Abstract
Clusterin is a secreted protein chaperone up-regulated in several pathologies, including cancer and neurodegenerative diseases. The present study shows that accumulation of aberrant proteins, caused by the proteasome inhibitor MG132 or the incorporation of the amino acid analogue AZC (L-azetidine-2-carboxylic acid), increased both clusterin protein and mRNA levels in the human glial cell line U-251 MG. Consistently, MG132 treatment was capable of stimulating a 1.3 kb clusterin gene promoter. Promoter deletion and mutation studies revealed a critical MG132-responsive region between −218 and −106 bp, which contains a particular heat-shock element, named CLE for ‘clusterin element’. Gel mobility-shift assays demonstrated that MG132 and AZC treatments induced the formation of a protein complex that bound to CLE. As shown by supershift and chromatin-immunoprecipitation experiments, CLE is bound by HSF1 (heat-shock factor 1) and HSF2 upon proteasome inhibition. Furthermore, co-immunoprecipitation assays indicated that these two transcription factors interact. Gel-filtration analyses revealed that the HSF1–HSF2 heterocomplexes bound to CLE after proteasome inhibition have the same apparent mass as HSF1 homotrimers after heat shock, suggesting that HSF1 and HSF2 could heterotrimerize. Therefore these studies indicate that the clusterin is a good candidate to be part of a cellular defence mechanism against neurodegenerative diseases associated with misfolded protein accumulation or decrease in proteasome activity.
Keywords: clusterin, glial cell, heat-shock factor (HSF), proteasome inhibition, proteotoxic stress, transcription
Abbreviations: AP-1, activator protein 1; AZC, L-azetidine-2-carboxylic acid; CAT, chloramphenicol acetyltransferase; CLE, clusterin element; ChIP, chromatin immunoprecipitation; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; ER, endoplasmic reticulum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HSE, heat-shock element; HSF, heat-shock factor; Hsp, heat-shock protein; sHsp, small Hsp; NRE-1, negative regulatory element 1; RT, reverse transcriptase; TBS, Tris-buffered saline; TK, thymidine kinase; UPR, unfolded protein response
INTRODUCTION
In the central nervous system, glial cells have a dramatic influence on neuron survival, by providing neurons with reparative proteins after stress or severe insults. Previous studies have indicated that clusterin may be one of these proteins [1]. Clusterin, also known as apolipoprotein J, is a secreted glycoprotein found in numerous organs and physiological fluids [2]. In the central nervous system, clusterin is mainly synthesized by glial cells [3] and it can be taken up by neurons, especially when neuronal insults occur [1,4]. Clusterin has also been associated with various neurodegenerative pathologies, such as Alzheimer's disease [5,6]. In these pathologies, the role of clusterin has given rise to much controversy. In some reports, the protein was described as a protective molecule involved in neuronal survival [7], whereas other studies have shown a cytotoxic effect of clusterin [5,8]. These dual and paradoxical roles of clusterin remain puzzling. The final action of the protein could depend on its location, extra- or intra-cellular, cytosolic or nuclear, and on its level of expression and accumulation [9].
The cytoprotective role of clusterin can be explained by its chaperone-like activity. It has been demonstrated, similar to αB-crystallin and other small Hsps (heat-shock proteins), to which clusterin is structurally related [10], clusterin binds to and stabilizes denatured proteins in a folding-competent state [11]. In turn, the cytotoxic action of clusterin may be due to its strong aggregative properties. When overexpressed in the cytosol, monomeric uncleaved clusterin or the cleaved α-subunit promotes aggresome formation [12]. Interestingly, the presence of clusterin in aggresomes has been observed in vivo in pathologies associated with abnormal protein deposits [13]. Moreover, clusterin can act in concert with apolipoprotein E on the fate of brain amyloid proteins by delaying the formation of extracellular protein deposits while increasing neurotoxicity [14]. Hence, identifying the transcriptional factors mediating clusterin expression in response to protein disorders would provide new therapeutic avenues for the prevention or the treatment of neurodegenerative diseases.
Clusterin expression is tightly regulated: whereas clusterin expression is low in most normal cells, it is strongly stimulated by various stresses, such as heat shock [15], oxidative stress [16] or ionizing radiation [17]. Given the close relationships between clusterin expression, cellular stress, disturbance of protein homoeostasiss and aggregative propensity of proteins in neurodegenerative diseases, we asked whether clusterin expression in glial cells could be up-regulated in response to unfolded protein accumulation. In the present study, we used the proteasome inhibitor MG132 or incorporation of the amino acid analogue AZC (L-azetidine-2-carboxylic acid) to induce unfolded protein accumulation. The present study shows that these two drugs increase the clusterin level and that clusterin induction has a transcriptional origin. Furthermore, we identified the transcription complex mediating this induction, made of a novel association between HSF1 (heat-shock factor 1) and HSF2.
MATERIALS AND METHODS
Plasmid constructs
The rat clusterin gene promoter reporter plasmids (pClust-1297bp-Luc; pClust-218bp-Luc; pClust-106bp-Luc; pClust-67bp-Luc; pClust-Δ609/-35-Luc; where Luc is luciferase) were a gift from Dr P. H. Howe (Department of Cell Biology, Cleveland Clinic Lemer College of Medicine, Cleveland Clinic Foundation, Case Western Reserve University of Cleveland, Cleveland, OH, U.S.A.). The pClust mut.CLE construct was generated by site-directed mutagenesis, starting from the pClust-218bp-Luc plasmid. A single mutation, deleting the second GAA recognition site of CLE (clusterin element), was created using the QuikChange® XL site-directed mutagenesis kit from Stratagene. The mutagenic primer 5′-AGGCTTCCAGATAGCTCC-3′ corresponds to the −127/−110 region of the promoter. The resulting mutated plasmid was confirmed by sequencing. The heterologous reporter plasmids containing one, two or four CLE elements in front of the TK (thymidine kinase) gene promoter (CLEx1-TK-CAT; CLEx2-TK-CAT; CLEx4-TK-CAT (chloramphenicol acetyltransferase), or two consensus HSEs (heat-shock elements) in front of a TATA box (HSEx2-TATA-Luc), were already described [15,18]. Dr A. Iwaki (Division of Disease Genes, Institute of Genetic Information, Kyushu University, Fukuoka, Japan) kindly provided the human αB-crystallin promoters fused to the CAT reporter gene (pαBcryst and pαBcryst mut.HSE).
Cell culture and transfections
The human astrocytoma cells, U-251 MG, were a gift from Dr Iwaki. Cells were grown in Dulbecco's modified Eagle's medium (Gibco/Life Technologies), supplemented with 10% (v/v) fetal calf serum, 1 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin and 0.25 μg/ml amphotericin (Gibco/Life Technologies). Transfections were performed using the calcium phosphate co-precipitation method, as previously described [19]. After transfection, cells were treated either with 5 μM MG132 (Sigma) for 16 h or with 25 mM AZC (Sigma), for 24 h. Luciferase assays were performed using the Promega luciferase assay system, according to the manufacturer's procedure. The CAT assays were carried out as previously described [19].
RT (reverse transcriptase)–PCR analyses
Total RNAs were prepared from cultured U-251 MG cells, using TRIzol® reagent (Gibco/Life Technologies). RNA (2 μg) from each sample was reverse-transcribed in the presence of 50 μM random hexamer primers. PCRs were performed as described previously [20]. The actin and the acidic ribosomal phosphoprotein PO mRNAs were used as internal controls for RT–PCR. The Hsp70 messenger amplification was used as the positive control of stress induction. Upstream (up) and downstream (down) primers were defined in different exons, as follows: clusterin: up 5′-AATGTGGGCTCCAAGCAGATC-3′ and down 5′-GAGATGTTCAGCATGTTCAGCAG-3′; αB-crystallin: up 5′-TTCTTCGGAGAGCACCTGTT-3′ and down 5′-TCCGGTACTTCCTGTGGAAC-3′; PO: up 5′-AAYGTGGGCTCCAAGCAGATG-3′ and down 5′-GAGATGTTCAGCATGTTCAGCAG-3′; actin: up 5′-GACAGGATGCAGAAGGAGAT-3′ and down 5′-TTGCTGATCCACATCTGCTG-3′; and Hsp70: up 5′-GGACATCAGCCAGAACAAGC-3′ and down 5′-GTGTAGAAGTCGATGCCCTC-3′.
Protein extract preparation and Western-blot analyses
Whole-cell extracts were obtained using an osmotic shock procedure. Dry cell pellets were resuspended in a buffer containing 10 mM Hepes (pH 7.9), 0.1 mM EGTA, 0.5 mM DTT (dithiothreitol), 5% (v/v) glycerol and protease inhibitor cocktail (Complete™ Mini EDTA-free; Roche). After 10 min of incubation on ice, the NaCl concentration was raised to 0.4 M and extracts were incubated for 10 min on ice. A 30 min centrifugation at 20000 g was then performed, and the supernatant was transferred to a fresh tube and kept at −80 °C. Nuclear extracts were prepared using the nuclear extract kit (Active Motif, Rixensart, Belgium) according to the manufacturer's procedure. All protein extracts were separated on a 7.5% (w/v) polyacrylamide gel and transferred on to a nitrocellulose membrane (Amersham Biosciences). Western blotting was performed as previously described [19]. Anti-clusterin (sc-6420), anti-HSF1 (sc-17756) and anti-HSF2 (sc-13056) antibodies were purchased from Santa Cruz Biotechnology. Anti-Hsp70 (MS-482-PO) and anti-β-tubulin (T4026) antibodies were obtained from LabVision and Sigma respectively. Primary antibodies were revealed using horseradish peroxidase-conjugated IgG (Amersham Biosciences), followed by enhanced chemiluminescence detection as recommended by the manufacturer's instructions (Amersham Biosciences).
Immunoprecipitation
For immunoprecipitation of HSF, 2×106 cells were lysed for 1 h on ice in 250 μl of Nonidet P40 lysis buffer (150 mM NaCl, 50 mM Tris/HCl, pH 8.0, and 1% Nonidet P40), with the protease inhibitor cocktail. The lysates were centrifuged at 20000 g for 30 min at 4 °C. Soluble cell extracts (30 μl) were used as input for Western-blot analyses. The rest of the soluble fractions were precleared at 4 °C for 30 min in 500 μl of TBS (Tris-buffered saline) containing 1% Triton X-100 (50 mM Tris/HCl, pH 7.5, 150 mM NaCl and 1% Triton X-100) with 50 μl of Protein A–Sepharose beads (Amersham Biosciences). Cell extracts were then incubated in the same buffer for 2 h at 4 °C with the antiHSF1 (sc-9144) or the anti-HSF2 (sc-13056) antibodies, followed by a 2 h incubation at 4 °C with Protein A–Sepharose beads. After extensive washing in TBS containing 1% Triton X-100 and then in TBS only, the immunocomplexes were analysed by immunoblotting as described above.
ChIP (chromatin immunoprecipitation) assays
For ChIP assays, U-251 MG cells, treated or not with MG132, were fixed in fixation buffer (PBS and 1.5%, v/v, formaldehyde) for 15 min to cross-link chromatin. Then, cells were sonicated three times for 15 s in lysis buffer (10 mM EDTA, 50 mM Tris/HCl, pH 8.0, 1% SDS and protease inhibitor cocktail). After centrifugation, 10 μl of the supernatants containing the genomic fragments was used as positive PCR controls, and the remaining amounts were subjected to immunoprecipitation with the antiHSF1 (sc-9144) or the anti-HSF2 (sc-13056) antibodies and Protein A–Sepharose beads. Immunoprecipitation was performed at 4 °C, for 16 h in immunoprecipitation buffer (0.5 mM DTT, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris/HCl, pH 8.1). After extensive washes, immunoprecipitated DNA–protein complexes were eluted by a brief incubation in extraction buffer (0.1 M NaHCO3 and 1% SDS) [21]. DNA fragments were then isolated from complexes by an overnight 65 °C incubation, followed by alcohol precipitation in 2 vol. of ethanol. Semi-quantitative PCRs were performed using the following upstream (up) and downstream (down) primers for clusterin, Hsp70 gene promoters and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) flanking region: up 5′-CGGTGCTGCACCGGCCC-3′, down 5′-CTGGGAGGCGCCGTATTTATAGC-3′ – up 5′-GGCACTCTGGCCTCTGATTGGT-3′, down 5′-TGAGCCAATCACCGAGCTCG-3′ – up 5′-ATGGTTGCCACTGGGGATCT-3′, down 5′-TGCCAAAGCCTAGGGGAAGA-3′ respectively.
EMSAs (electrophoretic mobility-shift assays)
Binding reactions for EMSA were carried out in 20 μl volumes containing 10 mM Tris/HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 5% glycerol, 1 μg of poly(dI-dC)·(dI-dC) and 1 ng of 32P-labelled double-stranded oligonucleotides. Whole-cell extracts (5 μg) were added and incubation continued for 30 min on ice. For supershift assay, 1 μl of anti-HSF1 (sc-9144) or anti-HSF2 (sc-13056) antibodies were added to the reaction mixture before incubation. The DNA–protein complexes were resolved on 5% polyacrylamide native gels in 45 mM Tris/borate, 20 mM EDTA and 2.5% glycerol buffer. Electrophoresis was run at 22 mA for 2 h. Gels were then fixed, dried and subjected to autoradiography. The sequences of the double-stranded DNA used as probes or competitors were: cons.HSE: 5′-tcgagcGAAtgTTC-taGAAac-3′; CLE: 5′-ggcTTCcaGAAagCTCcta-3′; mut.CLE: 5′-ggcTTCcaGATag-CTCcta-3′; GRE: 5′-accAGAACAtcaTG-TTCTgtactcatcAGAACAtcaTGTTCTgat-3′ (nucleotide motifs recognized by factors are underlined).
Gel filtration of DNA–protein complexes
Binding reactions were carried out with 100 μg of U-251 MG cell nuclear extract in 200 μl containing 10 mM Tris/HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 0.5% glycerol, 10 μg of poly(dI-dC)·(dI-dC) and 100 ng of 32P-labelled double-stranded oligonucleotides. Incubation was performed for 30 min on ice, then the DNA–protein complexes were separated from the free probe by gel filtration on a Superdex 200 HR 10/30 column with an FPLC apparatus (AKTA system; Amersham Biosciences). The samples were eluted at 0.3 ml/min with a buffer containing 10 mM Tris/HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.5 mM DTT and 0.5% glycerol. The 0.3 ml fractions were collected and added to 3 ml of aqueous counting scintillant (NACS104; Amersham Biosciences) to detect the radioactive probe. To identify the composition of the specific CLE-binding complexes, 150 μg of nuclear extract from control, heat-shocked or MG132-treated cells was used in gel-filtration experiments, as described above. The fractions containing the fast-eluted DNA-probe–protein complex (peak 1) were pooled, precipitated with trichloroacetic acid and analysed by Western blotting.
RESULTS
Proteotoxic stress enhances clusterin gene expression
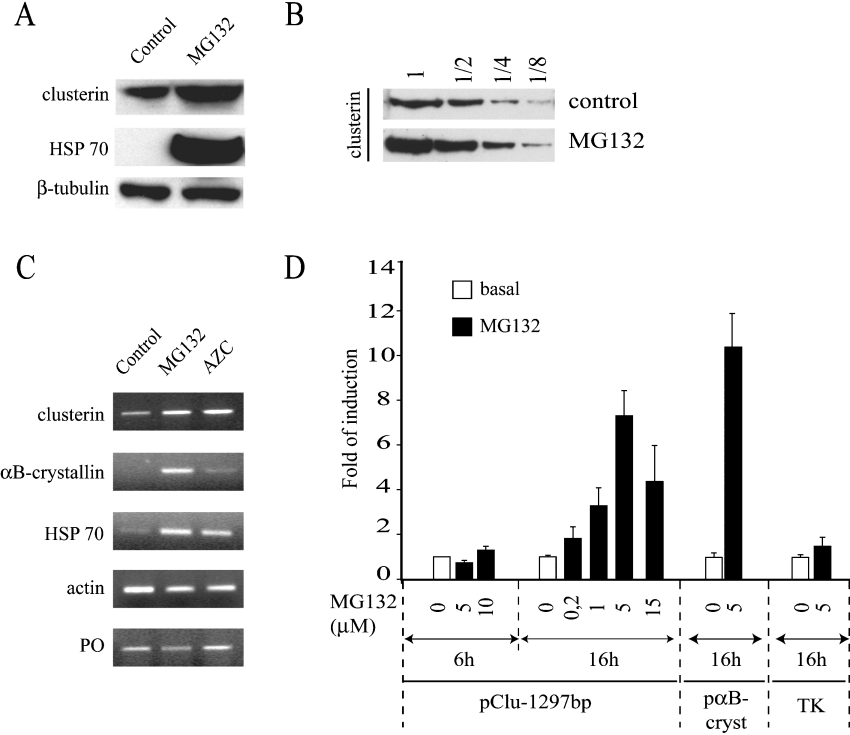
U-251 MG glial cells were treated with the proteasome inhibitor MG132, which is known to induce the intracellular accumulation of misfolded proteins that would otherwise be degraded [22]. Western-blot analysis using specific anti-clusterin antibodies showed that MG132 treatment significantly increased the levels of clusterin protein. Figure 1(A) shows the clusterin precursor that corresponds to the 64 kDa monomeric N-glycosylated clusterin, translocated into the ER (endoplasmic reticulum), but not yet cleaved [23]. Analysis of eight independent Western blots showed that the clusterin precursor level increased from a factor 2 to 5, with a mean of 3.6±1.6 (S.D.). The same MG132 treatment had no effect on the β-tubulin control protein level, whereas it also induced expression of the Hsp70 chaperone protein (Figure 1A), in agreement with previous reports [22,24]. To overcome film saturation artifacts, samples were loaded in three dilutions (2-, 4- or 8-fold) before Western-blot analysis. MG132 reproducibly increased the amount of intracellular clusterin by a factor 2–4 compared with control (Figure 1B). Dose effect and kinetic experiments showed that induction of the clusterin precursor required at least 16 h of treatment with 5 μM MG132 (results not shown). To verify that clusterin accumulation was not due to protein stabilization but to transcriptional activation, a new series of experiments were performed. First, clusterin mRNA content was assessed by RT–PCR. Cells were incubated with either MG132 or AZC, as shown in Figure 1(C). AZC is a competitively incorporated analogue of proline and its incorporation into proteins causes misfolding [25], providing an alternative method leading to misfolded protein accumulation. MG132 and to a lesser extent AZC treatment induced the clusterin, Hsp70 and αB-crystallin mRNAs, whereas it had no effect on the invariant controls (actin and the ribosomal phosphoprotein PO transcripts). Then, transfection experiments with clusterin and αB-crystallin promoter constructs confirmed that overexpression of both clusterin and αB-crystallin has a transcriptional origin (Figure 1D). As shown in the dose effect experiment, the maximal level of transcriptional activation was obtained with an overnight treatment of 5 μM MG132 (Figure 1D). The decrease in luciferase activity observed at 15 μM MG132 reflects likely the proteasome inhibitor toxicity. Nevertheless, cellular basal transcriptional activity was not affected at 5 μM MG132, as shown by the control reporter plasmid containing the viral TK promoter in front of luciferase gene. Interestingly, the transcriptional induction of clusterin needed a sustained MG132 treatment, since the increase in luciferase activity could not be detected after only 6 h of MG132 treatment (Figure 1D). Altogether, these results show that proteotoxic stress triggers transcriptional induction of clusterin and αB-crystallin genes.
Figure 1. Induction of clusterin gene expression by proteotoxic stress.
(A) Western-blot analysis. After a 16 h incubation with DMSO (control) or MG132 (5 μM), proteins were extracted and assayed for expression of clusterin protein precursor. β-Tubulin was used as a loading control and Hsp70 as a positive control of chaperone induction by MG132. (B) Western blotting showing clusterin protein precursor levels. Induction was analysed by comparison of dilution series (1, 1/2, 1/4 or 1/8) of cellular extracts obtained after a 16 h incubation of cells with DMSO (control) or MG132 (5 μM). (C) Semi-quantitative RT–PCR analysis. U-251 MG cells were treated with DMSO (control), with the proteasome inhibitor MG132 (5 μM) or with L-proline analogue AZC (25 μM). mRNA accumulation for the two internal controls, actin and PO, the positive control Hsp70, and the two members of the sHsp family, clusterin and αB-crystallin, were analysed. The data shown correspond to 28-cycle PCRs. The Figure presented is representative of three independent experiments. (D) Clusterin gene promoter activity. U-251 MG cells were transfected with clusterin (pClu-1297bp) or αB-crystallin (pαBcryst) gene promoter reporter plasmids and the control TK promoter. Cells were treated with increasing amounts of MG132 as indicated for various periods of time (6 or 16 h for clusterin), or 16 h at 5 μM for pαBcryst and TK. Folds of activation for CAT or luciferase activities were expressed as the means±S.E.M. (n=3–10).
CLE mediates clusterin induction by MG132
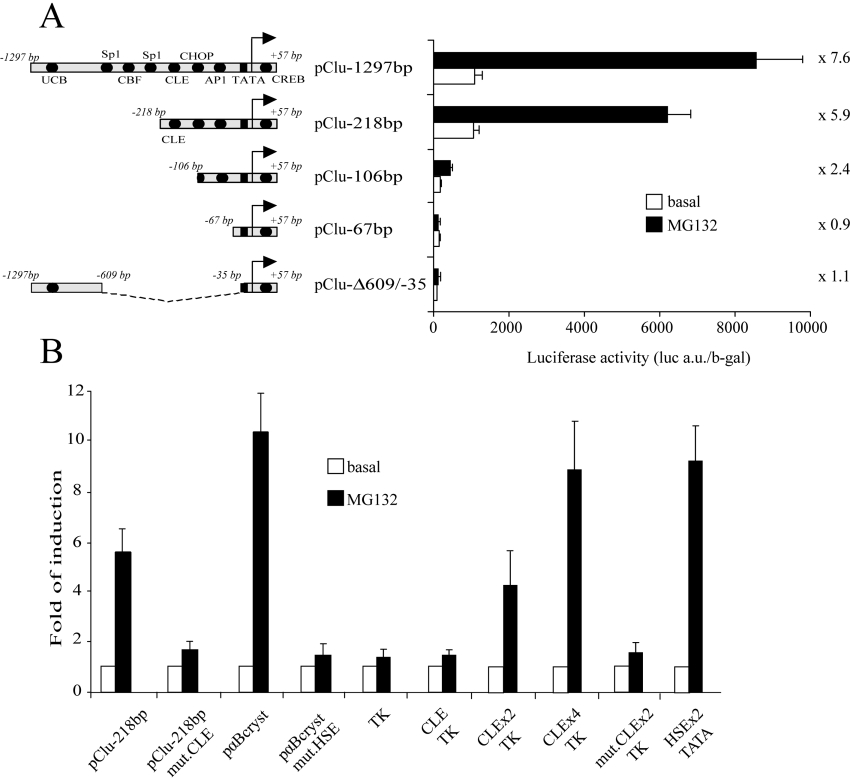
To determine which DNA element is involved in the MG132 induction of clusterin, various clusterin gene promoter constructs with 5′ termini at −1297, −218, −106 and −67 bp were tested by transfection in U-251 MG cells. Deletion of 1079 bp at 5′ termini did not significantly modify the transcriptional induction by MG132 treatment (Figure 2A, compare pClust-1297bp and pClust-218bp). However, deletion up to −106 bp nearly completely abolished the MG132 induction (Figure 2A). Interestingly, the −218/−106 bp region contains the highly conserved DNA element designated CLE. It corresponds to a non-consensus HSE, previously shown to mediate clusterin heat-shock response [15]. To test the role of this element in clusterin MG132 up-regulation, a reporter plasmid containing clusterin promoter with a mutation in the CLE was tested. Since a closely related sequence is found in the αB-crystallin gene promoter, equivalent experiments were carried out with plasmids containing αB-crystallin promoter [26]. For both promoters, mutation of these responsive elements nearly abolished MG132 transcriptional induction (Figure 2B). To determine if CLE is sufficient to mediate MG132 clusterin induction, we used chimaeric promoters containing CLE repeats in front of a minimum TK promoter. As shown in Figure 2(B), a single CLE was not able to significantly induce reporter expression. However, two or four CLE copies were able to confer MG132 sensitivity to our constructs. Altogether, these results show that a single CLE element is necessary to induce MG132 response, even if not sufficient by itself.
Figure 2. CLE is necessary to induce transcriptional response to MG132.
(A) U-251 MG cells were transfected with 5′-deleted fragment of the clusterin promoter, and treated for 16 h with MG132 (5 μM). Luciferase activities were measured and normalized to the β-galactosidase (b-gal) activities. The fold of MG132-induced transcription is indicated for each construct. a.u., arbitrary units. (B) U-251 MG cells were transfected with the pClu-218bp clusterin promoter or with the same DNA fragment containing a mutated CLE sequence (pClu-218bp mut.CLE). As control, constructs containing the αB-crystallin promoter were used, either with a wild-type (pαBcryst) or with a mutated HSE (pαBcryst mut.HSE). Heterologous promoters containing the wild-type (CLE) or the mutated (mut.CLE) CLE in front of the control promoter TK were also assayed. One, two or four repeats of CLE were inserted (CLEx1-TK, CLEx2-TK, mut.CLEx2-TK and CLEx4-TK respectively). As a positive control, we used a tandem repeat of a consensus HSE in front of a luciferase reporter gene (HSEx2-TATA). After transfection, cells were treated with MG132 or untreated. Results are shown as the fold induction and represent the means±S.E.M. for four to ten independent experiments.
HSF1 and HSF2 bind to CLE and form a heterocomplex upon MG132 treatment
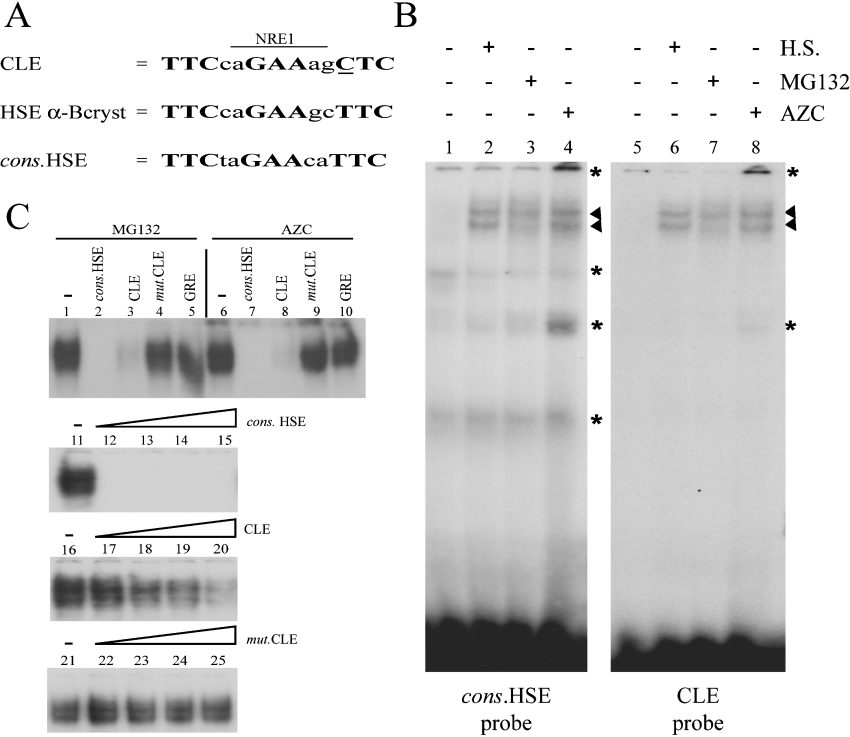
Sequence analysis of CLE reveals two overlapping putative responsive elements (Figure 3A). The first one corresponds to a degenerated HSE shown to be functional under conditions of heat-shock [15]. The second is closely related to an NRE-1 (negative regulatory element 1), involved in the irradiation-stress response for the MMTV (murine-mammary-tumour virus) genome [27]. Using EMSA, the DNA-binding activities obtained with probes containing either CLE or HSE consensus sequence (cons.HSE) lacking the overlapping NRE-1 were compared. Using U-251 MG cell extracts, the same pattern of shifted bands was observed with both probes (Figure 3B). Moreover, cell extracts from cells stressed by MG132, AZC or heat shock showed similar increases in CLE and HSE DNA-binding activities (Figure 3B, arrowheads). Competition experiments were then performed to assess the specificity and the affinity of the CLE-binding complex. A very low amount of unlabelled cons.HSE competitor was able to inhibit the CLE-binding complex formation (Figure 3C, lane 12). Furthermore, the CLE-binding complex had much more affinity for cons.HSE than for CLE, since a 10-fold excess of unlabelled CLE was not sufficient to fully compete with 32P-labelled CLE probe (Figure 3C, lane 20). The CLE-binding factor(s) showed no affinity for the mutant CLE (mut.CLE) competitor (Figure 3C, lanes 22–25). CLE differs from the ideal cons.HSE sequence by only one base in the third binding motif (Figure 3A, underlined nucleotide). This change may decrease CLE-binding complex affinity, and the second mutation in mut.CLE may abolish the sequence recognition. However, band-shift and competition experiments suggest that a similar type of protein complex can bind to both CLE and cons.HSE probes.
Figure 3. MG132 and AZC treatments increase protein DNA-binding activities on CLE.
(A) The conserved CLE sequence is a composite responsive element containing an NRE (NRE-1) overlapping a degenerate HSE. The three binding sites of HSEs are in boldface and the non-consensus nucleotide within CLE is underlined. The cons.HSE and αB-crystallin HSE are shown for comparison. (B) Gel-shift experiments with consensus HSE (lanes 1–4) or CLE (lanes 5–8) 32P-labelled double-stranded oligonucleotides as probes. The binding reaction was performed with cell extracts from U-251 MG control cells (lanes 1 and 5), 42 °C heat-shock (H.S.)-treated cells (lanes 2 and 6), 5 μM MG132-treated cells (lanes 3 and 7) and 25 μM AZC-treated cells (lanes 4 and 8). The position of the specific DNA–protein complexes are marked by arrowheads and the non-specific DNA–protein interactions by asterisks. (C) Competition experiments with 32P-labelled CLE oligonucleotides as probe and cell extracts from MG132- (lanes 1–5 and lanes 11–25) or AZC-treated U-251 MG cells (lanes 6–10). DNA–protein complexes involving specific interactions are shown. Unlabelled competitions were performed using a 25-fold molar excess of cons.HSE (lanes 2 and 7), CLE (lanes 3 and 8), mut.CLE (lanes 4 and 9) or the GRE (glucocorticoid responsive element) (lanes 5 and 10). DNA affinity was analysed using increasing amounts of unlabelled competitors (1-, 2-, 5- and 10-fold molar excess respectively). The unlabelled cons.HSE (lanes 12–15), CLE (lanes 17–20) and mut.CLE (lanes 22–25) oligonucleotides were used as competitors.
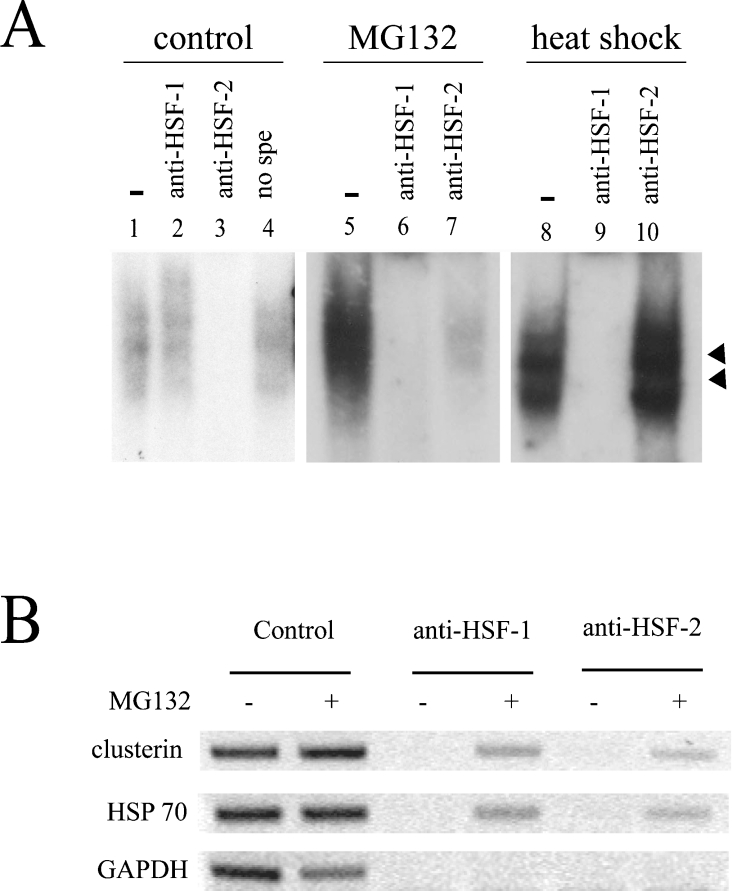
Our EMSA experiments yielded the same characteristic low-mobility band doublet (arrowheads in Figure 3B) as previously described [28,29], suggesting a binding of HSFs to the probes. As both HSF1 and HSF2 are known to bind to HSE, we performed supershift experiments to determine if they bind to CLE under proteasome inhibition. The specificity of each antibody was first tested by Western blotting using in vitro-translated mouse HSF1 or HSF2 (results not shown). Then, cell extracts from control, MG132-treated or heat-shock-stressed (42 °C) cells were compared in supershift assays. As shown in Figure 4(A), the polyclonal antibodies induced a specific disappearance of the shifted band. One can imagine that our antibodies interfere and abolish the HSF–DNA binding, for example, by disturbing the trimerization function. Interestingly, the weak CLE-binding complex from control cells was abolished only by the anti-HSF2 antibodies (Figure 4A, lane 3), whereas in MG132-treated cells, the CLE-binding complex was abolished by the two antibodies (Figure 4A, lanes 6 and 7), showing that both HSF1 and HSF2 are present. Detection of the two HSFs in the shifted bands was specific for the MG132 treatment, since the heat-shock-induced CLE-binding complex was only abolished by anti-HSF1, but not by anti-HSF2, antibodies (Figure 4A, lanes 9 and 10). Moreover, after MG132 treatment, almost 100% of the delayed band could be abolished by anti-HSF1 or anti-HSF2 antibodies (Figure 4A, lanes 6 and 7), strongly suggesting that HSF1 and HSF2 must be both present in the same CLE-binding complex.
Figure 4. HSF1 and HSF2 bind to CLE under MG132 treatment.
(A) EMSAs were performed with the 32P-labelled CLE probe and cell extracts from U-251 MG cells: 5 μg of total extracts from control cells (lanes 1–4), 2 μg of total extracts from MG132-treated cells (5 μM for 16 h, lanes 5–7), or 2 μg of total extracts from heat-shocked cells (lanes 8–10) were used. Only the specific DNA–protein complexes are shown (arrowheads). For supershift experiments, antibodies against HSF1 (lanes 2, 6 and 9), HSF2 (lanes 3, 7 and 10) or normal rabbit serum (no spe: lane 4) were added. Incubation without antibodies is shown in lanes 1, 5 and 8. (B) ChIP analysis of HSE binding activity. ChIP was performed on untreated (−) or MG132-treated (+) cells (5 μM, 16 h) using specific antibodies against HSF1 or HSF2. PCRs were carried out on material before (control) or after immunoprecipitation. Primers flanking CLE and HSE from clusterin and Hsp70 genes respectively were used. A GAPDH flanking region was used as a negative control. The Figure presented is representative of three independent experiments.
To confirm the dual binding of HSF1 and HSF2 to CLE, ChIP experiments were performed on U-251 MG cells. This technique allows the determination of which factors bind to the endogenous gene promoter in living cells. As shown in Figure 4(B) (top panel), CLE fragment was amplified by PCR after immunoprecipitation with either anti-HSF1 or anti-HSF2 antibodies in cells subjected to MG132 treatment. These results confirm that, upon proteasome inhibition, both factors are bound to the CLE element. Similar results were obtained using the Hsp70 promoter (Figure 4B, middle panel), showing that the recruitment of both HSFs to HSE was due to proteasome inhibition and was not specific to the clusterin gene promoter.
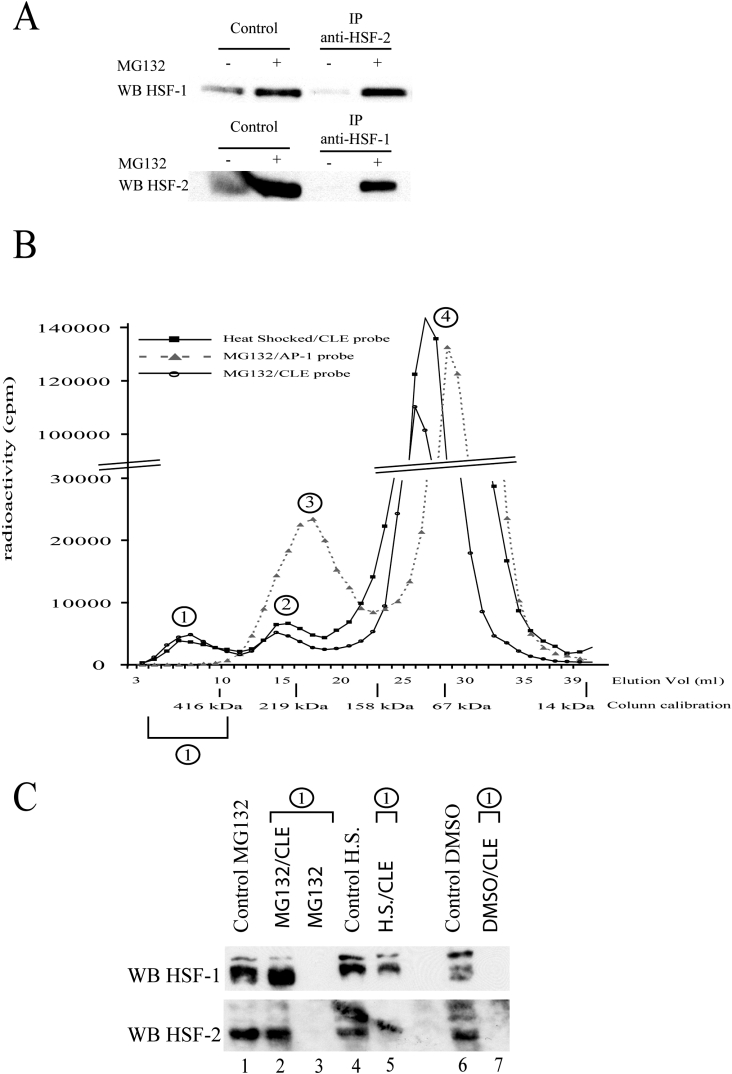
Co-immunoprecipitation experiments were then performed to verify if HSF1 and HSF2 physically interact to form a complex capable of binding to CLE. As shown in Figure 5(A), HSF1 and HSF2 were detected after HSF2 and HSF1 immunoprecipitations respectively. These co-immunoprecipitations were obtained after MG132 treatment, but not in non-treated cells (Figure 5A) or when using non-specific antibodies (results not shown). This demonstrates that proteasome inhibition induces changes in HSFs to allow their association. Together, our results suggest that HSF1 and HSF2 interact, form a complex and acquire DNA-binding activity upon proteasome inhibition. To verify if this HSF1–HSF2 complex corresponds to a novel heterotrimer or is the result of the association of HSF2 with a pre-existing HSF1 homotrimer, sizes of the CLE-binding complexes were measured by gel filtration. This technique has been used previously to determine the monomeric or trimeric nature of HSFs [30,31]. Radioactive CLE probe was incubated with nuclear extract and then passed through a Superdex 200 column, which permitted the elution of distinct radioactive peaks in addition to the free-probe peak (Figure 5B, peak 4). The specific CLE-binding complex was identified using specific (CLE) and non-specific [poly(dI-dC)·(dI-dC)] DNA competitors (results not shown). For the nuclear extracts from MG132 or heat-stressed cells, the specific HSF–CLE complexes were eluted in equivalent fractions (Figure 5B, peak 1), showing that the complex formed under proteasome inhibition is of equivalent size to the HSF1 homotrimer bound to CLE in heat-stressed cells. Unfortunately, this technique does not permit the calculation of the size of the HSF–CLE complex, as this complex contains DNA and probably assumes an extended conformation, which lowers the complex mobility as compared with the globular proteins used for calibration. Nevertheless, it allows comparison of the relative size of HSF–CLE complexes obtained after heat shock and containing only HSF1, or after MG132 treatment and containing both HSF1 and HSF2. As a control, the profile of elution of the AP-1 (activator protein 1)-binding complex (Figure 5B, peak 3), composed of a protein dimer, demonstrates that the volume of elution is a function of the DNA–protein complex size. The presence of both HSF1 and HSF2 in the fractions corresponding to peak 1 was confirmed by Western-blot analysis after trichloroacetic acid precipitation to concentrate proteins. As shown in Figure 5(C), HSF1 and HSF2 were both detected in peak 1 after gel-filtration chromatography in the presence of the CLE probe (Figure 5C, lane 2). Interestingly, in the absence of this probe (Figure 5C, lane 3) neither HSF1 nor HSF2 was detected in these fractions, showing that their elution at this level was due to the formation of a CLE–HSF complex. Nuclear extracts from untreated or heat-shocked cells were used as controls, showing that the formation of HSF1–HSF2 heterocomplex, which is capable of binding to CLE, specifically occurred after proteasome inhibition.
Figure 5. HSF1 and HSF2 form a heterocomplex under MG132 treatment.
(A) HSF1 and HSF2 co-immunoprecipitations. Cell extracts from untreated (−) or MG132-treated (+) cells (5 μM, 16 h) were used to detect HSF1 or HSF2 protein by Western blotting, before (control) or after immunoprecipitation (IP) with specific anti-HSF1 or anti-HSF2 antibodies. The Figure presented is representative of three independent experiments. (B) Estimation of the relative size of HSF–CLE complexes. 32P-labelled CLE or AP-1 probes were incubated with nuclear extract from U-251 MG cells treated as indicated. The sizes of the DNA–protein complexes were estimated by gel-filtration chromatography on a Superdex 200 column, which was calibrated using different globular proteins. Presence of DNA probe was determined by quantification of radioactivity (c.p.m.) in each fraction. Peak 1 corresponds to the specific HSF–CLE complex, whereas peaks 2, 3 and 4 correspond to non-specific complex, AP-1-binding complex and free probes respectively. Identities of each peak were determined by carrying out gel filtration with labelled DNA probe alone (peak 4), or with probe previously incubated with nuclear extract in the presence of high excess of non-specific DNA (peak 2 affected), or with high excess of specific CLE unlabelled competitor (peak 1 disappeared). Breaks in the y-axis scale are for better readability of the Figure. (C) Western-blot analysis of peak 1 contents. Western-blot analyses were carried out on nuclear extracts, before (control, lanes 1, 4 and 6) or after gel-filtration chromatography (lanes 2, 3, 5 and 7). Extracts from untreated cells (DMSO; lanes 6 and 7), MG132-treated cells (lanes 1–3) or from heat-shock-treated cells (H.S., lanes 4 and 5) were used. Gel filtration was performed as described in the Materials and methods section, in the presence (lanes 2, 5 and 7) or absence (lane 3) of CLE probe. Elution fractions corresponding to peak 1 were then pooled, precipitated and assayed for HSF1 and HSF2 presence using specific anti-HSF1 and anti-HSF2 antibodies (upper and lower panels respectively). An unspecific stain is visible in the lower panel, overlapping lanes 4 and 5.
DISCUSSION
Clusterin is a secreted protein chaperone that displays a tightly regulated gene expression. Its synthesis can be stimulated by physiological signals triggered by transforming growth factor-β [32], nerve growth factor or epidermal growth factor [33], but also by environmental stress conditions, such as heat shock [15] or ionizing radiation [17]. The present study shows that protein disorders can also lead to clusterin up-regulation. Accumulation of aberrant proteins within cells was achieved using two different approaches: proteasome inhibition and in corporation of the amino acid analogue AZC. Recently, using microarray analysis, Carreras et al. [34] also described a clusterin induction in lactacystin-treated neuroblastoma cells, but they did not explore the transcriptional pathways involved. Different transcriptional pathways related to UPR (unfolded protein response) have been described [35], depending on the location of misfolded proteins. The ER-specific pathway involves factors such as Ire1 (endoribonuclease, serine-threonine kinase, transmembrane protein 1)/XBP-1 (X-box binding protein 1), ATF6 (activating transcription factor 6) and CHOP [C/EBP (CCAAT/enhancer-binding protein) homology protein] [35]. A second possible pathway involves HSFs, and can also involve other types of cellular stress. Recently, proteasome inhibition was shown to induce the ER-specific pathway [36], and it has been known for a long time that cellular conditions affecting protein conformation can activate HSF DNA binding [37,38].
The clusterin gene promoter contains many putative responsive elements for the two major systems of UPR. Nevertheless, the precise transcription factors involved in the stimulation of the clusterin transcription during proteotoxic stress remained unknown. The present study shows that CLE is necessary for clusterin induction by MG132. CLE is strictly conserved in all homoeothermic vertebrate clusterin genes [15], and EMSA analysis clearly confirmed that this DNA sequence corresponds to a functional HSE which can be recognized by HSFs. Upon activation, HSFs homotrimerize and acquire DNA-binding ability [39]. In mammals, three members of the HSF gene family have been characterized [39,40] and some of them were shown to mediate specific responses to distinct forms of cellular stress. HSF1 is considered as the classical inducer of the heat-shock response, whereas HSF2 is generally believed to be a development-related transcription factor. Intriguingly, these two HSFs are able to recognize the same HSE, making difficult to explain their specificity of action. However, HSF1 binding was shown to utilize a higher degree of co-operativity than HSF2, which is more confined to short HSEs containing a small number of GAA blocks [41,42]. CLE is made of just two perfect and one mutated GAA blocks, which corresponds to the minimal binding site for one HSF trimer. CLE sequence fits to the HSF2 preference, which may explain why EMSA showed mainly an HSF2-binding activity in cell extracts from control cells. Interestingly, CLE is also closely similar to the αB-crytallin HSE (Figure 3A), which was recently found to bind to HSF2 after high potassium treatment [26], and can also mediate the action of MG132 (Figure 2B). Moreover, our results clearly show that dramatic change in CLE-binding activity occurs upon MG132 treatment. We present here complementary results for identifying the nature of the HSF–CLE complex: ChIP experiments demonstrated that both HSF1 and HSF2 bind to CLE; co-immunoprecipitation showed that these factors interact; then finally, supershift and Western blotting after gel filtration indicated that these two factors are present in the same HSF–CLE complex. Altogether, these results, add to the fact that CLE is just long enough to be bound by a single trimer of HSF, suggest that HSF1 and HSF2 can form a heterocomplex after MG132 treatment. HSF1 and HSF2 have already been shown to be capable of interacting together, and the coiled-coil domains of HSF1 and HSF2 were found necessary for this physical interaction, suggesting that HSF1–HSF2 complexes can be true heterotrimers [43,44]. However, the nature of this interaction remains a matter of debate and an important goal would be to distinguish true HSF1–HSF2 heterotrimers from the mere binding of HSF2 to the classical HSF1 homotrimers. The gel-filtration experiment further strengthens this possibility, by showing that the overall molecular mass of the HSF1–HSF2 complex formed after MG132 treatment is similar to that of HSF1 homotrimer obtained after heat shock. However, our experiments did not permit the precise determination of the HSF1–HSF2 stoichiometry, and we also do not know whether a true heterotrimer involving the classical oligomerization domains of HSFs is present. Proteasome inhibitors were already shown to induce both the HSF1 and HSF2 activation [38,45], but it was not known previously that such a treatment can trigger the HSF1–HSF2 heterocomplex formation. The exact molecular mechanisms leading to this association remained to be studied.
Clusterin expression was already found to be regulated by HSF1 following heat shock [15] or retinoid antagonist treatment [46]. In the present study, we show that clusterin is also the target of an original HSFs heterocomplex in response to aberrant protein accumulation, which may explain why clusterin is massively synthesized in neurodegenerative diseases associated with misfolded protein accumulation and impaired proteasome activity [47]. Secreted clusterin has been proposed to function as a holding chaperone, involved in the binding and the clearance of slowly aggregating proteins [10,48]. Clusterin is a suitable candidate to be part of a cellular defence mechanism favouring cell survival. Even if the relationships between all the factors of these pathologies still remain to be elucidated, the present study furthers our understanding of transcriptional adaptation involved in neurodegenerative diseases associated with misfolded protein accumulation and a decrease in proteasome activity.
Acknowledgments
This work was supported by the Association pour la Recherche Contre le Cancer (ARC; no. DM-4479), the Cancéropole Grand Ouest, no. 20048396, the Région Bretagne and Rennes Métropole. F.L. was supported by a grant from the Ministère de l'Education Nationale de La Recherche et de la Technologie.
References
- 1.Michel D., Moyse E., Trembleau A., Jourdan F., Brun G. Clusterin/ApoJ expression is associated with neuronal apoptosis in the olfactory mucosa of the adult mouse. J. Cell Sci. 1997;110:1635–1645. doi: 10.1242/jcs.110.14.1635. [DOI] [PubMed] [Google Scholar]
- 2.Jones S. E., Jomary C. Clusterin. Int. J. Biochem. Cell Biol. 2002;34:427–431. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- 3.Danik M., Chabot J. G., Hassan-Gonzalez D., Suh M., Quirion R. Localization of sulfated glycoprotein-2/clusterin mRNA in the rat brain by in situ hybridization. J. Comp. Neurol. 1993;334:209–227. doi: 10.1002/cne.903340205. [DOI] [PubMed] [Google Scholar]
- 4.Walton M., Young D., Sirimanne E., Dodd J., Christie D., Williams C., Gluckman P., Dragunow M. Induction of clusterin in the immature brain following a hypoxic-ischemic injury. Mol. Brain Res. 1996;39:137–152. doi: 10.1016/0169-328x(96)00019-8. [DOI] [PubMed] [Google Scholar]
- 5.DeMattos R. B., O'Dell M. A., Parsadanian M., Taylor J. W., Harmony J. A., Bales K. R., Paul S. M., Aronow B. J., Holtzman D. M. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtzman D. M. In vivo effects of ApoE and clusterin on amyloid-β metabolism and neuropathology. J. Mol. Neurosci. 2004;23:247–254. doi: 10.1385/JMN:23:3:247. [DOI] [PubMed] [Google Scholar]
- 7.Wehrli P., Charnay Y., Vallet P., Zhu G., Harmony J., Aronow B., Tschopp J., Bouras C., Viard-Leveugle I., French L. E., et al. Inhibition of post-ischemic brain injury by clusterin overexpression. Nat. Med. 2001;7:977–979. doi: 10.1038/nm0901-977. [DOI] [PubMed] [Google Scholar]
- 8.Han B. H., DeMattos R. B., Dugan L. L., Kim-Han J. S., Brendza R. P., Fryer J. D., Kierson M., Cirrito J., Quick K., Harmony J. A., et al. Clusterin contributes to caspase-3-independent brain injury following neonatal hypoxia-ischemia. Nat. Med. 2001;7:338–343. doi: 10.1038/85487. [DOI] [PubMed] [Google Scholar]
- 9.Trougakos I. P., Lourda M., Agiostratidou G., Kletsas D., Gonos E. S. Differential effects of clusterin/apolipoprotein J on cellular growth and survival. Free Radical Biol. Med. 2005;38:436–449. doi: 10.1016/j.freeradbiomed.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 10.Wilson M. R., Easterbrook-Smith S. B. Clusterin is a secreted mammalian chaperone. Trends Biochem. Sci. 2000;25:95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- 11.Humphreys D. T., Carver J. A., Easterbrook-Smith S. B., Wilson M. R. Clusterin has chaperone-like activity similar to that of small heat shock proteins. J. Biol. Chem. 1999;274:6875–6881. doi: 10.1074/jbc.274.11.6875. [DOI] [PubMed] [Google Scholar]
- 12.Debure L., Vayssiere J. L., Rincheval V., Loison F., Le Drean Y., Michel D. Intracellular clusterin causes juxtanuclear aggregate formation and mitochondrial alteration. J. Cell Sci. 2003;116:3109–3121. doi: 10.1242/jcs.00619. [DOI] [PubMed] [Google Scholar]
- 13.Ferrer I., Carmona M., Blanco R., Moreno D., Torrejon-Escribano B., Olive M. Involvement of clusterin and the aggresome in abnormal protein deposits in myofibrillar myopathies and inclusion body myositis. Brain Pathol. 2005;15:101–108. doi: 10.1111/j.1750-3639.2005.tb00504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMattos R. B., Cirrito J. R., Parsadanian M., May P. C., O'Dell M. A., Taylor J. W., Harmony J. A., Aronow B. J., Bales K. R., Paul S. M., et al. ApoE and clusterin cooperatively suppress Aβ levels and deposition: evidence that ApoE regulates extracellular Aβ metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 15.Michel D., Chatelain G., North S., Brun G. Stress-induced transcription of the clusterin/apoJ gene. Biochem. J. 1997;328:45–50. doi: 10.1042/bj3280045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viard I., Wehrli P., Jornot L., Bullani R., Vechietti J. L., Schifferli J. A., Tschopp J., French L. E. Clusterin gene expression mediates resistance to apoptotic cell death induced by heat shock and oxidative stress. J. Invest. Dermatol. 1999;112:290–296. doi: 10.1046/j.1523-1747.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- 17.Criswell T., Beman M., Araki S., Leskov K., Cataldo E., Mayo L. D., Boothman D. A. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J. Biol. Chem. 2005;280:14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- 18.Le Goff P., Le Drean Y., Le Peron C., Le Jossic-Corcos C., Ainouche A., Michel D. Intracellular trafficking of heat shock factor 2. Exp. Cell Res. 2004;294:480–493. doi: 10.1016/j.yexcr.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 19.Le Drean Y., Mincheneau N., Le Goff P., Michel D. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology. 2002;143:3482–3489. doi: 10.1210/en.2002-220135. [DOI] [PubMed] [Google Scholar]
- 20.Sourisseau T., Desbois C., Debure L., Bowtell D. D., Cato A. C., Schneikert J., Moyse E., Michel D. Alteration of the stability of Bag-1 protein in the control of olfactory neuronal apoptosis. J. Cell Sci. 2001;114:1409–1416. doi: 10.1242/jcs.114.7.1409. [DOI] [PubMed] [Google Scholar]
- 21.Metivier R., Stark A., Flouriot G., Hubner M. R., Brand H., Penot G., Manu D., Denger S., Reid G., Kos M., et al. A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol. Cell. 2002;10:1019–1032. doi: 10.1016/s1097-2765(02)00746-3. [DOI] [PubMed] [Google Scholar]
- 22.Bush K. T., Goldberg A. L., Nigam S. K. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 23.Lakins J., Bennett S. A., Chen J. H., Arnold J. M., Morrissey C., Wong P., O'Sullivan J., Tenniswood M. Clusterin biogenesis is altered during apoptosis in the regressing rat ventral prostate. J. Biol. Chem. 1998;273:27887–27895. doi: 10.1074/jbc.273.43.27887. [DOI] [PubMed] [Google Scholar]
- 24.Kim D., Kim S. H., Li G. C. Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem. Biophys. Res. Commun. 1999;254:264–268. doi: 10.1006/bbrc.1998.9840. [DOI] [PubMed] [Google Scholar]
- 25.Zagari A., Nemethy G., Scheraga H. A. The effect of the L-azetidine-2-carboxylic acid residue on protein conformation. I. Conformations of the residue and of dipeptides. Biopolymers. 1990;30:951–959. doi: 10.1002/bip.360300909. [DOI] [PubMed] [Google Scholar]
- 26.Sadamitsu C., Nagano T., Fukumaki Y., Iwaki A. Heat shock factor 2 is involved in the upregulation of αB-crystallin by high extracellular potassium. J. Biochem. (Tokyo) 2001;129:813–820. doi: 10.1093/oxfordjournals.jbchem.a002924. [DOI] [PubMed] [Google Scholar]
- 27.Giffin W., Torrance H., Rodda D. J., Prefontaine G. G., Pope L., Hache R. J. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature (London) 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 28.Mosser D. D., Theodorakis N. G., Morimoto R. I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol. 1988;8:4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirkkala L., Sistonen L. Antibody supershift assay is inadequate for determining HSF stoichiometry in HSE complexes. Cell Stress Chaperones. 1999;4:259–261. doi: 10.1379/1466-1268(1999)004<0259:asaiif>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakai A., Kawazoe Y., Tanabe M., Nagata K., Morimoto R. I. The DNA-binding properties of two heat shock factors, HSF1 and HSF3, are induced in the avian erythroblast cell line HD6. Mol. Cell. Biol. 1995;15:5268–5278. doi: 10.1128/mcb.15.10.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sistonen L., Sarge K. D., Morimoto R. I. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol. Cell. Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin G., Howe P. H. Regulation of clusterin gene expression by transforming growth factor β. J. Biol. Chem. 1997;272:26620–26626. doi: 10.1074/jbc.272.42.26620. [DOI] [PubMed] [Google Scholar]
- 33.Gutacker C., Klock G., Diel P., Koch-Brandt C. Nerve growth factor and epidermal growth factor stimulate clusterin gene expression in PC12 cells. Biochem. J. 1999;339:759–766. [PMC free article] [PubMed] [Google Scholar]
- 34.Carreras I., Garrett-Young R., Ullman M. D., Eisenhauer P. B., Fine R. E., Wells J. M., Conn K. J. Upregulation of clusterin/apolipoprotein J in lactacystin-treated SH-SY5Y cells. J. Neurosci. Res. 2005;79:495–502. doi: 10.1002/jnr.20374. [DOI] [PubMed] [Google Scholar]
- 35.Patil C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- 36.Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosser D. D., Kotzbauer P. T., Sarge K. D., Morimoto R. I. In vitro activation of heat shock transcription factor DNA-binding by calcium and biochemical conditions that affect protein conformation. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3748–3752. doi: 10.1073/pnas.87.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawazoe Y., Nakai A., Tanabe M., Nagata K. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur. J. Biochem. 1998;255:356–362. doi: 10.1046/j.1432-1327.1998.2550356.x. [DOI] [PubMed] [Google Scholar]
- 39.Pirkkala L., Nykanen P., Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 40.Morimoto R. I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 41.Kroeger P. E., Morimoto R. I. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol. Cell. Biol. 1994;14:7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manuel M., Rallu M., Loones M. T., Zimarino V., Mezger V., Morange M. Determination of the consensus binding sequence for the purified embryonic heat shock factor 2. Eur. J. Biochem. 2002;269:2527–2537. doi: 10.1046/j.1432-1033.2002.02917.x. [DOI] [PubMed] [Google Scholar]
- 43.Alastalo T. P., Hellesuo M., Sandqvist A., Hietakangas V., Kallio M., Sistonen L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 2003;116:3557–3570. doi: 10.1242/jcs.00671. [DOI] [PubMed] [Google Scholar]
- 44.He H., Soncin F., Grammatikakis N., Li Y., Siganou A., Gong J., Brown S. A., Kingston R. E., Calderwood S. K. Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. J. Biol. Chem. 2003;278:35465–35475. doi: 10.1074/jbc.M304663200. [DOI] [PubMed] [Google Scholar]
- 45.Mathew A., Mathur S. K., Jolly C., Fox S. G., Kim S., Morimoto R. I. Stress-specific activation and repression of heat shock factors 1 and 2. Mol. Cell. Biol. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayon Y., Ortiz M. A., Lopez-Hernandez F. J., Howe P. H., Piedrafita F. J. The retinoid antagonist MX781 induces clusterin expression in prostate cancer cells via heat shock factor-1 and activator protein-1 transcription factors. Cancer Res. 2004;64:5905–5912. doi: 10.1158/0008-5472.CAN-03-3657. [DOI] [PubMed] [Google Scholar]
- 47.Fortun J., Li J., Go J., Fenstermaker A., Fletcher B. S., Notterpek L. Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. J. Neurochem. 2005;92:1531–1541. doi: 10.1111/j.1471-4159.2004.02987.x. [DOI] [PubMed] [Google Scholar]
- 48.Bartl M. M., Luckenbach T., Bergner O., Ullrich O., Koch-Brandt C. Multiple receptors mediate apoJ-dependent clearance of cellular debris into nonprofessional phagocytes. Exp. Cell Res. 2001;271:130–141. doi: 10.1006/excr.2001.5358. [DOI] [PubMed] [Google Scholar]