. J.P. is the recipient of a National Research Council Fellowship. R.N.H. was supported in part by NIH grant AI46499.
REFERENCES
- 1.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., T. K. Sato, L. M. Banta, and S. D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukreyev, A. A., V. E. Volchkov, V. M. Blinov, S. A. Dryga, and S. V. Netesov. 1995. The complete nucleotide sequence of the Popp (1967) strain of Marburg virus: a comparison with the Musoke (1980) strain. Arch. Virol. 140:1589-1600. [DOI] [PubMed] [Google Scholar]
- 6.Carter, C. A. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203-205. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmann, H., H. D. Klenk, and A. Sanchez. 1993. Molecular biology and evolution of filoviruses. Arch. Virol. Suppl. 7:81-100. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature 381:744-745. [DOI] [PubMed] [Google Scholar]
- 14.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Justice, P. A., W. Sun, Y. Li, Z. Ye, P. R. Grigera, and R. R. Wagner. 1995. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J. Virol. 69:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 23.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenk, H. D. 2000. Will we have and why do we need an Ebola vaccine? Nat. Med. 6:1322-1323. [DOI] [PubMed] [Google Scholar]
- 25.Kolesnikova, L., H. Bugany, H. D. Klenk, and S. Becker. 2002. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 76:1825-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondo, E., F. Mammano, E. A. Cohen, and H. G. Gottlinger. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 69:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, M. A., and M. Huang. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Biol. Chem. 270:23883-23886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 30.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patnaik, A., and J. W. Wills. 2002. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 76:2789-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile: assimilation of cellular machinery by HIV-1. Immunity 15:687-690. [DOI] [PubMed] [Google Scholar]
- 36.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada, A., and Y. Kawaoka. 2001. The pathogenesis of Ebola hemorrhagic fever. Trends Microbiol. 9:506-511. [DOI] [PubMed] [Google Scholar]
- 42.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of ebola virus matrix protein VP40. Virology 283:1-6. [DOI] [PubMed] [Google Scholar]
- 43.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]