Abstract
The V protein of the paramyxovirus simian virus 5 blocks interferon (IFN) signaling by targeting STAT1 for proteasome-mediated degradation. Here we report on the isolation of human cell lines that express the V protein and can no longer respond to IFN. A variety of viruses, particularly slow-growing wild-type viruses and vaccine candidate viruses (which are attenuated due to mutations that affect virus replication, virus spread, or ability to circumvent the IFN response), form bigger plaques and grow to titers that are increased as much as 10- to 4,000-fold in these IFN-nonresponsive cells. We discuss the practical applications of using such cells in vaccine development and manufacture, virus diagnostics and isolation of newly emerging viruses, and studies on host cell tropism and pathogenesis.
Interferons (IFNs) are produced in response to virus infections and are powerful biological mediators, inducing an antiviral state in cells and influencing the subsequent immune response. Alpha and beta IFN (IFN-α/β) are produced by many cell types and have particular importance as an early line of innate immune defense; it is this type of IFN that is produced by nonlymphoid tissue culture cells. Gamma IFN (IFN-γ) is produced by subsets of lymphocytes and plays a more prominent role in regulating the adaptive immune response. To survive in nature, it appears that all viruses must have some strategy for circumventing the IFN response, particularly the innate antiviral defense induced by IFN-α/β. Viruses usually achieve this by producing proteins which either interfere with the ability of IFNs to induce an antiviral state within cells or block the activity of antiviral enzymes which have the potential to inhibit virus replication (2, 10, 11). Many paramyxoviruses at least partially circumvent the IFN response by blocking IFN-induced intracellular signaling and/or IFN production. For example, simian virus 5 (SV5) blocks IFN signaling by targeting STAT1 (a host cell transcription factor essential for both IFN-α/β and IFN-γ signaling) for proteasome-mediated degradation (7, 8). Since this is a property solely of the V protein, it is possible to make cells insensitive to IFN by constitutively expressing the V protein of SV5 (1).
Vaccines have proved extremely successful in controlling many virus infections. However, vaccines still have to be developed against many viruses, including, among the negative-strand RNA viruses, respiratory syncytial virus (RSV), the parainfluenza viruses, Ebola virus, and members of the Bunyaviridae family, including Hantavirus. It would also be desirable to develop improved vaccines for a number of negative-strand RNA viruses, including the measles, mumps, and influenza viruses. One of the most successful approaches to producing virus vaccines has been the generation of attenuated viruses, which are administered to mimic natural infection and induce protective immunity without causing disease. In the past, the generation of attenuated viruses has been empirical. However, with the advent of recombinant technology, the possibility of designing attenuated viruses that have been engineered to possess specific phenotypes has become a reality. One general approach to producing attenuated viruses would be to engineer viruses so as to disable their capacity to circumvent the IFN response (7, 9). This is feasible because viral anti-IFN proteins are usually dispensable for virus replication in cell culture, and viruses in which the genes encoding these proteins have been knocked out are attenuated in vivo. However, one problem which arises from knocking out viral IFN resistance genes is that it can be difficult to grow such viruses to high titer in tissue culture cells which produce and respond to IFN. Normally, IFN-sensitive viruses are grown in Vero cells, which are African green monkey fibroblasts that have lost the ability to produce IFN due to spontaneous gene deletions (6, 12). However, not all viruses grow well in Vero cells, presumably because of other host cell constraints on virus replication. Here we demonstrate how cells, including MRC5 cells (which are human diploid cells suitable for the production of attenuated viruses for use in humans), can simply and easily be engineered to express the V protein of SV5 and thus to no longer respond to IFN. We demonstrate that RSVs from which the NS1, NS2, SH, or glycoprotein G gene has been deleted (5, 17, 19, 20), a Bunyamwera (BUN) virus from which the NSs gene has been deleted (4), and a recombinant SV5 virus (SV5VΔC; generated so as to make a truncated version of the V protein and thus to be sensitive to IFN [10a]) form larger plaques and can be grown to much higher titers in these IFN-nonresponsive cells than in the parental cells. Furthermore, many of the wild-type (wt) RNA and DNA viruses that were evaluated formed significantly larger plaques in the V-expressing cells.
MATERIALS AND METHODS
Cells and viruses.
Human 2fTGH cells (14), MRC5 cells, and HEp2 cells and their derivatives were grown as monolayers in 25- or 75-cm2 tissue culture flasks, in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. All cell lines were negative for mycoplasmas as screened by 4′,6′-diamidino-2-phenylindole (DAPI) staining. Cells expressing the V protein of SV5 were isolated as previously described (5). Briefly, cells were transfected with the plasmids pEF.SV5-V.IRES.neo and cultured in the presence of 400 μg of Geneticin (G418; Sigma)/ml, and resistant colonies were isolated. The colonies were then screened for expression of the V protein by immunofluorescence, and only colonies in which the V protein could be detected in 100% of the cells were selected. All viruses used in this study were grown and titrated under appropriate conditions. The details of the recombinant RSV, Bunyamwera, and SV5 viruses have been described elsewhere (4, 5, 17, 18, 20; He et al., submitted).
Immunofluorescence.
Cells to be stained for immunofluorescence were grown on 10-mm-diameter coverslips (General Scientific Co. Ltd.). The cells were treated and stained with the anti-Pk monoclonal antibody (15). Antibody binding was detected by indirect immunofluorescence by using a secondary goat anti-mouse immunoglobulin Texas Red-conjugated antibody (catalog number SBA 1010-02; Seralab). Following staining for immunofluorescence, the monolayers of cells were examined by using a Nikon Microphot-FXA immunofluorescence microscope.
RESULTS
We speculated that in cells that produce and respond to IFN, the efficiency of replication of a virus is strongly influenced by two opposing processes: on the one hand, the relative kinetics of IFN production and induction of an antiviral state, and, on the other hand, the speed of virus replication and virus-induced inhibition of the IFN response. As a consequence, cells which cannot respond to IFN may be better able to support the replication of viruses, particularly those which are sensitive to IFN or which replicate slowly due to innate properties or due to the presence of attenuating mutations. To test this, we compared the yields of several wt and attenuated viruses, namely, BUNwt, BUNΔNSs (in which the NSs gene has been deleted and which has increased sensitivity to IFN [19]), human RSVwt, and derivatives in which the NS1, NS2, or SH gene has been deleted (RSVΔNS1, RSVΔNS2, and RSVΔSH, respectively) (5, 17, 20). These viruses were compared in human fibrosarcoma cells (2fTGH [13]) and 2f/SV5-V cells, which are derivatives of 2fTGH cells that cannot respond to IFN because they express the V protein of SV5 (1), as well as in Vero cells. It was found that whereas there was no major difference between the yield of BUNwt grown in 2fTGH and that grown in 2f/SV5-V cells, there was more than a 3 log increase in the yield of BUNΔNSs in 2f/SV5-V cells compared to that in 2fTGH cells at 3 days postinfection (p.i.) (Table 1). Similarly, RSVΔNS1, RSVΔNS2, and RSVΔSH grew to significantly higher titer on 2f/SV5-V cells than on 2fTGH cells. It is also of note that RSVwt grew to slightly higher titers on 2f/SV5-V cells than on 2fTGH cells, both of which produced more virus than Vero cells by this time p.i. (Table 1).
TABLE 1.
Amount of infectious virus released into the medium by 3 days p.i. from cells infected at a multiplicity of infection of 0.01a
| Virus | Amt of virus (PFU/cell) released from:
|
||
|---|---|---|---|
| 2fTGH cells | 2f/SV5-V cells | Vero cells | |
| BUNwt | 45 | 50 | 6 |
| BUNΔNSs | 0.002 | 2 | 3 |
| RSVwt | 13 | 90 | 0.1 |
| RSVΔNS1 | <0.01 | 3 | 0.01 |
| RSVΔNS2 | <0.01 | 0.2 | <0.01 |
| RSVΔSH | 0.02 | 7 | <0.01 |
Monolayers of cells growing in 25-cm2 flasks were infected at a multiplicity of infection of 0.01 PFU/cell for 1 h, after which time the inoculum was replaced with 10 ml of fresh maintenance medium. The cells were then incubated at 37°C with continuous rocking. At 3 days p.i., the medium was harvested and the amount of infectious virus released into the medium was immediately titrated by plaque assays on Vero cells.
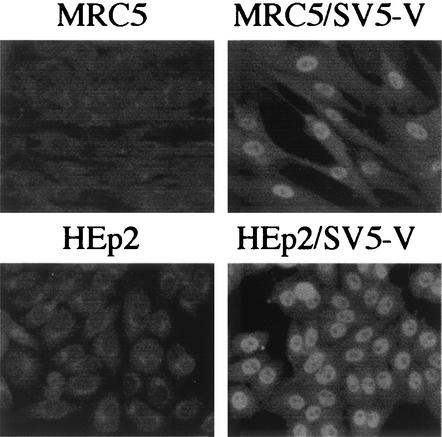
These results established the principle that significant increases in the yields of viruses may be achieved by growth in cells which cannot respond to IFN. However, 2fTGH cells have not been licensed for use in vaccine production and are not used in diagnostic laboratories for the isolation of viruses from clinical samples. We therefore decided to develop versions of two commonly used cell lines, namely, MRC5 and HEp2, that express the V protein of SV5. These engineered cell lines, termed MRC5/SV5-V and HEp2/SV5-V, were isolated by previously described methods (1). It was confirmed (i) by immunofluorescence that all the cells expressed the V protein (Fig. 1), (ii) by immunoblot analysis that STAT1 could not be detected, and (iii) by IFN reporter gene assays that the cells could not signal in response to IFN (data not shown).
FIG. 1.
Photomicrographs showing the intracellular localization of the V protein of SV5 in MRC5/SV5-V and HEp2/SV5-V cells. Note that all the cells are positive for V and that V has a primarily nuclear distribution.
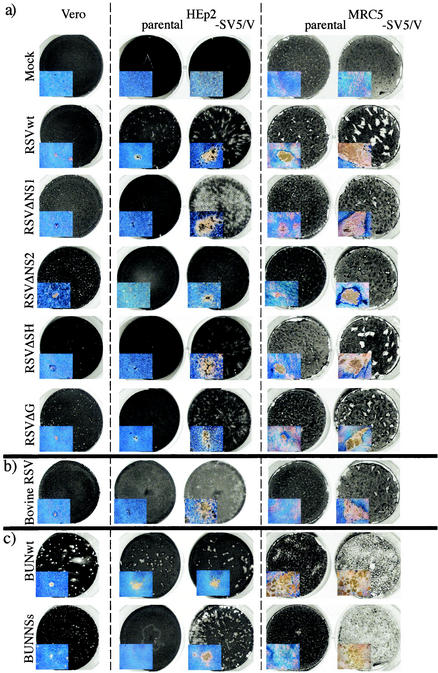
Initially, to test the potential of these cells for growing mutant viruses, BUNwt, BUNΔNSs, RSVwt, RSVΔNS1, RSVΔNS2, RSVΔSH, and a derivative of RSV in which the G gene had been deleted (RSVΔG) were grown on all three cells types. In addition, bovine RSV also was grown on these cells, as it had previously been shown that bovine RSV is more sensitive to the IFN response in human cells than in bovine cells. Plaques were allowed to develop for 6 days, and the results revealed large increases in plaque size for most of the viruses in IFN nonresponder cells (Fig. 2). Thus, by 6 days p.i., RSVwt formed visible plaques on HEp2 and MRC5 cells, but the plaques were considerably larger on HEp2/SV5-V and MRC5/SV5-V cells (3). This indicated that although RSV has a mechanism for circumventing the IFN response that relies on the function of the NS1 and NS2 genes (16), its effect is apparently incomplete and can be augmented by expression of the SV5 V protein in trans. It also is noteworthy that RSVwt formed only pinpoint plaques on Vero cells by 6 days p.i. despite the absence of an IFN response in these cells. This indicates the presence of cell-specific differences in the ability of Vero cells to support RSV growth that are separate from the IFN system, and it illustrates the value of being able to readily convert desirable cell lines to be IFN nonresponsive.
FIG.2.
Photographs of plaques of RSVwt, RSVΔNS1, RSVΔNS2, RSVΔSH, and RSVΔG (a), bovine RSV (b), and BUNwt and BUNΔNSs (c) formed on Vero, HEp2, HEp2/SV5-V, MRC5, and MRC5/SV5-V cells grown in six-well petri dishes. The cells were fixed at 6 days p.i., or at 9 days p.i. in the case of cells infected with RSVΔNS2, and stained with Coomassie brilliant blue. The inserts are photographs of single plaques taken on an inverted microscope at 4× magnification.
The increase in plaque size achieved in the IFN nonresponder cells with RSVΔNS1, RSVΔSH, and RSVΔG was even more striking. Each of these viruses produced pinpoint plaques on Vero and HEp2 cells and somewhat larger plaques on MRC5 cells. In contrast, each of these three RSV deletion mutants produced very large plaques on HEp2/SV5-V and MRC5/SV5-V cells. The situation with RSVΔNS2 was slightly different in that, although the plaques that formed on HEp2/SV5-V and MRC5/SV5-V cells were significantly larger than those formed on HEp2 or MRC5 cells, it took longer for the plaques to develop (note for Fig. 2 that the RSVΔNS2 samples were fixed at 9 days p.i. as opposed to 6 days p.i. for all the other viruses). This suggests that while knocking out the NS2 gene reduced its ability to circumvent the IFN response, it also is deleterious to virus replication by an additional mechanism that likely does not involve IFN.
A striking increase in plaque size on the IFN nonresponder cells also was seen with bovine RSV. While bovine RSV produced pinpoint plaques at 6 days p.i. in Vero cells and did not produce detectable plaques in either HEp2 or MRC5 cells, very large plaques developed in both HEp2/SV5-V and MRC5/SV5-V cells. In contrast, BUNwt formed plaques of similar size on Vero cells and on HEp2 cells that did or did not express the V protein of SV5. However, the plaques formed by BUNwt on MRC5 cells were extremely diffuse, and in this experiment BUNwt clearly spread more rapidly in MRC5/SV5-V cells, destroying most of the monolayer by 6 days p.i. BUNΔNSs formed pinpoint plaques on Vero cells and failed to form plaques on HEp2 cells but formed large plaques on HEp2/SV5-V cells that were similar in size to those of the BUNwt virus. BUNΔNSs also failed to form plaques on MRC5 cells but destroyed the MRC5/SV5-V monolayer of cells.
Since clear differences were observed between the abilities of the RSV and BUN mutant viruses, as well as those of wt human and bovine RSV, to form plaques on HEp2/SV5-V and MRC5/SV5-V cells and their abilities to form plaques on HEp2 and MRC5 cells, it was decided to compare the amounts of infectious virus released into the medium following infection of these cells with selected viruses (BUNwt and BUNΔNSs, RSVwt, RSVΔNS1, RSVΔSH, and bovine RSV) (Table 2). In general agreement with the results obtained by comparing the yields of these viruses on 2fTGH and 2f/SV5-V cells (Table 1), there was no significant difference between the yields of BUNwt virus in HEp2 and MRC5 cells and the yields in HEp2/SV5-V and MRC5/SV5-V cells. In contrast, there was a 1,000- to 4,000-fold increase in the yield of BUNΔNSs in MRC5/SV5-V and HEp2/SV5-V cells compared to that in MRC5 and HEp2 cells, respectively. Similarly, there was an approximately 200-fold increase in the yield of RSVΔNS1 and RSVΔNS2 in HEp2/SV5-V cells compared to that in HEp2 cells, and there was a 500-fold increase the yield in MRC5/SV5-V cells compared to that in MRC5 cells. The increase in RSVΔSH and RSVΔG was not quite as dramatic, with a 10-fold difference observed between yields in HEp2/SV5-V and HEp2 cells and no difference between yields in MRC5/SV5-V and MRC5 cells. Interestingly, HEp2/SV5-V cells were also better able to support the replication of RSVwt than HEp2 cells (an eightfold difference) and were clearly the best cells for growing RSVwt under these conditions. Furthermore, the yields of bovine RSV at 6 days p.i. in HEp2/SV5-V and MRC5/SV5-V were significantly higher than those in the parental cell lines. This was most marked with MRC5 cells, whose parental line produced hardly any infectious virus. In contrast, the yield of bovine RSV was 2 PFU per cell from MRC5/SV5-V cells. These latter results are thus in general agreement with those recently published by Bossert and Conzelmann (3), who presented data which showed that the NS1 and NS2 proteins of bovine RSV, which act in concert to circumvent the IFN response (16), work poorly in human cells. In contrast, the NS1 and NS2 proteins of human RSV would be more effective in blunting the IFN response in the homologous human HEp2 and MRC5 cells, thus accounting for the higher titers obtained for human RSVwt in the parental cells and the lesser increase in titer conferred by expression of the SV5 V protein.
TABLE 2.
Amount of infectious virus released into the medium by 3 days p.i. from cells infected at a multiplicity of infection of 0.01a
| Virus | Amt of virus (PFU/cell) released from the indicated cells
|
||||
|---|---|---|---|---|---|
| Vero | HEp2 | HEp2/V | MRC5 | MRC5/V | |
| RSVwt | 0.2 | 6 | 50 | 1 | 1 |
| RSVΔNS1 | 0.2 | 0.03 | 7 | 0.001 | 0.5 |
| RSVΔSH | 1 | 5 | 50 | 1 | 1 |
| RSVΔNS2 | 0.2 | 0.03 | 7 | 0.001 | 0.5 |
| RSVΔG | 1 | 5 | 50 | 1 | 1 |
| Bovine RSV | 6 | 0.6 | 20 | 0.00003 | 2 |
| BUNwt | 3 | 0.5 | 0.2 | 10 | 10 |
| BUNΔNSs | 1 | 0.0001 | 0.4 | 0.001 | 1 |
Monolayers of cells growing in 25-cm2 flasks were infected at a multiplicity of infection of 0.01 PFU/cell for 1 h, after which time the inoculum was replaced with 10 ml of fresh maintenance medium. The cells were then incubated at 37°C with continuous rocking. At 3 days p.i., the medium was harvested and the amount of infectious virus released into the medium was estimated by plaque assays on HEp2/SV5-V cells.
To further examine how the ability to produce and respond to IFN influences the replication of viruses, a variety of human and animal RNA and DNA viruses were assayed for plaque formation on the various cell lines described (Fig. 3). In no situation did expression of the SV5 V protein in cells decrease plaque size, and in most cases the IFN-nonresponsive cells provided increased growth, as assayed by plaque size. A wt measles isolate formed similarly sized plaques on HEp2 and HEp2/SV5-V cells but formed significantly larger plaques on MRC5/SV5-V cells than on MRC5 cells (it should be noted that the efficiency of plaque formation was lower on MRC5 and MRC5/SV5-V cells than on Vero or HEp2 cells). A closely related animal morbillivirus, canine distemper virus (CDV), formed large plaques on Vero cells and formed pinpoint plaques on HEp2 cells but failed to form plaques on MRC5 cells. CDV formed bigger plaques on HEp2/SV5-V cells than on HEp2 cells and formed large plaques on MRC5/SV5-V cells.
FIG. 3.
Photographs of plaques of a variety of RNA and DNA viruses on Vero, HEp2, HEp2/SV5-V, MRC5, and MRC5/SV5-V cells grown in six-well petri dishes. The cells were fixed at various days p.i. (indicated in parentheses) and stained with Coomassie brilliant blue. Photographs were taken on an inverted microscope at 4× magnification. The viruses used in the plaque assays were measles virus (MeV, a wt isolate), canine distemper virus (CDV), SV5 (strain W3), hPIV2 (a wt and a recent clinical isolate, 5234), mumps virus (a wt and the Enders strain [att]), hPIV3 (PIV3), Theiler's virus, adenovirus type 2, HSV (strain STH2), and vaccinia virus (VV).
Mumps, human parainfluenza virus type 2 (hPIV2), and SV5 all belong to the Rubulavirus genus of paramyxoviruses. Whereas wt SV5 formed small plaques on MRC5 cells, the recombinant virus SV5VΔC that encodes only the N-terminal domain of V and does not block IFN signaling (10a, 13a) failed to form plaques on MRC5 cells but formed large plaques on MRC5/SV5-V cells. Mumps and hPIV2 (both a laboratory-adapted strain [wt] and a recent clinical isolate [5234]) also failed to form plaques on MRC5 cells but formed large plaques on MRC5/SV5-V cells. Since each of these wt viruses blocks IFN signaling and reduces IFN production (1, 7, 8, 10a, 13a, 21), the natural block is shown to be leaky and can be supplemented by SV5 V protein expressed in trans. hPIV3, a member of the Respirovirus genus, formed slightly larger plaques on HEp2/SV5-V than on HEp2 cells, and while it caused clear plaques on MRC5 cells, the plaques were extremely large on MRC5/SV5-V cells.
Theiler's virus, a rodent picornavirus, failed to form plaques in Vero cells and formed small plaques on HEp2, slightly larger plaques on HEp2/SV5-V cells, large plaques on MRC5 cells, and extremely large plaques on MRC5/SV5-V cells. Of the DNA viruses examined, vaccinia virus formed plaques on all cells, although the plaques were slightly larger on HEp2/SV5-V cells than on HEp2 cells. Herpes simplex virus (HSV) grew equally well in Vero, MRC5, and MRC5/SV5-V cells but formed only pinpoint plaques in HEp2 and HEp2/V cells. By 8 days p.i., adenovirus type 2 had failed to form plaques on Vero cells and formed pinpoint plaques on HEp2 and MRC5 cells, but it formed larger plaques on HEp2/SV5-V and MRC5/SV5-V cells. It is also of note that certain viruses failed to form plaques on Vero cells (Theiler's virus and adenovirus type 2) or HEp2/V cells (HSV), illustrating that there are host cell constraints other than the IFN response which may limit virus growth, although the IFN response may amplify the effects of these constraints.
DISCUSSION
We engineered cell lines that are commonly used in virus diagnostics and vaccine manufacture to be nonresponsive to IFN by the constitutive expression of the SV5 V protein, a viral protein that promotes degradation of STAT1 and thereby blocks IFN signaling. These engineered lines supported an increase in plaque size and virus yield for most of the viruses tested from a large, diverse panel. The effects were particularly striking for viruses that replicate inefficiently in vitro, whether due to intrinsic properties or to the presence of mutations or defects that adversely affect virus replication, virus spread, or the ability to circumvent the IFN response.
One unexpected finding was that the effect of improved growth appeared to be a general one. Thus, it was not restricted to viruses with defects in genes known to be involved in circumventing the IFN response, such as the RSVΔNS1, RSVΔNS2, BUNΔNSs, and SV5VΔC viruses, although the effects on these viruses were particularly striking; rather, the effect also applied to viruses that had mutations in genes that do not appear to be directly relevant to IFN responses, such as the RSVΔG and RSVΔSH viruses. Furthermore, for the majority of wt viruses tested, it is clear that their ability to circumvent the IFN response is not absolute, as they formed significantly larger plaques on IFN-nonresponsive cells. This supports the idea that viruses in general face a race between virus growth and the establishment of an IFN-mediated antiviral state. Thus, any viral defect, be it in attachment, entry, RNA synthesis, or packaging, has the potential to tip the balance in this race against the virus. This implies that in a normal acute infection, the IFN response likely constitutes a constant selective pressure that keeps viruses maximally fit in terms of replication speed and competence, as well as in the maintenance of mechanisms to circumvent the IFN response. It also suggests that the efficiency by which a given virus overcomes the IFN response may be extremely important in terms of its pathogenesis and host range.
These findings have direct relevance for vaccine development. By generating cell lines which cannot respond to IFN, it is possible to significantly increase the yield of viruses, especially those which grow poorly due to mutations such as those which adversely effect virus replication, spread, or ability to circumvent the IFN response. In addition, we and others (7, 9) suggested that a general approach to making attenuated virus vaccines would be to knock out the genes that allow them to circumvent the IFN response. This strategy would likely necessitate that such IFN-sensitive viruses be grown in IFN nonresponder cells. However, as noted above, any defect that reduces the replication competence of a virus makes it a candidate to exhibit improved growth in an IFN-nonresponsive cell line. While Vero cells may be used for growing IFN-sensitive viruses, from the results presented here, it is clear that alternative IFN-nonresponsive cells can be generated relatively easily and may be substantially better able to support the growth of some attenuated virus vaccines. Indeed, one of the reasons for making MRC5/SV5-V cells was because MRC5 cells are diploid cells and are one of the few human cell lines that have been approved for vaccine production. Given that MRC5/SV5-V cells appear better able to support the growth of viruses than MRC5 cells, it may therefore be possible, following proper regulatory certification, to substitute MRC5 cells with MRC5/SV5-V cells (or their equivalent) in current manufacturing practices to enhance the production of certain attenuated virus vaccines.
MRC5 and HEp2 cells are often the cells of choice for isolating viruses from clinical samples. Since viruses which replicate poorly in MRC5 and HEp2 cells may form plaques much more efficiently in the MRC5/SV5-V or HEp2/SV5-V cells, these IFN nonresponder cells might be of great value in diagnostic laboratories for the rapid isolation of known and/or unknown viruses. Furthermore, in situations of cross-species infection, the IFN response is a major constraint that may prevent animal viruses from replicating efficiently in human cells, in which the operation of their IFN resistance genes can be suboptimal due to host differences (7). This was further illustrated in the present study by the greater sensitivity of bovine RSV, compared to human RSV, to the IFN system of the human cells. Thus, IFN nonresponder cell lines likely will provide a more sensitive method of isolating nonhuman viruses that have the potential to be pathogenic in humans. In addition, by using IFN-nonresponsive human cells, it will also be possible to assess the ability of animal viruses to replicate in human cells and thereby pose a potential threat to cross into the human population (if they adapt to overcome the human IFN response).
Following the themes developed above, another potential application for IFN nonresponder cell lines is in the cultivation of viruses which have previously been refractory to replication in vitro, such as certain hepatitis viruses or caliciviruses. While it is likely that these viruses do not grow in tissue culture cells due to factors in addition to the IFN system, it might be that elimination of the IFN antiviral defense will be sufficient to allow suboptimal growth to become significant. Similarly, the general approach of making IFN-nonresponsive cells by expressing IFN antagonists, such as the V protein of SV5, may be extended to animal cells to improve the growth of animal viruses which grow poorly in the currently available human or animal cell lines.
Finally, there has been some concern about the generation of certain novel recombinant viruses, such as ones in which genes have been swapped. Such debate would now have to include the exchange of virus genes which encode IFN antagonists. The perceived danger is that such viruses may have altered tissue tropism and/or pathogenicity (14). However, a general strategy to ensure that any recombinant virus generated is attenuated would be to create each recombinant by using an IFN-sensitive viral backbone and to grow the recombinant viruses in IFN-nonresponsive cells. For example, if gene manipulation were performed on bunyaviruses to study the function of the glycoprotein gene, it would be possible to undertake such studies in the IFN-sensitive NSs deletion virus and to grow and evaluate the recombinant viruses in IFN-nonresponsive cells, secure in the knowledge that any virus generated would be attenuated. Furthermore, IFN-sensitive vectors expressing foreign genes could also be used as safe recombinant virus vaccines.
Acknowledgments
L. Andrejeva and D. F. Young are supported by grants from the BBSRC and Wellcome Trust. This work was supported in part by research grant AI-23173 from the National Institute of Allergy and Infectious Disease (R.A.L.). R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gammaI interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron, C. A., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-349. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 3.Bossert, B., and K. K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol 76:4287-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, P. L., and B. R. Murphy. 2002. Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 296:204-211. [DOI] [PubMed] [Google Scholar]
- 6.Desmyter, J., J. L. Melnick, and W. E. Rawls. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 10.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 10a.He, B., G. Reay, R. G. Paterson, N. Stock, J. Durbin, R. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-β induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 11.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and the mechanisms of viral evasion. Cytokines Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 12.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 14.Randall, R. E. 1998. Genetic rescue of enveloped RNA viruses: potentials and consequences. ASM News 64:547-548. [Google Scholar]
- 15.Randall, R. E., D. Young, K. K. A. Goswami, and W. C. Russell. 1987. Isolation and characterisation of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 16.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize the alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng, M. N., S. S. Whitehead, A. Bermingham, M. St. Claire, W. R. Elkins, B. R. Murphy, and P. L. Collins. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or the M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus (RSV) G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 189:283-296. [DOI] [PubMed] [Google Scholar]
- 19.Weber, F., A. Bridgen, J. K. Fazakereley, H. Stritenfeld, N. Kessler, R. E. Randall, and R. A. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead, S. S., M. G. Hill, C. Y. Firestone, M. St. Claire, W. R. Elkins, B. R. Murphy, and P. L. Collins. 1999. Recombinant respiratory syncytial virus (RSV) bearing a deletion in either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]