Abstract
Cellular immune responses play a critical role in the control of human immunodeficiency virus type 1 (HIV-1); however, the breadth of these responses at the single-epitope level has not been comprehensively assessed. We therefore screened peripheral blood mononuclear cells (PBMC) from 57 individuals at different stages of HIV-1 infection for virus-specific T-cell responses using a matrix of 504 overlapping peptides spanning all expressed HIV-1 proteins in a gamma interferon-enzyme-linked immunospot (Elispot) assay. HIV-1-specific T-cell responses were detectable in all study subjects, with a median of 14 individual epitopic regions targeted per person (range, 2 to 42), and all 14 HIV-1 protein subunits were recognized. HIV-1 p24-Gag and Nef contained the highest epitope density and were also the most frequently recognized HIV-1 proteins. The total magnitude of the HIV-1-specific response ranged from 280 to 25,860 spot-forming cells (SFC)/106 PBMC (median, 4,245) among all study participants. However, the number of epitopic regions targeted, the protein subunits recognized, and the total magnitude of HIV-1-specific responses varied significantly among the tested individuals, with the strongest and broadest responses detectable in individuals with untreated chronic HIV-1 infection. Neither the breadth nor the magnitude of the total HIV-1-specific CD8+-T-cell responses correlated with plasma viral load. We conclude that a peptide matrix-based Elispot assay allows for rapid, sensitive, specific, and efficient assessment of cellular immune responses directed against the entire expressed HIV-1 genome. These data also suggest that the impact of T-cell responses on control of viral replication cannot be explained by the mere quantification of the magnitude and breadth of the CD8+-T-cell response, even if a comprehensive pan-genome screening approach is applied.
Accumulating data indicate a central role of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses in controlling viral replication (13, 29, 31, 35, 37, 38, 41, 47). Studies in the simian immunodeficiency virus (SIV)-macaque model show that removal of CD8+ cells in vivo results in a rapid increase in the steady-state level of SIV viremia (27, 43). In humans, HIV-1-specific CD8+-T-cell responses have been associated with the decline in viremia during acute HIV-1 infection (13, 31). The selective immune pressure mediated by CD8+ T cells on the viral genome has been further supported by studies demonstrating viral escape from CD8+-cytotoxic T-cell (CTL)-mediated immune pressure in HIV-1 and SIV infection (3, 9, 14, 23, 24, 40). Strong virus-specific CTL responses have furthermore been demonstrated in HIV-1-infected individuals with long-term nonprogressive disease (26) and have been inversely correlated with HIV-1 viremia in some studies (19, 37). However, other studies did not detect an inverse association between viral load and HIV-1-specific CD8+ cell responses (11), and the matter remains controversial. In addition, to date most studies investigating HIV-1-specific T-cell responses have focused either on single peptides (37), restricted panels of optimal CTL epitopes (17, 18), or a limited selection of HIV proteins (19), mainly structural in nature. No studies have examined the full breadth of CD8-T-cell responses at the individual epitope level, or even at the protein subunit level, in a large number of infected individuals, and thus, the relationship between breadth of responses and viral load remains undefined.
In this report we comprehensively characterized HIV-1-specific T-cell responses in 57 HIV-1-infected individuals at different stages of HIV-1 infection, using a set of 504 overlapping peptides spanning all HIV-1 proteins (p17-Gag, p24-Gag, p15-Gag, protease, reverse transcriptase (RT), integrase, Vif, Vpr, Vpu, Tat, Rev, gp41-Env, gp120-Env, and Nef), and determined the individual regions containing T-cell epitopes for each individual. To facilitate this analysis, we adapted and evaluated a peptide pool matrix-based screening approach that allows for a comprehensive characterization of T-cell responses using a limited number of peripheral blood mononuclear cells (PBMC) in a gamma interferon (IFN-γ) enzyme-linked immunospot (Elispot) assay. Our results demonstrate that with sensitive assays, HIV-1-specific T-cell responses are detectable in all infected study subjects, and all HIV-1 proteins and in fact all protein subunits can serve as targets for the virus-specific immune response. Both the magnitude and breadth of responses for each individual displayed a wide spectrum and differed significantly among study groups. Although immunodominant proteins and epitope-rich regions within the HIV-1 genome were readily apparent, no correlation between viral load and breadth of cellular immune responses was found.
MATERIALS AND METHODS
Subjects studied.
Blood samples from 57 HIV-1-infected individuals at different stages of HIV-1 infection were obtained and investigated in this study. These included 22 study subjects diagnosed and treated during acute or early HIV-1 infection, 11 of whom received continuous highly active antiretroviral therapy and 11 of whom had undergone one or two supervised treatment interruptions (STI). Of the remaining 35 chronically HIV-1-infected individuals, 23 were untreated and 12 received antiretroviral therapy. Study subjects were recruited from the Massachusetts General Hospital (MGH), the Fenway Community Health Center, and the Lemuel Shattuck Hospital in Boston. Relevant clinical and demographic data of all study subjects are summarized in Table 1. Ten HIV-1-seronegative individuals were studied as control subjects. The study was approved by the respective institutional review boards, and all subjects gave written informed consent.
TABLE 1.
Clinical and demographical information about the study subjectsa
| Subject | Eth- nicity | Gender | Risk factor | Plasma viral load (RNA copies/ml)b | CD4-cell counts (cells/mm3)b | Treatment |
|---|---|---|---|---|---|---|
| AC 21 | C | m | msm | <50 | 990 | HAART |
| AC 31 | C | m | msm | <50 | 676 | HAART |
| AC 32 | C | m | msm | <50 | 467 | HAART |
| AC 34 | C | m | msm | <50 | 304 | HAART |
| AC 43 | C | m | msm | <50 | 530 | HAART |
| AC 52 | C | m | msm | <50 | 367 | HAART |
| AC 68 | C | m | msm | <50 | 389 | HAART |
| AC 85 | C | m | msm | <50 | 1,023 | HAART |
| AC 88 | C | m | msm | <50 | 365 | HAART |
| AC 100 | C | m | msm | 1570 | 801 | HAART |
| AC 101 | C | m | msm | 3,580 | 426 | HAART |
| AC 04 | C | m | msm | <50 | 799 | STI/on HAART |
| AC 05 | C | m | msm | 14,200 | 465 | STI/off HAART |
| AC 06 | C | m | msm | 63,380 | 605 | STI/off HAART |
| AC 14 | C | m | msm | 1,510 | 1,017 | STI/on HAART |
| AC 15 | C | m | msm | 80 | 555 | STI/on HAART |
| AC 16 | C | m | msm | 196 | 1,004 | STI/off HAART |
| AC 25 | C | m | msm | 7,680 | 520 | STI/off HAART |
| AC 26 | C | m | msm | <50 | 770 | STI/on HAART |
| AC 34 | C | m | msm | 505 | 631 | STI/off HAART |
| AC 45 | C | m | msm | 81 | 477 | STI/on HAART |
| AC 46 | C | m | msm | <50 | 772 | STI/on HAART |
| CHR-1 | C | m | msm | <50 | 823 | HAART |
| CHR-2 | C | m | msm | <50 | 828 | HAART |
| CHR-3 | C | m | msm | <50 | 597 | HAART |
| CHR-4 | C | m | msm | <50 | 239 | HAART |
| CHR-5 | C | m | msm | 73 | 89 | HAART |
| CHR-6 | C | m | msm | <50 | 707 | HAART |
| CHR-7 | C | f | hetero | <50 | 561 | HAART |
| CHR-8 | C | m | msm | <50 | 852 | HAART |
| CHR-9 | C | f | hetero | 80 | 385 | HAART |
| CHR-10 | C | m | msm | <50 | 246 | HAART |
| CHR-11 | C | m | msm | <50 | 396 | HAART |
| CHR-12 | C | m | msm | <50 | 430 | HAART |
| CHR-13 | C | m | msm | 7190 | 770 | None |
| CHR-14 | AA | m | hetero | 141,000 | 142 | None |
| CHR-15 | C | m | msm | 3,900 | 549 | None |
| CHR-16 | C | m | msm | 7,980 | 641 | None |
| CHR-17 | H | m | msm | 6,200 | 653 | None |
| CHR-18 | C | m | msm | 97,400 | 813 | None |
| CHR-19 | AA | m | hetero | 3,960 | 449 | None |
| CHR-20 | C | f | hetero | 41,600 | 410 | None |
| CHR-21 | H | f | hetero | 49,200 | 730 | None |
| CHR-22 | C | m | msm | 187,000 | 253 | None |
| CHR-23 | C | f | hetero | 73,700 | 259 | None |
| CHR-24 | C | m | msm | 53,863 | 186 | None |
| CHR-25 | C | m | hetero | 300,000 | 43 | None |
| CHR-26 | C | f | ivdu | 2,230 | 420 | None |
| CHR-27 | C | m | hemo | <50 | 523 | None |
| CHR-28 | C | m | msm | <50 | 875 | None |
| CHR-29 | C | m | msm | 876 | 843 | None |
| CHR-30 | C | m | msm | 1,412 | 1,020 | None |
| CHR-31 | C | m | msm | 132 | 930 | None |
| CHR-32 | C | m | msm | 800 | 670 | None |
| CHR-33 | C | m | msm | 980 | 720 | None |
| CHR-34 | C | m | msm | 738 | 597 | None |
| CHR-35 | C | m | msm | <50 | 540 | None |
AC, acute; CHR, chronic; C, caucasian; AA, African American; H, Hispanic; m, male; f, female; msm, men having sex with men; hetero, heterosexual contact; ivdu, IV drug use; hemo, hemophiliac; HAART, highly active antiretroviral therapy.
Measured at time of HIV-1-specific T-cell analysis.
Lymphocyte separation.
Fresh PBMC were separated from whole blood by Ficoll-Hypaque (Sigma, St. Louis, Mo.) density gradient centrifugation. For substudies investigating CD4 or CD8 dependence of HIV-1-specific responses, RosetteSep CD4 and CD8 reagents were used according to the manufacturer's directions (StemCell Technologies, Vancouver, Canada).
Synthetic HIV-1 peptides.
Five hundred four peptides, overlapping by 10 amino acids, which were 13 to 18 amino acids in length and spanned the entire expressed HIV-1 clade B genome (Gag, Pol, Vif, Vpr, Vpu, Rev, Tat, Env, and Nef) were synthesized at the MGH Peptide Core Facility on an automated peptide synthesizer (MBS 396; Advanced Chemtech, Louisville, Ky.) using Fmoc chemistry.
Design of peptide matrices.
All 504 overlapping peptides were included in five different peptide matrix systems, with Gag, Pol, Env, and Nef as individual peptide matrices while Rev, Tat, Vif, Vpr, and Vpu were pooled into a combined accessory and regulatory protein (Acc/Reg) peptide matrix. Within a given protein matrix, each peptide was represented in two different peptide pools, allowing for the identification of the respective peptide by responses in the two corresponding pools. This is exemplified for the identification of the Nef-4 peptide in Table 2 (e.g., Pool G = peptides Nef-1 to Nef-6; Pool D = Nef-4, Nef-10, Nef-16, Nef-22, Nef-28, Nef-34, and Nef-40) (28). The number of pools per matrix and the number of peptides per pool depended on the total number of peptides spanning each protein and are summarized in Table 3. The final concentration of each peptide within a peptide pool was 200 μg/ml.
TABLE 2.
Example of peptide matrix setup for Nefa
| Pool A | Pool B | Pool C | Pool D | Pool E | Pool F | |
|---|---|---|---|---|---|---|
| Pool G | Nef-1 | Nef-2 | Nef-3 | Nef-4 | Nef-5 | Nef-6 |
| Pool H | Nef-7 | Nef-8 | Nef-9 | Nef-10 | Nef-11 | Nef-12 |
| Pool I | Nef-13 | Nef-14 | Nef-15 | Nef-16 | Nef-17 | Nef-18 |
| Pool J | Nef-19 | Nef-20 | Nef-21 | Nef-22 | Nef-23 | Nef-24 |
| Pool K | Nef-25 | Nef-26 | Nef-27 | Nef-28 | Nef-29 | Nef-30 |
| Pool L | Nef-31 | Nef-32 | Nef-33 | Nef-34 | Nef-35 | Nef-36 |
| Pool M | Nef-37 | Nef-38 | Nef-39 | Nef-40 | Nil | Nil |
Example, shown in bold: a positive response to peptide Nef-4 would be reflected in positive responses in pools D and G.
TABLE 3.
Composition of protein matrix and number of peptides per pool
| Protein | No. of peptide pools | No. of peptides per pool |
|---|---|---|
| Gag | 20 | 9-10 |
| Pol | 23 | 11-12 |
| Acc/Reg | 24 | 11-12 |
| Env | 20 | 10-11 |
| Nef | 13 | 5-7 |
Elispot assays.
Elispot assays were performed as described previously (2). Briefly, fresh PBMC were plated in 96-well polyvinylidene plates (Millipore, Bedford, Mass.) that had been precoated with 0.5 g of anti-IFN-γ monoclonal antibody, 1-DIK (Mabtech, Stockholm, Sweden)/ml. PBMC were added at a concentration of 50,000 to 100,000 cells per well in a volume of 100 μl of R10 medium (RPMI 1640 [Sigma], 10% fetal calf serum [Sigma], 10 mM HEPES buffer [Sigma]) with antibiotics (2 mM l-glutamine, 50 U of penicillin-streptomycin/ml). The final concentration of the peptides in the well was 20 μg/ml. Plates were incubated overnight at 37°C, 5% CO2, and developed as described previously (2, 6). Wells containing PBMC and R10 medium were used as negative controls and were run in triplicate on each plate. Wells containing PBMC and phytohemagglutinin (PHA) served as positive controls. The numbers of spots per well were counted using an automated Elispot plate reader (AID EliSpot reader system; Autoimmune Diagnostika GmbH, Strassberg, Germany), and the number of specific T cells was calculated by subtracting the negative control values. The background was <30 per million PBMC (three spots/well at 100,000 PBMC/well) in all cases. Responses were regarded as positive if they had at least three times the mean number of SFC in the three control wells and had to be >50 SFC/106 PBMC.
Statistical analysis.
Statistical analysis and graphical presentation was done using SigmaPlot 5.0 (SPSS Inc., Chicago, Ill.) and GraphPad Prism 3.0. Results are given as means with standard deviations or medians with ranges. Statistical analysis of significance (P values) was based on two-tailed t tests. Comparisons were made using the Spearman rank correlation test, and linear regression analysis was used to determine the slope. In order to avoid overestimation of the total breadth of the HIV-1-specific CD8 response, we abstained from simply summarizing all peptide responses for a given individual but evaluated responses to two adjacent overlapping peptides as responses to one epitopic region, since some T-cell epitopes can be located in the overlapping region of two adjacent peptides, resulting in responses to both overlapping peptides. Similarly, in the analysis of the total magnitude of HIV-1-specific CD8-T-cell response, we also considered only the higher response of two adjacent overlapping peptides for final calculations. While avoiding overestimation of responses, this conservative evaluation of data can potentially underestimate the total breadth and magnitude of responses, since more than one epitope can be contained within one overlapping peptide (6, 25, 49).
RESULTS
Efficient assessment of HIV-1-specific T-cell responses using a peptide matrix-based IFN-γ Elispot assay.
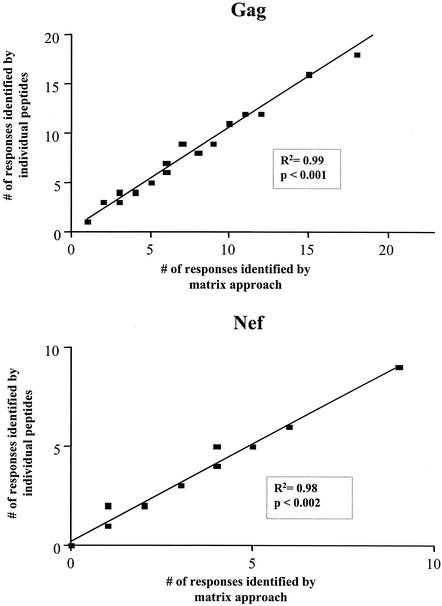
The comprehensive analysis of immune responses to individual HIV-1 peptides spanning the entire HIV genome requires large numbers of PBMC. The use of peptide pools for initial screening offers the possibility of a lower level of specimen usage. We therefore first evaluated a peptide matrix-based approach, using peptide matrices for HIV-1 Nef and Gag. Matrices consisted of pools of peptides in which each peptide was present in two separate pools (see Table 2 for an example), thus offering internal positive controls. These were tested in Elispot assays on 20 HIV-1 infected individuals in parallel to Elispot assays using the individual 40 Nef peptides and 94 Gag peptides in separate wells as the current “gold standard.” The sensitivity of the matrix approach was calculated by dividing the number of responses detected in the peptide matrix by the number of responses detected by use of individual peptides. Specificity was calculated by division of the expected number of positive peptide pools based on the peptide responses identified using individual peptides by the number of positive peptide pools in the matrix approach. The use of peptide matrices for the detection of peptide-specific T-cell responses had a high sensitivity, 94 and 98% for Gag and Nef matrices, respectively (98 and 100% for responses of greater than 200 SFC/106 PBMC, respectively). The specificity was found to be 97 and 98.5% for Gag and Nef, respectively. Furthermore, the number of T-cell responses to individual peptides detected via peptide matrix correlated well with the number of responses detected by use of individual peptides as depicted for the Gag and Nef matrices in Fig. 1.
FIG. 1.
Correlation between peptide matrix and individual peptide approach. The number of responses to peptides detected by the matrix approach on the x axis is compared to the number of responses detected by the use of individual peptides as the current gold standard (y axis) for Gag (upper panel) and Nef (lower panel). Linear regression lines, correlation coefficients, and P values are given in the plots as calculated by linear regression analysis (n = 20).
This matrix method was subsequently evaluated for the total set of peptides spanning the entire HIV-1 genome and showed equally high sensitivity and specificity for the Env (94 and 98%, respectively), Pol (93 and 97%, respectively) and Rev/Tat/Vpr/Vpu/Vif matrices (97 and 99%, respectively). No HIV-1 T-cell responses were detectable for the 10 HIV-1-negative individuals screened by either of the two methods (data not shown). CD4+- and CD8+-T-cell depletion studies showed that more than 95% of responses were CD8+ T cell dependent (data not shown). In addition to the high sensitivity and specificity, the matrix approach also limited the number of cells required. Screening for T-cell responses against the entire expressed HIV-1 genome with individual peptides (including positive and negative controls) required 51 × 106 cells per individual (100,000 PBMC/well), and this did not include cells needed for confirmation of positive responses. Using the Elispot-based peptide matrix approach reduced the number of PBMC needed for a comprehensive screening (including reconfirmation of positive responses with individual peptides) to a median of 14.5 × 106 cells (range, 11.8 × 106 to 22.4 × 106 PBMC), representing the number of cells that would typically be obtainable from 20 ml of blood.
These data indicate that the peptide matrix approach allows for a highly sensitive and specific detection of HIV-1-specific T-cell responses against the entire expressed genome using a limited number of PBMC and that pooled peptides can accurately detect polyclonal CD8-T-cell responses.
The majority of HIV-1 peptides and all HIV-1 protein subunits are targeted by virus-specific CD8+-T-cell responses.
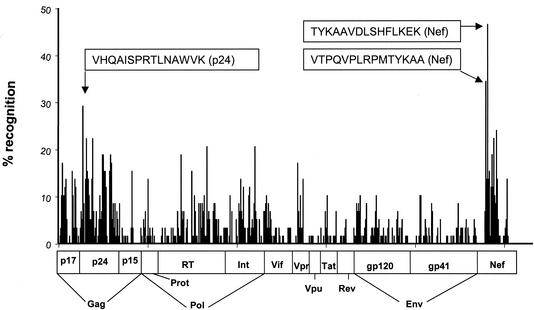
Using the above-described peptide matrix approach, we screened a total of 57 HIV-1-infected individuals for CD8+-T-cell responses against the entire expressed HIV-1 genome in order to comprehensively assess the total breadth and magnitude of virus-specific responses on the single peptide level. All T-cell responses to peptides identified by the matrix approach were subsequently reconfirmed individually in a second assay. These studies revealed that all HIV-1 proteins and protein subunits can serve as targets for HIV-1-specific CD8-T-cell responses (Fig. 2). Of the 504 overlapping peptides, 320 (63%) were targeted by T cells in this study, indicating that at least two-thirds of the expressed HIV-1 genome can be immunogenic. The percentage of peptides targeted within the individual proteins displayed a large variation, with 91% (40 of 44) of p24-Gag peptides recognized at least once, while only 21% (3 of 14) of Vpu peptides served as targets for HIV-1-specific CD8 T cells (Table 4). The mean magnitude of responses to individual targeted peptides was 430 (range, 50 to 2,300 SFC/106 PBMC).
FIG. 2.
Peptide recognition across the entire expressed HIV-1 genome. The 504 individual overlapping peptides are represented on the x axis, and the corresponding percentage of study subjects with a response to the individual peptide are represented on the y axis. The horizontal bar indicates the corresponding regions for the individual peptides. The sequences of the three most frequently targeted peptides, located in Nef and p24Gag, are shown in the text boxes.
TABLE 4.
Number of peptides recognized per HIV-1 protein subunit
| Protein | No. of overlapping peptides recognized per protein subunit (%) |
|---|---|
| p17-Gag | 20/24 (83) |
| p24-Gag | 40/44 (91) |
| p15-Gag | 10/26 (39) |
| Protease | 9/13 (69) |
| RT | 50/83 (60) |
| Integrase | 26/37 (70) |
| Vif | 20/35 (57) |
| Vpr | 9/17 (53) |
| Vpu | 3/14 (21) |
| Tat | 7/15 (47) |
| Rev | 10/21 (48) |
| gp-41-Env | 34/68 (50) |
| gp-120-Env | 43/67 (64) |
| Nef | 27/40 (68) |
The individual peptides were targeted at different frequencies. While some peptides were only recognized by one study subject, others were targeted by several individuals independent of their HLA types (Fig. 2). The three most frequently recognized peptides were located in Nef, TYKAAVDLSHFLKEK (27 of 57 [47%]) and VTPQVPLRPMTYKAA (20 of 57 [35%]), and p24-Gag, VHQAISPRTLNAWVK (17 of 57 [30%]), respectively (Fig. 2 and Table 5). These regions also represented very highly conserved regions of the HIV-1 genome with 90, 94, and 97% sequence conservation among published clade B sequences (obtained from hiv-web.lanl.gov/cgi-bin/EPILIGN). The distribution of the individual peptide responses and their frequency of recognition in the total study cohort of 57 HIV-1-infected individuals across the entire expressed HIV-1 genome is summarized in Fig. 2. Nef and p24-Gag emerged as the proteins containing the most widely targeted peptides; however, peptides recognized by more than 20% of study subjects were also found within RT and integrase (Fig. 2 and Table 5).
TABLE 5.
Summary of the most frequently recognized peptides in this study cohort
| Protein | Amino acid position | Peptide sequence | Study subjects with response (%) |
|---|---|---|---|
| Nef | 80-94 | TYKAAVDLSHFLKEK | 47 |
| Nef | 70-84 | VTPQVPLRPMTYKAA | 34 |
| p24 | 11-25 | VHQAISPRTLNAWVK | 29 |
| Nef | 130-144 | GPGVRYPLTFGWCYK | 24 |
| p24 | 31-44 | AFSPEVIPMFSALS | 22 |
| p24 | 63-80 | AAMQMLKETINEEAAEW | 22 |
| Nef | 115-129 | YHTQGYFPDWQNYTP | 22 |
| RT | 373-390 | QKIATESIVIWGKTPKFK | 21 |
| Int | 218-235 | TKIQNFRVYYRDSRDPLW | 21 |
| p24 | 121-135 | NPPIPVGEIYKRWII | 19 |
| p24 | 126-140 | VGEIYKRWIILGLNK | 19 |
| p24 | 165-177 | VDRFYKTLRAEQAS | 19 |
| RT | 151-168 | QGWKGSPAIFQSSMTKIL | 19 |
| Nef | 105-119 | RRQDILDLWIYHTQG | 19 |
| p17 | 16-30 | WEKIRLRPGGKKKYK | 17 |
| p24 | 169-184 | YKTLRAEQASQDVKNWM | 17 |
| Vpr | 25-40 | ELKNEAVRHFPRIWLH | 17 |
| p17 | 71-85 | GSEELRSLYNTVATL | 16 |
| p24 | 35-48 | EVIPMFSALSEGATP | 16 |
| p24 | 131-145 | KRWIILGLNKIVRMY | 16 |
| p24 | 136-150 | LGLNKIVRMYSPTSI | 16 |
| p24 | 161-174 | FRDYVDRFYKTLRA | 16 |
| p15 | 66-80 | RQANFLGKIWPSYKG | 16 |
| RT | 240-257 | TVQPIVLPEKDSWTVNDI | 16 |
| Nef | 90-105 | FLKEKGGLEGLIHSQ | 16 |
Taken together, these data demonstrate that HIV-1-specific T cells can target all HIV-1 proteins and that at least two-thirds of the expressed HIV-1 clade B sequence can be immunogenic.
Dominant recognition of Nef and p24-Gag by HIV-1-infected persons at differing disease stages.
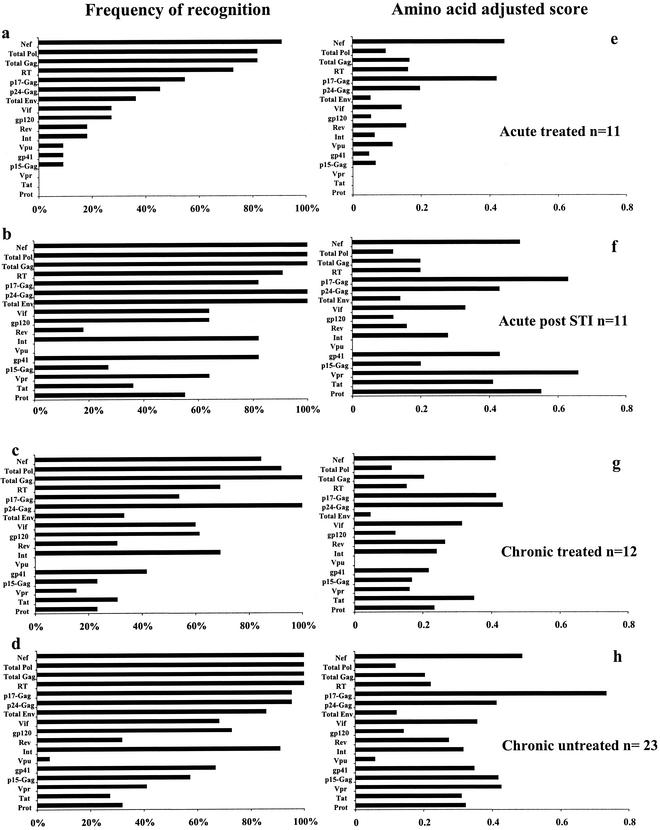
All study subjects had detectable T-cell responses to at least one HIV-1 protein. In the entire group of 57 HIV-1-infected individuals studied, Nef and p24-Gag were the most frequently recognized protein subunits. These were targeted by 95% and 88% of the tested individuals, respectively, followed by RT and p17-Gag (86 and 75%, respectively). Considering whole proteins rather than protein subunits, Gag (p15, p17, and p24), Nef, and Pol (Int, RT, and Prot) dominated the frequency of recognition, with 96, 95 and 95% of study subjects targeting these proteins, respectively. When adjusted to amino acid length, p17-Gag, Nef, and p24-Gag emerged as the most frequently targeted antigens. The subgroup analysis showed that irrespective of the status of HIV-1 disease, p24-Gag, RT, and Nef were always among the four most frequently targeted HIV-1 proteins (Fig. 3a to d). Adjusted to the amino acid length of the corresponding protein subunit, p24-Gag and Nef were still among the four most frequently recognized proteins in all study groups, while RT was less frequently targeted (Fig. 3e to h). In contrast, p17-Gag was by far the most frequently recognized protein subunit after adjusting for protein length in the chronic untreated subgroup and emerged with the second-highest recognition scores in all other groups (Fig. 3). Concordant with previous reports, when results were adjusted to amino acid length of the protein, Vpr was the most frequently targeted protein in the subgroup of individuals treated during acute infection and undergoing STI (5).
FIG. 3.
Frequency of recognition of the individual proteins and protein subunits. Panels a to d summarize the percentages of individuals with responses to at least one peptide for the individual protein or subunit, stratified by study groups as indicated. Panels e to h show amino acid-adjusted scores for protein recognition. The amino acid-adjusted score is defined as the frequency of recognition of the individual protein divided by protein length in amino acids. (a and e) Acute treated; (b and f) acute post-STI; (c and g) chronic treated; (d and h) chronic untreated.
Overall these data show that all study subjects, regardless of disease stage or treatment history, had detectable HIV-1-specific T-cell responses and that Nef and p17-Gag emerged as the most frequent targets when adjusted to amino acid length of the protein subunit, followed by p24-Gag, while Vpu was the least-frequently recognized protein.
Breadth and magnitude of the total HIV-1-specific CD8+-T-cell response are lowest in individuals treated during acute HIV-1 infection and highest in chronically infected untreated subjects.
We next analyzed the breadth of the CD8+-T-cell response against all HIV-1 protein subunits. None of the study subjects had detectable responses to all 14 HIV-1 protein subunits; however, the number of protein subunits targeted per patient ranged from 1 of 14 for an individual treated during chronic HIV-1 infection to 13 of 14 for an untreated chronically infected individual with long-term nonprogressive disease. Individuals with untreated chronic HIV-1 infection and acutely treated patients after one or two cycles of STI recognized significantly more protein subunits (median, 9; range, 5 to 11; and median, 9; range, 6 to 13, respectively; P = 0.73) than individuals with treated acute infection (median, 4; range, 2 to 7; P = 0.001 and 0.001, respectively) and treated chronic infection (median, 6 range, 1 to 10; P = 0.001 and 0.001, respectively).
In order to take this analysis to higher resolution, we next investigated how many individual epitopic regions within the HIV-1 proteins and subunits were targeted by each study subject. The number of epitopes targeted per study subject ranged from 2 to 42 (median, 14) in this cohort. In the subgroup analysis, once again chronic untreated individuals and the acute/STI group recognized the highest number of epitopic regions, with medians of 18.5 (range, 8 to 42) and 18 (range, 9 to 26), respectively (Fig. 4a). The individual with the highest number of peptides targeted was an HIV-1 long-term nonprogressor with a viral load below the limits of detection (<50 HIV-1 RNA copies/ml) in the context of untreated chronic HIV-1 infection.
FIG. 4.
Comparison of breadth and magnitude between the different study groups. Panel a summarizes the number of epitopic regions targeted per individual in the different study groups, while panel b indicates the total magnitude of the CD8-T-cell response to the entire expressed genome per individual (measured by Elispot and given in SFC/106 PBMC). For both panels the median number of regions targeted and median magnitude for each group are indicated as horizontal black bars. P values were calculated using a two-tailed t test.
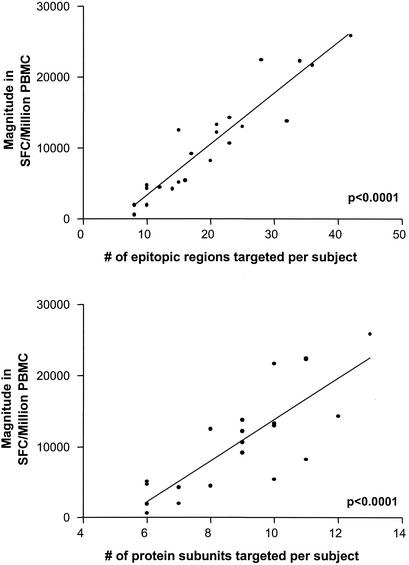
We next examined the total magnitude of responses against the entire expressed HIV-1 genome. In order to avoid overestimation of the total CD8+-T-cell response, responses to two adjacent overlapping peptides were counted only once, as outlined in Materials and Methods. The magnitude of responses ranged from 280 SFC/106 PBMC in an individual treated during acute HIV-1 infection to 25,860 SFC/106 PBMC in an individual with untreated chronic nonprogressive HIV-1 infection mentioned above (median, 4,245 SFC/106 PBMC) (Fig. 4b). In the subgroup analysis, significantly lower-magnitude responses were found with subjects with treated acute infection (median, 890 SFC/106 PBMC; range, 280 to 3,490) than in chronically infected treated subjects (median, 2,568 SFC/106 PBMC; range, 480 to 10,110; P = 0.001), the acute/STI group (median, 5,340 SFC/106 PBMC; range, 2,710 to 14,730; P = 0.023), and chronically infected untreated individuals (median, 10,640 SFC/106 PBMC; range, 1930 to 25,860 [P < 0.0001]) (Fig. 4b). Not surprisingly, the magnitude of the total HIV-1-specific CD8-T-cell responses correlated significantly with the breadth of responses as measured by both epitopic regions and targeted protein subunits (P <0.001 and 0.001, respectively) (Fig. 5).
FIG. 5.
Correlation between total magnitude and breadth of the HIV-1-specific CD8+-T-cell response. The top panel shows the correlation between total magnitude of the HIV-1-specific CD8+-T-cell response in SFC/106 PBMC (as measured by Elispot) to the total number of epitopic regions targeted per individual. The bottom panel shows the association between the total magnitude of the HIV-1-specific CD8+-T-cell response and the number of protein subunits recognized per study subject.
Taken together, these data indicate that T-cell responses in individuals on antiretroviral treatment, and particularly in individuals treated during acute HIV-1 infection, are significantly lower and more narrowly directed than in individuals with untreated or intermittently treated HIV-1 infection. They also demonstrate that CD8+-T-cell responses can be very broadly directed in the setting of uncontrolled viremia.
Differences in protein targeting in acute and chronic HIV-1 infection.
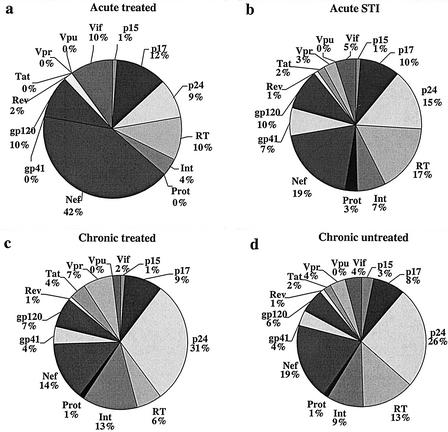
We next assessed the contribution of the individual proteins to the total magnitude of the HIV-1-specific CD8 responses in the four subgroups. In both subgroups of individuals treated during primary HIV-1 infection (with or without subsequent STI), HIV-1 Nef was the HIV-1 protein that contributed most importantly to the total HIV-1-specific responses, with 42 and 19%, respectively (Fig. 6a and b). Among the individuals with treated or untreated chronic HIV-1 infection, p24-Gag dominated the relative magnitude of the virus-specific CD8 response, with 31 and 26%, respectively (Fig. 6c and d). This decrease in the relative contribution of Nef to the total magnitude of HIV-1-specific response in chronically infected individuals does not reflect decreasing Nef responses in absolute numbers, but rather the increased contribution of the other proteins, particularly p24-Gag, to the total response in the chronic phase of infection. In contrast, for all subgroups the regulatory proteins (Rev and Tat) and accessory proteins (Vif, Vpr, and Vpu) on average contributed least to the total magnitude of the CD8-T-cell response. In the analysis of whole proteins rather than subunits, total Gag contributed most importantly to the total magnitude of responses for both chronic and untreated individuals, followed by Pol, Nef, and Env. In contrast, for individuals with acute treated HIV-1 infection, Nef remained the dominant contributor to the total magnitude of response, followed by total Gag, Pol, and Env.
FIG. 6.
Relative contributions of individual protein subunits to the total magnitude of the HIV-1-specific CD8-T-cell response. Each pie chart shows the mean contribution of the individual protein subunits to the total magnitude of response directed against all of HIV-1 for one of the four study groups.
Taken together, these data suggest differences in patterns of protein recognition between the acute and chronic phase of HIV-1 infection, with preferential targeting of HIV-1 Nef during acute infection. These data also demonstrate that for the whole study cohort as well as in the subgroup analysis, >70% of the total magnitude of responses is directed against the three HIV-1 proteins Gag, Pol, and Nef.
Plasma viral load does not correlate with breadth and magnitude of total HIV-1-specific CD8-T-cell responses.
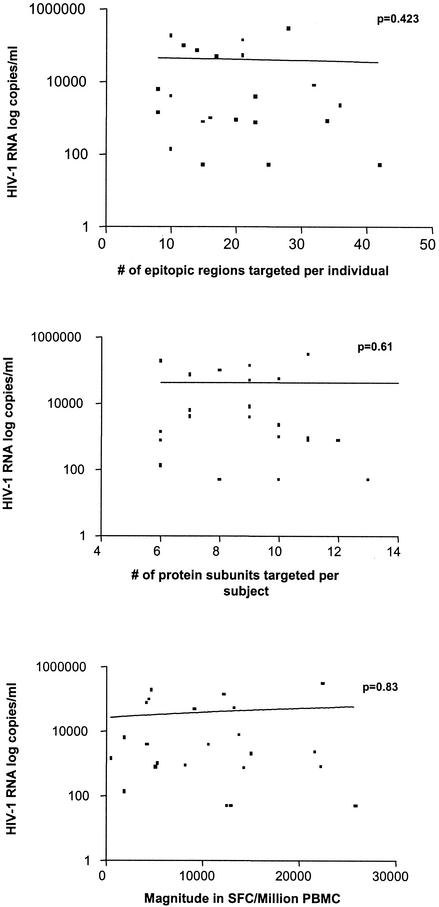
A proposed correlation between viral load and HIV-1-specific T-cell responses remains controversial. While some previous studies have suggested that the number of HIV-1-specific CD8 T cells inversely correlates with the plasma viral load (19, 37, 46), others have found no such correlation (16, 21), and more comprehensive studies have found a positive correlation between specific CD8+-T-cell frequency and viral load (11). None of these studies examined responses at the individual peptide level. We investigated the potential association between plasma viral load and three entities: magnitude of the HIV-1-specific CD8-T-cell response, number of protein subunits targeted, and number of individual epitopic regions targeted in the subset of 23 individuals with untreated chronic HIV-1 infection. The results indicate that there is no correlation between viral load and either breadth or magnitude of the total virus-specific CD8-T-cell response (Fig. 7). We also performed the analyses regarding breadth and magnitude for all 14 individual subunits as well as Gag (p17, p24, and p15), Pol (Prot, RT, and Int), and Env (gp120 and gp41) as total protein entities and failed to detect a statistically significant correlation with plasma viral load (data not shown).
FIG. 7.
Correlation between viral load and breadth and magnitude of the total HIV-1-specific CD8-T-cell response. The association between HIV-1 RNA and the number of epitopic regions targeted per individual (top panel), the number of protein subunits recognized (middle panel), and the total HIV-1-specific magnitude as measured by Elispot (in SFC/million PBMC) (bottom panel) is depicted. Viral load in plasma (HIV-1 RNA copies/ml) was determined at the time of T-cell analysis for each individual. P values shown in each panel were determined by the Spearman rank correlation. The solid line represents a regression line, and all graphic representation was performed using Graphpad Prism 3. Plasma viral loads of <50 were expressed as 49 for statistical analysis.
These data suggest that the influence of CD8+-T-cell responses on plasma viral load is not be explained by the mere quantification of magnitude and breadth of the T-cell responses, even if a comprehensive pan-genome screening approach is applied.
DISCUSSION
Cellular immune responses are believed to play a pivotal role in the control of HIV-1 infection; however, the total magnitude and breadth of virus-specific responses directed against the entire expressed genome of HIV-1 has never been assessed at the level of the responses to individual epitopes. Recent advances in methodology, in particular flow-based intracellular cytokine staining and the Elispot assay, now allow for a more comprehensive and precise analysis of HIV-1-specific T-cell responses. In the present study, we used an IFN-γ Elispot assay and 504 overlapping peptides spanning all HIV-1 proteins to comprehensively analyze HIV-1-specific T-cell responses in 57 HIV-1-infected individuals at different stages of HIV-1 infection. Our data demonstrate that all HIV-1 proteins and protein subunits are targeted by HIV-1-specific CD8 T cells, even in persons who fail to control infection. The magnitude and breadth of virus-specific T-cell responses varied significantly among individuals at different stages of infection, with the broadest and strongest responses detectable for individuals with untreated chronic infection, while responses in treated acute and chronic HIV-1 infection were more narrowly directed. However, despite the detailed pan-genome analysis and broad and high-frequency responses detected in this study, no significant correlation to markers of disease progression was observed. These data also show that the use of peptide matrices for the comprehensive pan-genome assessment of virus-specific T-cell responses is highly sensitive, specific, and specimen saving, making it an attractive tool for large-scale clinical or vaccine trials.
Recent vaccine studies in the nonhuman primate model have shown encouraging results regarding attenuation of disease progression following induction of robust SIV-specific CD8+-T-cell responses (7, 10, 44). These vaccine candidates and others will be moving into human clinical trials in the near future, and a comprehensive analysis of HIV-1-specific T-cell responses directed against the entire viral genome will be needed to evaluate the immunogenicity of these and other vaccines in humans. Hence, rapid and practicable screening methods applicable for use in larger scale trials, preferably also adaptable to resource poor settings, need to be devised. The IFN-γ Elispot assay is a simple and reliable method that has been widely used for the analysis of T-cell responses in viral and parasitic infections (20, 32-34, 39, 48) and malignant diseases (30, 45) and has been successfully utilized in resource-poorer settings with more basic laboratory facilities (22). In the present study we demonstrate that the Elispot assay can easily be adapted to perform whole HIV-1 genome T-cell analysis with high sensitivity and specificity, allowing a detailed assessment of the exact regions targeted and the number of epitopes recognized using only limited numbers of PBMC. This may make this approach a practical tool for vaccine studies and clinical trials.
Most previous studies on HIV-1-specific CD8-T-cell responses have focused on either single epitopes (37), panels of optimal epitopes (17), single HIV-1 proteins (50), or a limited selection of HIV-1 proteins (1, 2, 4, 5, 19), often structural in nature. The comprehensive approach used in this study showed that all HIV-1 proteins serve as targets for HIV-1-specific CD8+ T cells, displaying an unprecedented breadth and magnitude of the total HIV-1-specific T-cell response. To our knowledge only one other report in the literature has assessed HIV-1-specific T-cell responses directed against the whole expressed HIV-1 genome in a larger study population (11). In that study, Betts et al. used pools of pan-genome-spanning overlapping peptide pools in intracellular cytokine staining assays to assess the total magnitude of T-cell responses to all HIV-1 proteins in untreated HIV-1-infected individuals. In line with our findings, all of their untreated study subjects had detectable HIV-1-specific CD8-T-cell responses to HIV-1 Gag, and all HIV-1 proteins served as targets for HIV-1-specific T cells, with HIV-1 Gag, Nef, and Pol being among the most frequently targeted proteins and Vpu being the least immunogenic HIV-1 protein. However, while the study of Betts et al. focused on the magnitude of responses to whole proteins and measured breadth as the number of proteins recognized per individual, our study takes these findings a step further and assesses the individual peptides containing CD8+-T-cell epitopes targeted within all HIV-1 proteins. This more detailed study demonstrates that a very high number of epitopic regions (up to 42) can be recognized by HIV-1-specific T cells in a single individual and frequencies of total HIV-1-specific CD8+ T cells as high as 25,860 SFC/Mill PBMC can be detected. However, of note is the finding that the persons with the greatest magnitude of responses included one long-term nonprogressor with an undetectable viral load, as well as a progressor with a high viral load.
These data emphasize again that the response to a single HIV-1 epitope is not predictive of the entire response within an infected individual, since multiple HIV-1 proteins are recognized in every HIV-1-infected patient, as indicated in previous reports (12, 18). However, breadth and magnitude of virus-specific T-cell responses differed significantly among individuals at different stages of HIV-1 infection. While individuals with treated acute and chronic infection had narrowly directed HIV-1-specific responses with low magnitudes, chronically infected untreated individuals and individuals after supervised treatment interruptions had broader responses of higher magnitude, suggesting a broadening of the HIV-1-specific T-cell response with continuous exposure to antigen (5).
In the detailed analysis of epitopic regions the most frequently targeted peptides were detected in HIV-1 Nef, with up to 45% of individuals recognizing the sequence TYKAAVDLSHFLEK, followed by the Nef peptide VTPQVPLRPMTYKAA (34%). HIV-1 Nef was also the most frequently recognized protein and contributed most importantly to the total HIV-1-specific response in individuals with acute HIV-1 infection. These data suggest that Nef may play a privileged role in the immunodominance of HIV-1-specifc T-cell responses. A recent report by Yusim et al. suggests that frequency of recognition of a response is in part a function of sequence variability with HIV-1-specific CD8+-T-cell responses clustering within more conserved regions of the virus (51). In our study, indeed all peptides targeted by >20% of individuals (Table 5) were highly conserved among published clade B sequences. In contrast, the more variable HIV-1 proteins Vpu, Tat, and Rev (51) were overall the least frequently targeted proteins, as described previously (1, 2, 4, 46). A limitation of current studies of virus-specific T-cell responses is that the sets of overlapping peptides used for the assessment of responses are based on sequences of primary HIV-1 isolates or clade B consensus sequences. The use of such sequences is likely to favor detection of responses in well-conserved areas of the genome, since differences between the autologous virus sequence and the peptides used for the assays are smallest within these regions. In contrast, there is strong evidence from the SIV-macaque model, using peptide sequences based on the infecting virus for the assessment of T-cell responses, that variable proteins, such as Tat, are consistently detected in early infection and may contain immunodominant responses (3, 36). Therefore, studies comparing consensus or primary strain sequences versus autologous virus sequences for the analysis of HIV-1-specific T-cell responses need to be performed in order to evaluate potential discrepancies. Studies are currently under way to address this issue using autologous versus consensus sequences (M. Altfeld and M. M. Addo, unpublished data).
Despite the impressive breadth and magnitude of responses, no significant correlation between virus-specific T-cell responses and HIV-1 viral load in plasma was observed in this study. While some individuals with good control of viremia were found to have broadly directed HIV-1-specific CD8-T-cell responses of high magnitude, similar breadth and magnitudes of responses were found in individuals with high viral replication. The absence of a negative correlation between viral load and IFN-γ production of HIV-1-specific T cells is in line with the studies of Betts et al., but in contrast to studies assessing responses to a more limited number of epitopes or HIV-1 proteins that did show inverse correlations (19, 37, 46). The restriction of the analysis to responses directed against particular regions of the HIV-1 genome in different studies as well as differences in the methods used and the population studied may have contributed to these discrepancies among the different study results. The absence of an association between HIV-1-specific CD8+-T-cell responses and viral load in the more comprehensive studies suggests that the sole measurement of IFN-γ production of T cells after stimulation with HIV-1 peptides that are not based on autologous sequences may not be the adequate approach to identifying the correlates of control of viral replication. Future studies, such as the investigation of phenotypic and functional determinants and the “quality” of HIV-1-specific T-cell responses (8, 15, 42), as well as the assessment of responses against the autologous virus, are therefore needed to identify the correlates of immune-mediated control of HIV-1 replication.
Taken together, this study provides a comprehensive analysis of HIV-1-specific T-cell responses and a detailed dissection of the exact epitopic regions targeted and the magnitude of the total HIV-1-specific CD8-T-cell response, using a highly sensitive and specific peptide matrix system. Despite the detection of broad and strong HIV-1-specific T-cell responses, no correlation to HIV-1 plasma viral load was observed in this study, emphasizing the need for studies assessing additional functional and phenotypic parameters as well as the true breadth and magnitude of responses directed against the autologous virus. The finding of very broadly directed and often high-magnitude CD8+-T-cell responses against HIV-1 in persons who are unable to contain HIV-1 infection suggests that effective therapeutic augmentation of HIV-1-specific T-cell immunity will be a formidable challenge.
Acknowledgments
We thank all study participants and the dedicated clinical research staff at the collaborating sites.
This study was supported by the German Research Council (DFG; Emmy Noether grant AD-171) (M.M.A.), the American Foundation for AIDS Research (AmfAR) (M.M.A.), Concerned Parents for AIDS Research (CPFA) (M.M.A.), the Doris Duke Charitable Foundation (M.A., E.S.R., and B.D.W.), the National Institutes of Health (R37 AI128568, R01 AI30914, R01 AI44656, P30AI42851, and R01 AI50429), and the Partners/Fenway/Shattuck Center for AIDS Research (X.G.Y.). P.J.R.G. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation. B.D.W. is the recipient of a Doris Duke Distinguished Clinical Scientist Award.
REFERENCES
- 1.Addo, M. M., M. Altfeld, A. Rathod, M. Yu, X. G. Yu, P. J. R. Goulder, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 Vpu represents a minor target for cytotoxic T lymphocytes (CTL) in HIV-1-infection. AIDS 16:1071-1073. [DOI] [PubMed] [Google Scholar]
- 2.Addo, M. M., M. Altfeld, E. S. Rosenberg, R. L. Eldridge, M. N. Philips, K. Habeeb, A. Khatri, C. Brander, G. K. Robbins, G. P. Mazzara, P. J. Goulder, and B. D. Walker. 2001. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc. Natl. Acad. Sci. USA 98:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 4.Altfeld, M., M. M. Addo, R. L. Eldridge, X. G. Yu, S. Thomas, A. Khatri, D. Strick, M. N. Phillips, G. B. Cohen, S. A. Islam, S. A. Kalams, C. Brander, P. J. Goulder, E. S. Rosenberg, and B. D. Walker. 2001. Vpr is preferentially targeted by CTL during HIV-1 infection. J. Immunol. 167:2743-2752. [DOI] [PubMed] [Google Scholar]
- 5.Altfeld, M., E. S. Rosenberg, R. Shankarappa, J. S. Mukherjee, F. M. Hecht, R. L. Eldridge, M. M. Addo, S. H. Poon, M. N. Phillips, G. K. Robbins, P. E. Sax, S. Boswell, J. O. Kahn, C. Brander, P. J. Goulder, J. A. Levy, J. I. Mullins, and B. D. Walker. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193:169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altfeld, M. A., A. Trocha, R. L. Eldridge, E. S. Rosenberg, M. N. Phillips, M. M. Addo, R. P. Sekaly, S. A. Kalams, S. A. Burchett, K. McIntosh, B. D. Walker, and P. J. Goulder. 2000. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J. Virol. 74:8541-8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 8.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. [DOI] [PubMed] [Google Scholar]
- 9.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 10.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 11.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betts, M. R., J. P. Casazza, B. A. Patterson, S. Waldrop, W. Trigona, T. M. Fu, F. Kern, L. J. Picker, and R. A. Koup. 2000. Putative immunodominant human immunodeficiency virus-specific CD8+-T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J. Virol. 74:9144-9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 15.Champagne, P., G. S. Ogg, A. S. King, C. Knabenhans, K. Ellefsen, M. Nobile, V. Appay, G. P. Rizzardi, S. Fleury, M. Lipp, R. Forster, S. Rowland-Jones, R. P. Sekaly, A. J. McMichael, and G. Pantaleo. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106-111. [DOI] [PubMed] [Google Scholar]
- 16.Dalod, M., M. Dupuis, J. C. Deschemin, D. Sicard, D. Salmon, J. F. Delfraissy, A. Venet, M. Sinet, and J. G. Guillet. 1999. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8+ responses in HIV type 1-infected patients: comparison with anti-Epstein-Barr virus responses and changes during antiretroviral therapy. J. Virol. 73:7108-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalod, M., M. Harzic, I. Pellegrin, B. Dumon, B. Hoen, D. Sereni, J. C. Deschemin, J. P. Levy, A. Venet, and E. Gomard. 1998. Evolution of cytotoxic T lymphocyte responses to human immunodeficiency virus type 1 in patients with symptomatic primary infection receiving antiretroviral triple therapy. J. Infect. Dis. 178:61-69. [DOI] [PubMed] [Google Scholar]
- 18.Day, C. L., A. K. Shea, M. A. Altfeld, D. P. Olson, S. P. Buchbinder, F. M. Hecht, E. S. Rosenberg, B. D. Walker, and S. A. Kalams. 2001. Relative dominance of epitope-specific cytotoxic T-lymphocyte responses in human immunodeficiency virus type 1-infected persons with shared HLA alleles. J. Virol. 75:6279-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+-T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 76:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flanagan, K. L., E. A. Lee, M. B. Gravenor, W. H. Reece, B. C. Urban, T. Doherty, K. A. Bojang, M. Pinder, A. V. Hill, and M. Plebanski. 2001. Unique T cell effector functions elicited by Plasmodium falciparum epitopes in malaria-exposed Africans tested by three T cell assays. J. Immunol. 167:4729-4737. [DOI] [PubMed] [Google Scholar]
- 21.Gea-Banacloche, J. C., S. A. Migueles, L. Martino, W. L. Shupert, A. C. McNeil, M. S. Sabbaghian, L. Ehler, C. Prussin, R. Stevens, L. Lambert, J. Altman, C. W. Hallahan, J. C. de Quiros, and M. Connors. 2000. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 165:1082-1092. [DOI] [PubMed] [Google Scholar]
- 22.Goulder, P. J., M. M. Addo, M. A. Altfeld, E. S. Rosenberg, Y. Tang, U. Govender, N. Mngqundaniso, K. Annamalai, T. U. Vogel, M. Hammond, M. Bunce, H. M. Coovadia, and B. D. Walker. 2001. Rapid definition of five novel HLA-A∗3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by Elispot and intracellular cytokine staining assays. J. Virol. 75:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 24.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 25.Goulder, P. J., Y. Tang, S. I. Pelton, and B. D. Walker. 2000. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J. Virol. 74:5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrer, T., E. Harrer, S. A. Kalams, T. Elbeik, S. I. Staprans, M. B. Feinberg, Y. Cao, D. D. Ho, T. Yilma, A. M. Caliendo, R. P. Johnson, S. P. Buchbinder, and B. D. Walker. 1996. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable nonprogressing HIV type 1 infection. AIDS Res. Hum. Retrovir. 12:585-592. [DOI] [PubMed] [Google Scholar]
- 27.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern, F., I. P. Surel, N. Faulhaber, C. Frommel, J. Schneider-Mergener, C. Schonemann, P. Reinke, and H. D. Volk. 1999. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 73:8179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konopitzky, R., U. Konig, R. G. Meyer, W. Sommergruber, T. Wolfel, and T. Schweighoffer. 2002. Identification of HLA-A∗0201-restricted T Cell epitopes derived from the novel overexpressed tumor antigen calcium-activated chloride channel 2. J. Immunol. 169:540-547. [DOI] [PubMed] [Google Scholar]
- 31.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsson, M., D. Messmer, S. Somersan, J. F. Fonteneau, S. M. Donahoe, M. Lee, P. R. Dunbar, V. Cerundolo, I. Julkunen, D. F. Nixon, and N. Bhardwaj. 2000. Requirement of mature dendritic cells for efficient activation of influenza A-specific memory CD8+ T cells. J. Immunol. 165:1182-1190. [DOI] [PubMed] [Google Scholar]
- 33.Larsson, M., D. T. Wilkens, J. F. Fonteneau, T. J. Beadle, M. J. Merritt, R. G. Kost, P. A. Haslett, S. Cu-Uvin, N. Bhardwaj, D. F. Nixon, and B. L. Shacklett. 2002. Amplification of low-frequency antiviral CD8 T cell responses using autologous dendritic cells. AIDS 16:171-180. [DOI] [PubMed] [Google Scholar]
- 34.Lauer, G. M., K. Ouchi, R. T. Chung, T. N. Nguyen, C. L. Day, D. R. Purkis, M. Reiser, A. Y. Kim, M. Lucas, P. Klenerman, and B. D. Walker. 2002. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J. Virol. 76:6104-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMichael, A. J., and S. L. Rowland-Jones. 2001. Cellular immune responses to HIV. Nature 410:980-987. [DOI] [PubMed] [Google Scholar]
- 36.Mothe, B. R., H. Horton, D. K. Carter, T. M. Allen, M. E. Liebl, P. Skinner, T. U. Vogel, S. Fuenger, K. Vielhuber, W. Rehrauer, N. Wilson, G. Franchini, J. D. Altman, A. Haase, L. J. Picker, D. B. Allison, and D. I. Watkins. 2002. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. J. Virol. 76:875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 38.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5:518-525. [DOI] [PubMed] [Google Scholar]
- 39.Pittet, M. J., A. Zippelius, D. E. Speiser, M. Assenmacher, P. Guillaume, D. Valmori, D. Lienard, F. Lejeune, J. C. Cerottini, and P. Romero. 2001. Ex vivo IFN-gamma secretion by circulating CD8 T lymphocytes: implications of a novel approach for T cell monitoring in infectious and malignant diseases. J. Immunol. 166:7634-7640. [DOI] [PubMed] [Google Scholar]
- 40.Price, P., R. P. Johnson, D. T. Scadden, C. Jassoy, T. Rosenthal, S. Kalams, and B. D. Walker. 1995. Cytotoxic CD8+ T lymphocytes reactive with human immunodeficiency virus-1 produce granulocyte/macrophage colony-stimulating factor and variable amounts of interleukins 2, 3, and 4 following stimulation with the cognate epitope. Clin. Immunol. Immunopathol. 74:100-106. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 42.Rowland-Jones, S. L., S. Pinheiro, R. Kaul, P. Hansasuta, G. Gillespie, T. Dong, F. A. Plummer, J. B. Bwayo, S. Fidler, J. Weber, A. McMichael, and V. Appay. 2001. How important is the ′quality' of the cytotoxic T lymphocyte (CTL) response in protection against HIV infection? Immunol. Lett. 79:15-20. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 44.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 45.Stern, B. V., B. O. Boehm, and M. Tary-Lehmann. 2002. Vaccination with tumor peptide in CpG adjuvant protects via IFN-gamma-dependent CD4 cell immunity. J. Immunol. 168:6099-6105. [DOI] [PubMed] [Google Scholar]
- 46.van Baalen, C. A., O. Pontesilli, R. C. Huisman, A. M. Geretti, M. R. Klein, F. de Wolf, F. Miedema, R. A. Gruters, and A. D. Osterhaus. 1997. Human immunodeficiency virus type 1 Rev- and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J. Gen. Virol. 78:1913-1918. [DOI] [PubMed] [Google Scholar]
- 47.Walker, B. D., S. Chakrabarti, B. Moss, T. J. Paradis, T. Flynn, A. G. Durno, R. S. Blumberg, J. C. Kaplan, M. S. Hirsch, and R. T. Schooley. 1987. HIV-specific cytotoxic T lymphocytes in seropositive individuals. Nature 328:345-348. [DOI] [PubMed] [Google Scholar]
- 48.Yang, J., V. M. Lemas, I. W. Flinn, C. Krone, and R. F. Ambinder. 2000. Application of the ELISPOT assay to the characterization of CD8+ responses to Epstein-Barr virus antigens. Blood 95:241-248. [PubMed] [Google Scholar]
- 49.Yu, X. G., M. M. Addo, E. S. Rosenberg, W. R. Rodriguez, P. K. Lee, C. A. Fitzpatrick, M. N. Johnston, D. Strick, P. J. Goulder, B. D. Walker, and M. Altfeld. 2002. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1(HIV-1)-specific CD8(+)T-cell responses following acute HIV-1 infection. J. Virol. 76:8690-8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, X. G., H. Shang, M. M. Addo, R. L. Eldridge, M. N. Phillips, M. E. Feeney, D. Strick, C. Brander, P. J. Goulder, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2002. Important contribution of p15 Gag-specific responses to the total Gag-specific CTL responses. AIDS 16:321-328. [DOI] [PubMed] [Google Scholar]
- 51.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1(HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]