Abstract
GnRH (gonadotropin-releasing hormone) plays a pivotal role in the regulation of reproduction in vertebrates through interaction with a specific receptor. Previously, we isolated a GnRH homo-logue, oct-GnRH, from the common octopus (Octopus vulgaris). In the present study, we have identified a GnRH receptor (oct-GnRHR) specific for oct-GnRH from Octopus brain. Oct-GnRHR includes domains and motifs typical of vertebrate GnRH receptors. The intron-inserted positions are conserved between oct-GnRHR and the chordate GnRHR genes. The oct-GnRHR expressed in Xenopus (South African clawed frog) oocytes was responsive to oct-GnRH, but not to any other HPLC fractions of the Octopus brain extract. These results show that oct-GnRHR is an authentic receptor for oct-GnRH. Southern blotting of reverse-transcription PCR products revealed that the oct-GnRHR mRNA was widely distributed in the central and peripheral nervous systems and in several peripheral tissues. In situ hybridiz-ation showed that oct-GnRHR mRNA was expressed in some regions involved in autonomic functions, feeding, memory and movement. Oct-GnRH was shown to induce steroidogenesis of testosterone, progesterone and 17β-oestradiol in Octopus ovary and testis, where oct-GnRHR was abundantly expressed. These results suggest that oct-GnRH, like its vertebrate counterparts, acts as a multifunctional neurotransmitter, neuromodulator and hormone-like factor, both in Octopus central nervous system and peripheral tissues, and that both structure and functions of the GnRH family are, at least partially, evolutionarily conserved between octopuses and chordates.
Keywords: gonadotropin-releasing hormone (GnRH), invertebrate, G-protein-coupled receptor (GPCR), molecular evolution, reproduction
Abbreviations: AKH, adipokinetic hormone; cGnRH, chicken gonadotropin-releasing hormone; CNS, central nervous system; DIG, digoxigenin; GnRHR, GnRH receptor; GPCR, G-protein-coupled receptor; IC, intracellular loop; mGnRH, mammalian GnRH; nano ESI-TOF-MS, nanoflow electrospray-ionization–time-of-flight MS; ORF, open reading frame; RACE, rapid amplification of cDNA ends; RT, reverse transcription; TFA, trifluoroacetic acid; TM, transmembrane region; UTR, untranslated region
INTRODUCTION
GnRH (gonadotropin-releasing hormone) is responsible for various physiological actions, including reproductive development and function. GnRH is a hypothalamic hormone that is secreted into the hypothalamic–hypophysial portal system to regulate the synthesis and release of pituitary gonadotropins which, in turn, trigger steroidogenesis and stimulate gonadal maturation [1,2]. Furthermore, GnRH is distributed over a wide range of tissues in vertebrates and has diverse neuroendocrine, paracrine, autocrine and neurotransmitter/neuromodulatory roles in the central and peripheral nervous systems [1,3].
GnRHs exert their actions through interactions with specific receptors (GnRHRs) that belong to the rhodopsin-like GPCR (G-protein-coupled receptor) family. GnRHRs are localized in gonadotrope membranes of the anterior pituitary and/or peri-pheral tissues [3,4]. GnRHRs have been isolated from several mammalian and non-mammalian vertebrate species, and the inter-action with GnRH activates phospholipase C β-isoforms via the G protein Gq/G11 resulting in an increased phospholipid turnover and the formation of Ins(1,4,5)P3 and diacylglycerol and elevation of intracellular Ca2+ [1]. Notably, the mammalian GnRHRs have a truncated cytoplasmic C-terminal tail, whereas several non-mammalian GnRHRs are tailed by C-terminal extentions. On the basis of increasing evidence for the occurrence of varied GnRH ligand forms in one organism and the distribution of GnRH-bind-ing sites [1,5], it has been suggested that multiple subtypes of GnRHR are present in individual vertebrate species.
Molecular forms and activities of protostome GnRHs have been little understood. Octopuses are one of the most advanced invert-ebrates in the aspects of dioecism, intelligence, sensory systems and physical ability [6], suggesting that octopuses have some unique neuropeptidergic regulation comparable with those of vertebrates. Previously, we showed that the common octopus (Octo-pus vulgaris) has a unique form of GnRH, oct-GnRH (Table 1), with structural features similar to those of chordate GnRHs and showed luteinizing-hormone-releasing activity in quail (Coturnix coturnix japonica) anterior pituitary cells [7]. Oct-GnRH immunoreactive fibres and oct-GnRH mRNA-expressing cell bodies were distributed in all the neurophils of the CNS (central nervous system) lobes and peripheral tissues, and modulatory effects of oct-GnRH on the contractions of the heart and the oviduct were demonstrated, suggesting that oct-GnRH acts as a modulatory factor in controlling brain functions [8]. We therefore anticipated that investigation of an oct-GnRHR would provide crucial clues to a clarification of biological roles of protostome GnRH. In the present study, we identified a novel GnRHR in Octopus, namely oct-GnRHR. Sequence identity, structural organization, localization and physiological functions of oct-GnRHR provided evidence that oct-GnRHR is the Octopus counterpart of the chordate GnRHRs and that oct-GnRH is in-volved in the regulation of neuronal and reproductive processes, as well as in other physiological functions in Octopus.
Table 1. Primary structures of GnRH-family peptides (a) and two synthetic analogues (b).
| (a) GnRH-family peptides | |
|---|---|
| Species | Primary structure |
| Octopus | pGlu-Asn-Tyr-His-Phe-Ser-Asn-Gly-Trp-His-Pro-Gly-NH2 |
| Mammalian | pGlu-His-Trp-Ser-Tyr-Gly-Leu-Arg-Pro-Gly-NH2 |
| Guinea-pig | pGlu-Tyr-Trp-Ser-Tyr-Gly-Val-Arg-Pro-Gly-NH2 |
| Chicken I | pGlu-His-Trp-Ser-Tyr-Gly-Leu-Gln-Pro-Gly-NH2 |
| Chicken II | pGlu-His-Trp-Ser-His-Gly-Trp-Tyr-Pro-Gly-NH2 |
| Frog | pGlu-His-Trp-Ser-Tyr-Gly-Leu-Trp-Pro-Gly-NH2 |
| Salmon | pGlu-His-Trp-Ser-Tyr-Gly-Trp-Leu-Pro-Gly-NH2 |
| Whitefish | pGlu-His-Trp-Ser-Tyr-Gly-Met-Asn-Pro-Gly-NH2 |
| Sea bream | pGlu-His-Trp-Ser-Tyr-Gly-Leu-Ser-Pro-Gly-NH2 |
| Medaka | pGlu-His-Trp-Ser-Phe-Gly-Leu-Ser-Pro-Gly-NH2 |
| Catfish | pGlu-His-Trp-Ser-His-Gly-Leu-Asn-Pro-Gly-NH2 |
| Herring | pGlu-His-Trp-Ser-His-Gly-Leu-Ser-Pro-Gly-NH2 |
| Dogfish | pGlu-His-Trp-Ser-His-Gly-Trp-Leu-Pro-Gly-NH2 |
| Lamprey I | pGlu-His-Tyr-Ser-Leu-Glu-Trp-Lys-Pro-Gly-NH2 |
| Lamprey III | pGlu-His-Trp-Ser-His-Asp-Trp-Lys-Pro-Gly-NH2 |
| Tunicate I | pGlu-His-Trp-Ser-Asp-Tyr-Phe-Lys-Pro-Gly-NH2 |
| Tunicate II | pGlu-His-Trp-Ser-Leu-Cys-His-Ala-Pro-Gly-NH2 |
| (b) Synthetic analogues | |
| Analogue | Primary structure |
| Des-NY-oct-GnRH | pGlu-His-Phe-Ser-Asn-Gly-Trp-His-Pro-Gly-NH2 |
| NY-cGnRH-II | pGlu-Asn-Tyr-His-Trp-Ser-Tyr-Gly-Trp-Tyr-Pro-Gly-NH2 |
EXPERIMENTAL
Animals
Adult octopuses (body weight approx. 2 kg), Octopus vulgaris, were purchased from a local fish shop and kept in artificial sea-water at 18 °C.
Total RNA and mRNA preparation
Total RNA was extracted from various tissues using Sepasol-RNA I Super (Nacalai tesque, Kyoto, Japan) and mRNA was prepared using an Oligotex™-dT30 mRNA Purification Kit (Takara-bio, Kyoto, Japan) according to the manufacturer's instructions.
Oligonucleotide primers
Oligonucleotide primers were purchased from Operon Techno-logies (Tokyo, Japan) and Proligo (Kyoto, Japan). Degenerate primers, GnRHR Fw1, GnRHR Rev1 and GnRHR Rev2, were designed based on sequences in the second and sixth trans-membrane domains respectively, both of which are conserved among the GnRHR family.
Cloning of the partial-length cDNA
All PCR amplifications were carried out in a reaction mixture containing Ex Taq™ polymerase (Takara-bio) and 200 μM dNTP in a PerkinElmer GeneAmp PCR System 9700 thermal cycler (Applied Biosystems Japan, Tokyo, Japan). Total RNA was iso-lated from various tissues of Octopus and reverse-transcribed using oligo(dT)12–18 primer and Superscript III supplied in the SuperScript III First-Strand Synthesis System for RT (reverse transcription)-PCR (Invitrogen, Carlsbad, CA, U.S.A.). The first round of PCR was performed using GnRHR Fw1 [5′-GCIG(C/T)-ITGGAA(C/T)(A/G)(C/T)I(A/G)TI(C/G)A(A/G)TGG-3′; I represents inosine] and GnRHR Rev1 [5′-GGICI(A/G)AACCA(A/G)TA-CCAIA(A/G/T)ICC-3′] under the following conditions: 5 min at 94 °C, 40 cycles of 30 s at 94 °C, 30 s at 52 °C, 1 min at 72 °C (3 min for the last cycle). The second round of PCR was per-formed using GnRHR Fw1 and GnRHR Rev2 [5′-CCIA-(A/G)IA(A/C/G)(A/G)(A/T)A(A/G)TAIGGIG(C/T)CA(A/G)CA-3′] under the following conditions: 94 °C for 5 min; 30 cycles of 30 s at 94 °C, 30 s at 52 °C, 1 min at 72 °C (3 min for the last cycle). The methods for subcloning and sequencing of PCR products were the same as those previously described [9]. Universal M13 primers or gene-specific primers were used to sequence both strands.
3′-RACE (3′-rapid amplification of cDNA ends) and 5′-RACE
The transcriptional start site was determined by oligo-capping RACE methods by use of the GeneRacer kit (Invitrogen). First-strand cDNA was synthesized from mRNA with the GeneRacer Oligo dT Primer supplied in the GeneRacer kit (Invitrogen) according to the manufacturer's instructions. The first round of PCR was performed using the GeneRacer 3′ Primer and GnRHR 3′-1F (5′-CGAGACAGTTTCACGATACTCC-3′) under the fol-lowing conditions: 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 52 °C, 2 min at 72 °C (7 min for the last cycle). The second round of PCR was performed using the GeneRacer 3′ Nested Primer and GnRHR 3′-2F (5′-TGCAGTGATTGTCGCAGCTTTC-3′) under the following conditions: 94 °C for 5 min; 35 cycles of 30 s at 94 °C, 30 s at 52 °C, 2 min at 72 °C (7 min for the last cycle). The second PCR products were subcloned and sequenced as described above. The 5′-ends of the cDNAs were deter-mined as follows: the first template was amplified using the GeneRacer 5′ Primer and GnRHR 5′-1R (5′-CCAACGCTATGCT-TACTGTTAT-3′). Each of the first PCR products was re-amplified using the GeneRacer 5′ Nested Primer and GnRHR 5′-2R (5′-TGTAAGATACTTCATAATCCTGCAC-3′). Both the first and second rounds of PCR were performed for 5 min at 94 °C, 35 cycles for 30 s at 94 °C, 30 s at 52 °C, and 2 min at 72 °C (7 min for the last cycle). The second PCR products were subcloned and sequenced as described in the above subsection.
Analysis of genomic organization
The methods for isolation of high-molecular-mass DNA and construction of a genomic DNA library were the same as those previously described [9]. Octopus genome DNA digested with DraI, EcoRV, PvuII and StuI was subjected to PCR amplification in the 5′ and 3′ directions using adaptor primers and gene-speci-fic primers according to the manufacturer's instructions. The amplified products were subcloned and sequenced using several gene-specific primers.
Molecular phylogenetic analysis
The deduced amino acid sequences of oct-GnRHR were aligned with the amino acid sequence of GnRHRs and related GPCRs from other animals using the ClustalW program. Amino acid sequences of rat oxytocin receptor was included in the alignment as one group. A neighbour-joining tree was constructed on the basis of alignment by the ClustalW program. The evolutionary distances were estimated using Kimura's [9a] empirical method. The sequences used were as follows: Rattus norvegicus (rat) GnRHR, S59525, Mus musculus (mouse) GnRHR, L01119; Ovis orientalis aries (sheep) GnRHR, L22215; Homo sapiens (human) GnRHR, (I) L03380, (II) AY077708; Rana catesbeiana (bullfrog) GnRHR, (I) AF144063, (II) AF153913, (III) AF144062; Gallus gallus (chicken) GnRHR, AJ304414; Oryzias latipes (Japanese medaka) GnRHR, (I) AB057675, (II) AB057674; Anguilla japonica (Japanese eel), AB041327; Ciona intestinalis (an ascidian) GnRHR, (I) AB103333, (II) AB103334; Rattus norvegicus (rat) oxytocin receptor, P70536; Drosophila melanogaster (fruitfly) AKH (adipokinetic hormone) receptor, AF077299.
Expression of oct-GnRHR in Xenopus oocytes
The ORF (open reading frame) region of oct-GnRHR cDNA was amplified and inserted into a pSP64 poly(A) vector (Promega, Madison, WI, U.S.A.). The plasmid was linearized with EcoRI, and cRNA was prepared using SP6 RNA polymerase (Ambion, Austin, TX, U.S.A.). The method for assay in oocytes of Xenopus laevis was the same as those previously described [10]. oct-GnRH, des-NY-oct-GnRH (des-Asn2-Tyr3-oct-GnRH) and NY-cGnRH-II (endo-Asn2a-Tyr3a-chicken GnRH-II) were synthesized with a solid-phase peptide synthesizer (model 433A; Applied Biosystems Japan) using standard FastMoc™ chemistry. mGnRH (mammalian GnRH), cGnRH-I (chicken GnRH-I) and cGnRH-II (chicken GnRH-II) were purchased from the Peptide Institute (Osaka, Japan). AKH I and II, and corazonin, were purchased from American Peptide (Sunnyvale, CA, U.S.A.).
Isolation and structure determination of an endogenous ligand of oct-GnRHR from Octopus brain
The brain (40 g) was boiled in water and extracted with 3% (v/v) acetic acid. The extract was applied to a solid-phase extraction column (Sep-pak Vac C18; Waters, Milford, MA, U.S.A.). The retained material was eluted with 60% (v/v) acetonitrile in 0.1% TFA (trifluoroacetic acid) and condensed in vacuo. The residue was dissolved in 0.1% TFA and fractionated using a linear gradient elution of 0–36% acetonitrile in 0.1% TFA at a flow rate of 1.5 ml/min on a reversed-phase HPLC with a C18 column (Capcell pak C18; UG80; 10 mm diameter×250 mm long; Shiseido, Tokyo, Japan). Fractions were collected every 2 min. A 1/100 aliquot of each fraction was condensed to dryness in vacuo and dissolved in ND96 buffer [10], then assayed in the Xenopus oocytes that expressed oct-GnRHR. The active fractions were further purified by using the following columns: a cation-exchange column (TSKgel SP-5PW, 7.5 mm×75 mm; Tosoh, Tokyo, Japan; 0–0.6 M NaCl/60 min linear gradient in 20 mM phosphate buffer, pH 7.0; flow rate 1.5 ml/min; fractions collected every 2 min), a C18 column (Capcell pak C18; UG80; 4.6 mm×250 mm; Shiseido; 18–30, 15–27 and 16.8% acetonitrile in 0.1% TFA/40 min linear gradient; flow rate 1.0 ml/min; fractions collected every 1 min). In the assay of cation-exchange HPLC fractions, a 1/100 aliquot of each fraction was diluted in ND96 buffer and applied to the assay chamber.
The molecular mass was determined by a nano ESI-TOF-MS (nanoflow electrospray-ionization–time-of-flight MS) (Q-TOF; Micromass UK, Wythenshawe, Manchester, U.K.). The sequence of oct-GnRH was further confirmed by nano ESI-TOF-MS/MS analysis. The capillary voltage was optimized at 1200 V and the cone voltage was set at 50 V. Argon was used as the collision gas and the energy was set at 35 V.
RT-PCR Southern-blot analysis
Each of total RNAs (1 μg) extracted from various tissues was reverse-transcribed using Superscript III (Invitrogen) and oligo(dT)12–18 primer. The PCR was performed using GnRHR FwA (5′-CAGGATACTCTAATGACATCTACGC-3′) and GnRHR 5′-1R under the following conditions: 5 min at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 52 °C, 1 min at 72 °C (3 min for the last cycle). The PCR products were separated by 1.5%-agarose-gel electrophoresis, and then transferred on to Hybond-N+ membrane (Amersham Bioscience, Piscataway, NJ, U.S.A.) and cross-linked by UV irradiation. Digoxigenin (DIG)-labelled oligonucleotide probes, DIG-GnRHR 5′-2R, were hybridized with the oct-GnRHR cDNA at 50 °C. The method for detection was the same as those previously described [10]. The expression of Octopus β-actin (AB053937) was also tested as an internal control. As a negative control, the extracted total RNAs, which were not reverse transcribed, were used as templates for PCR. Thus, we confirmed that there was no amplification of traces of the genomic DNA (results not shown).
In situ hybridization
DIG-labelled oct-GnRHR antisense and sense RNA probes were synthesized using pCRII-TOPO-oct-GnRHR plasmid and DIG RNA labelling kit (Roche Applied Science, Basel, Switzerland) according to the manufacturer's instructions. The whole brain of Octopus was fixed in a solution of 4% (v/v) paraformaldehyde in PBS, pH 7.4, at 4 °C overnight. After washes with PBS to remove the fixative, the fixed brain was dehydrated in ethanol and xylene, then embedded in paraffin. Serial sections of 5 μm thickness were made and treated as previously described [11]. The methods for detection were carried out according to the DIG SYSTEM protocol (Roche Applied Science).
Biological assay using radula retractor muscle
One end of the radula retractor muscle was tied with a cotton thread to the experimental chamber and the other end was con-nected with a cotton thread to a force-displacement transducer. The tension change was recorded as previously described [12,13]. The train of electrical pulses of stimulation (20 V, 1 ms, 0.2 Hz, five pulses) was applied at 8 min intervals until the muscle responded to the stimulation with a uniform train pattern of twitch contractions.
Steroidogenesis assay
A 50 mg portion of the follicle and of spermatozoa were excised from Octopus. The follicle and spermatozoa were washed with culture medium [L15 medium (Invitrogen) adjusted to seawater salt concentration (460 mM NaCl/10 mM KCl/55 mM MgCl2/11 mM CaCl2), 10 mM Hepes (pH 7.6), 0.1% BSA, and 10 units/ml penicillin/streptomycin] and transferred to sterile 24-well plates (Iwaki, Tokyo, Japan) with each well containing 1 ml of culture medium at 16 °C. The tissues were precultured on the plate for 90 min, and oct-GnRH at indicated concentrations was added to the medium. Medium was collected after the treatment of oct-GnRH at 16 °C and centrifuged at 10000 g for 5 min. The supernatants were used for ELISA of sex steroids. Progesterone, testosterone (R&D systems, Minneapolis, MN, U.S.A.), and 17β-oestradiol (Cayman Chemical Company, Ann Arbor, MI, U.S.A.) were measured using hormone kits according to the manu-facturer's instructions. Results are shown as the mean±S.E.M. Concentration–response studies on in vitro assay were analysed by the one-way ANOVA with Dunnett error protection. Differ-ences were accepted as significant when P<0.05.
RESULTS
Cloning and structure of the oct-GnRHR candidate
The second and sixth transmembrane domains are highly con-served among the known GnRHR family. To identify receptors for oct-GnRH in Octopus, three degenerate primers were designed on the basis of the conserved regions and were applied for RT-PCR of first-strand cDNA prepared from various tissues. BLAST searches of the PCR product sequence showed the high homology with GnRH, corazonin and AKH receptors. A full-length cDNA sequence (1835 bp) encoding the putative octopus GnRH receptor (oct-GnRHR) was determined by the 5′/3′-RACE methods from the Octopus brain. It has an ORF of 1224 bp and 568 bp of 5′-UTR (5′-untranslated region) and 43 bp of 3′-UTR. Multiple sets of clones in every PCR were analysed and gave identical nucleotide sequences.
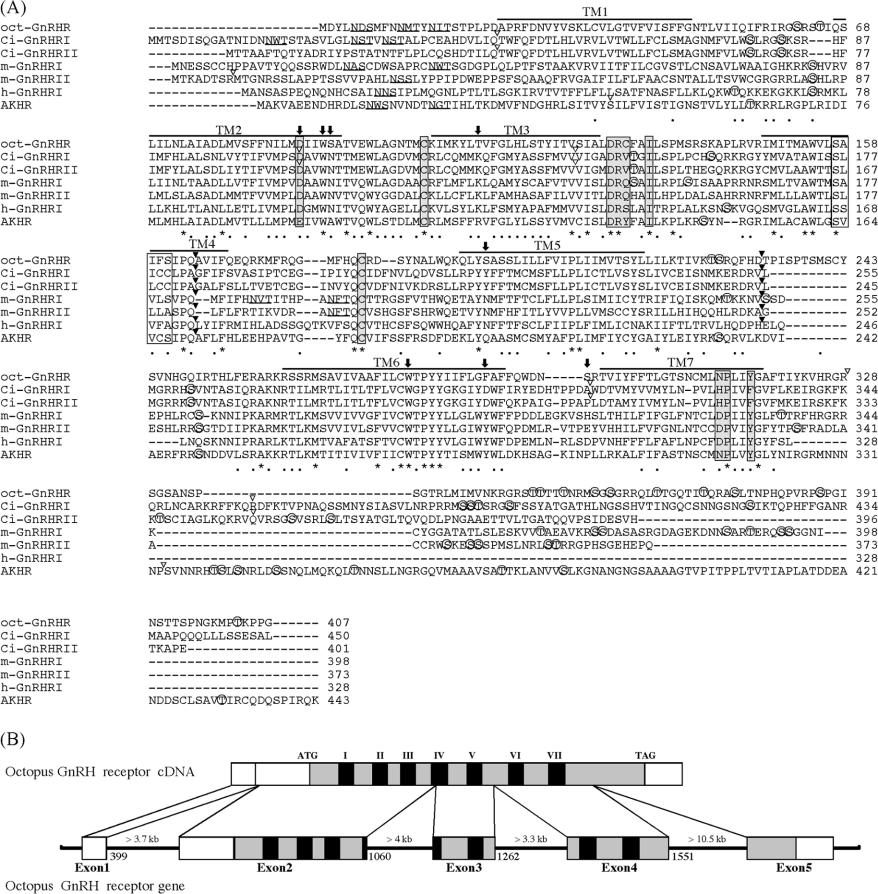
The sequence showed the presence of the seven hydrophobic TMs (transmembrane regions) that are the most typical charac-teristic of GPCRs. As shown in Figure 1(A), the cloned receptor contains several potential sites for N-linked glycosylation and phosphorylation as follows: two sites of consensus sequences for N-linked glycosylation sites (N-X-S/T) in the extracellular N-terminal domain, ten sites of consensus sequences for phos-phorylation by protein kinase A [K/R-X-(X)-S/T] and three sites of phosphorylation by protein kinase C (S/T-X-K/R). Phosphorylation is involved in the modulation of G-protein coupling and receptor function [14]. Putative phosphorylation sites were located exclusively in the third intracellular loop (IC) and the C-terminal region. The GnRHRs are classified as a typical member of the rhodopsin-type (class I) GPCR family. In the class I GPCR family, an aspartic acid residue in TM2 and the consensus tripeptide (E/D-R-Y) at the interface of TM3 are believed to play a key role in the receptor activation [14], whereas this motif is replaced by DRS, DRQ or DRH in vertebrate GnRHRs [15]. The cloned receptor had a Asp77 in TM2 and a DRC sequence at the interface of TM3. Two cysteine residues responsible for a disulphide bridge in most GPCRs were present (at positions 104 and 183) in the first and second extracellular loops in the cloned receptor. The microdomain DRXXXI/V at the junction of TM3 and IC2, and NPX2–3Y in TM7, which are critical for the agonist-induced re-ceptor activation and signal transduction in the class I GPCR family [16,17], were conserved as DRCFAI and NPLIY in the cloned receptor. It is believed that TM4 of a number of GPCRs, including GnRHRs, contain the G/SXXXG/S motif, which is in-volved in homo- or hetero-dimerization of GPCRs via helix–helix association [1]. The cloned receptor was also found to harbour SAIFS motif in TM4. These results indicated that the cloned receptor belongs to the class I GPCR family.
Figure 1. Sequence alignment of oct-GnRHR and other GnRHRs.
(A) The amino acid sequence of oct-GnRHR is aligned with those of the Ciona intestinalis GnRH receptor (Ci-GnRHR I and II [18]), the medaka GnRH receptors (m-GnRHR I and II [19]), human GnRH receptor I (h-GnRHR I [20]) and Drosophila AKH receptor [21]. Amino acid residues conserved in all receptors are indicated by an asterisk. N-linked glycosylation sites are underlined. Bars indicate the seven putative transmembrane domains. Potential phosphorylated serine or threonine residues are marked by open circles. Amino acid residues in grey boxes are believed to play a pivotal role in the GPCR activation, whilst those in white boxes are thought to favour TM helix–helix association [1]. Residues in the agonist-binding pocket described in the text are shown by arrows. The conserved introns in the corresponding genes are indicated the positions by black arrowheads above the sequences; open arrowheads indicate non-conserved introns. (B) Structure organization of the Octopus oct-GnRHR gene. Exons are white boxes, ORFs are grey and TMs are black.
Comparative analysis of amino acid sequences and exon/intron structures between the cloned receptor and the GnRH receptor family
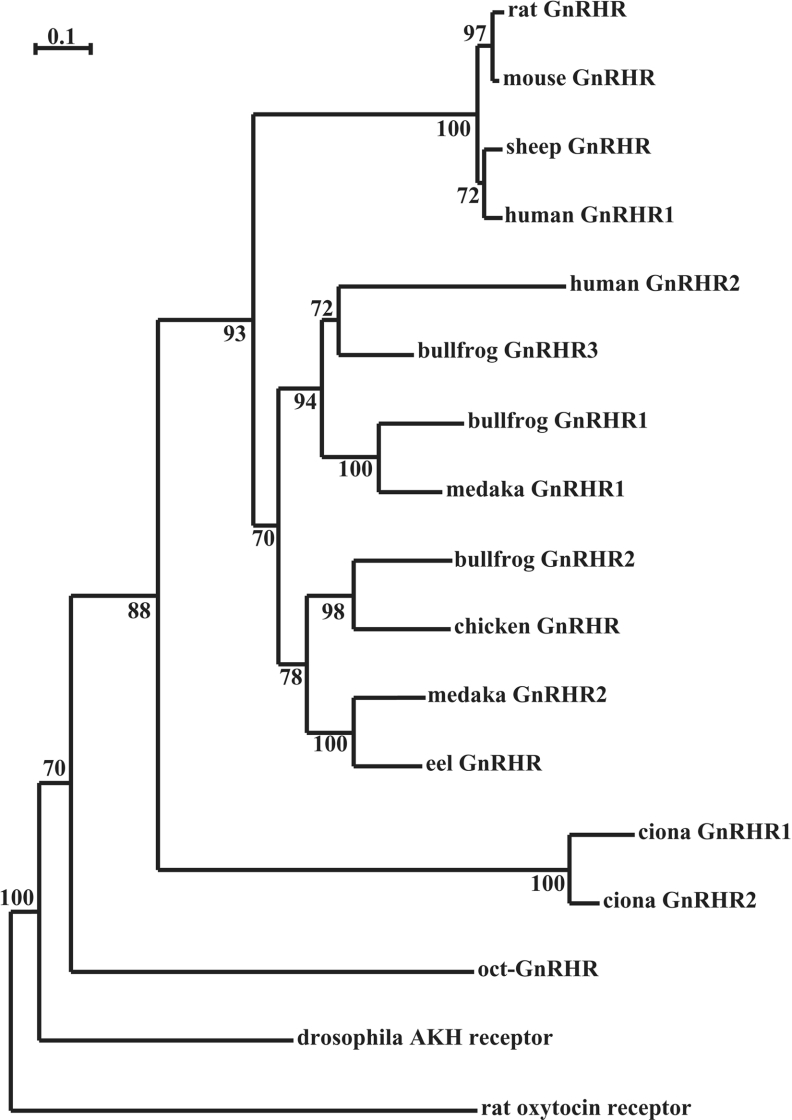
The sequence encoding the intracellular, extracellular and trans-membrane domains of the cloned receptor showed high amino acid sequence similarity (26.7–37.4%) to those of the other GnRHR family members (Table 2). Molecular phylogenetic analysis of GnRHR sequences showed that vertebrate GnRHRs can be classified into three groups, and the cloned receptor was found to belong to a separate sister group of the clade among chordate GnRHRs (Figure 2). On the basis of these results, the cloned receptor is suggested to be a novel orthologue of the GnRHR.
Table 2. Identity of sequence encoding the intracellular, extracellular and transmembrane domains of oct-GnRHR to those of other GnRHRs, AKH and corazonin receptor (A) and that encoding each transmembrane domain (I–VII).
| Percentage sequence identity versus Octopus GnRHR | ||||||||
|---|---|---|---|---|---|---|---|---|
| Species | A | I | II | III | IV | V | VI | VII |
| Human I | 27.6 | 20.8 | 22.2 | 30.4 | 45.5 | 33.3 | 31.0 | 38.1 |
| Human II | 27.9 | – | – | – | – | – | – | – |
| Rat | 28.8 | – | – | – | – | – | – | – |
| Medaka I | 35.9 | 22.2 | 40.7 | 21.7 | 50.0 | 33.3 | 55.2 | 42.1 |
| Medaka II | 33.6 | 7.1 | 40.7 | 39.2 | 36.8 | 33.3 | 44.8 | 33.3 |
| Chicken | 29.8 | – | – | – | – | – | – | – |
| Eel | 33.1 | – | – | – | – | – | – | – |
| Ciona I | 26.7 | 20.8 | 22.2 | 21.7 | 36.4 | 29.2 | 34.5 | 20.0 |
| Ciona II | 26.9 | 20.8 | 25.9 | 21.7 | 31.8 | 29.2 | 34.5 | 20.0 |
| Drosophila AKH | 33.3 | 29.2 | 40.7 | 47.8 | 50.0 | 33.3 | 44.8 | 47.4 |
| Corazonin | 37.4 | – | – | – | – | – | – | – |
Figure 2. Molecular phylogenetic tree of GnRHR family.
A phylogenetic tree was inferred from the amino acid sequences by the neighbour-joining method. A total of 1000 bootstrap trials were run. The scale bar indicates 0.1 amino acid replace-ments per site. The numbers at each branch node represent the percentage values given by booststrap. Rat oxytocin receptor was used as an outgroup.
Although many GPCR genes have an intronless gene structure, the GnRHR genes [18–20] contain introns at the same location in TM4 and IC3. Thus we determined the exon/intron structure of the cloned receptor. The cloned receptor gene consisted of five exons and four introns. The introns longer than 3.7, 4, 3.3 and 10.5 kb were inserted at positions 399–400, 1060–1061, 1262–1263, and 1551–1552 respectively in the cloned receptor (Figure 1B), which was supported by the presence of the GT-AG splicing consensus at exon/intron junctions. The cloned receptor was also found to harbour introns in TM4 and IC3 (Figure 1A), which were located at the same positions as in chordate GnRHRs. This comparative analysis suggested that the chordate GnRHR genes and the cloned receptor gene were derived from a common ancestral gene, and these results allowed us to conclude that this GPCR is a receptor for oct-GnRH and to designate the GPCR as oct-GnRHR.
Functional expression of oct-GnRHR in Xenopus oocytes and isolation of an endogenous ligand from the octopus brain
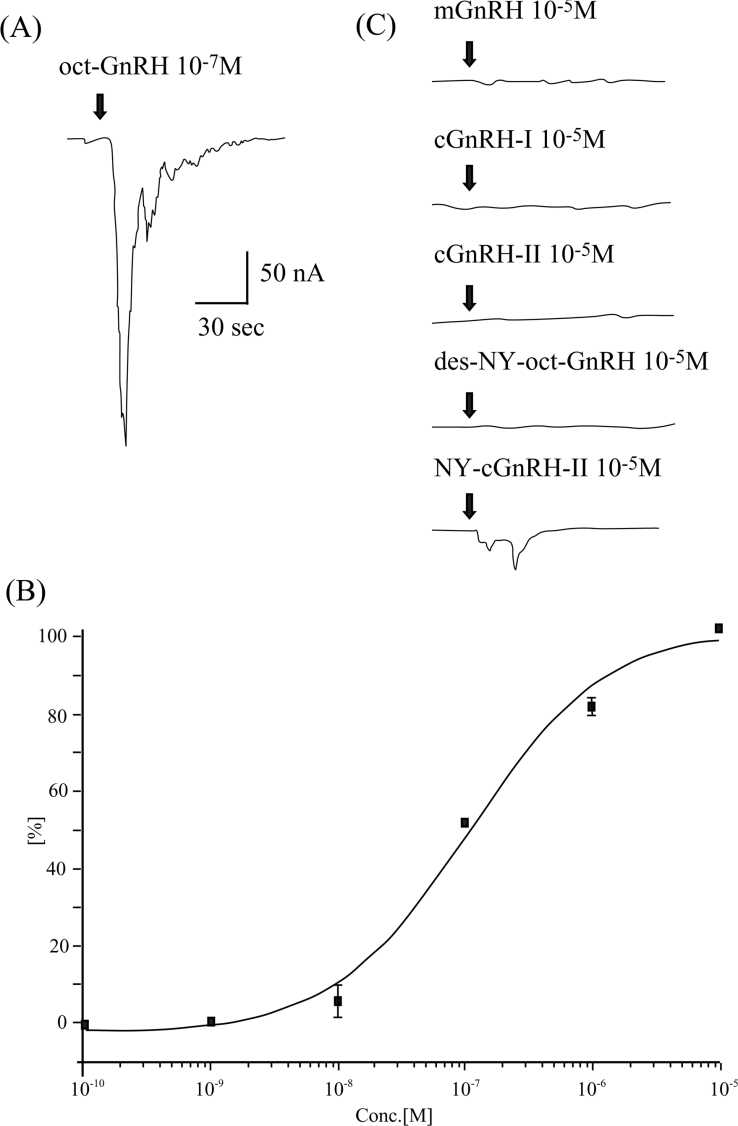
To evaluate binding affinity and selectivity of oct-GnRHR to a putative ligand oct-GnRH, the activation of GnRHRs expressed in Xenopus oocytes by GnRHs was evaluated by monitoring an induction of membrane Cl− currents coupled to the inositol phos-phate/Ca2+ pathway [3,18]. The voltage-clamped oocytes express-ing oct-GnRHR displayed typical inward membrane currents upon application of oct-GnRH at 10−7 M (Figure 3A). The EC50 was estimated to be 97.9±4.3 nM from the concentration–response curve (Figure 3B).
Figure 3. Functional expression of oct-GnRHR in Xenopus oocytes.
(A) Traces of membrane current induced by oct-GnRH at 10−7 M in an oocyte expressing oct-GnRHR. (B) Concentration–response curve over the concentration range of 10−10–10−5 M oct-GnRH in oct-GnRHR. Maximum membrane currents elicited by the ligands are plotted. Error bars denote S.E.M. (n=5). (C) Traces of membrane current induced by mGnRH, cGnRH-I, cGnRH-II, des-NY-oct-GnRH, and NY-cGnRH-II at 10−5 M in an oocyte expressing oct-GnRHR.
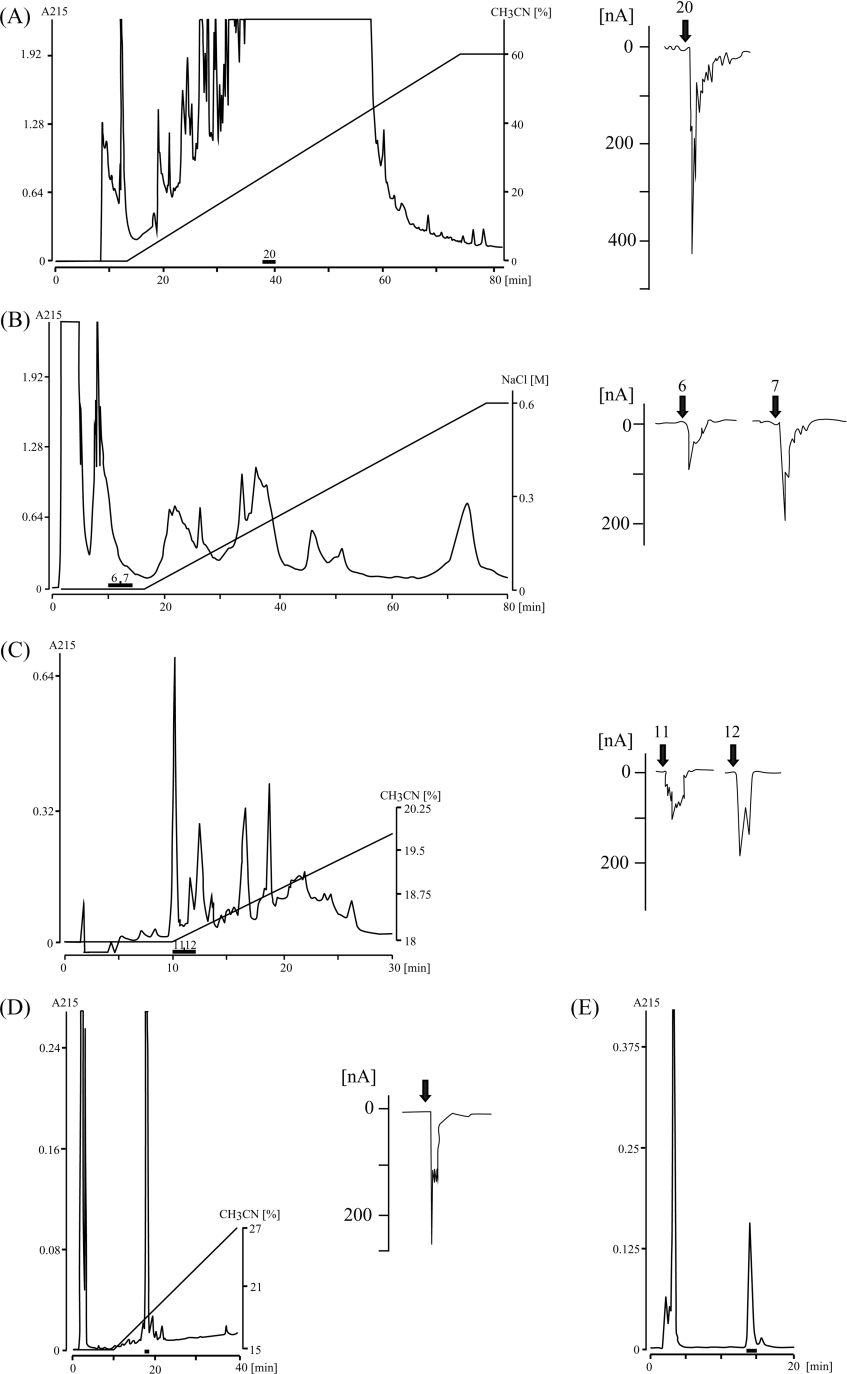
Most vertebrate species possess at least two, and usually three, molecular forms of GnRH, and seven GnRHs were found in ascidians by in silico analysis [1]. To investigate whether Octopus possesses other endogenous ligands for oct-GnRHR, Octo-pus brain extract was fractionated by HPLC and the resultant fractions were assayed in Xenopus oocytes expressing oct-GnRHR. Five steps of reversed-phase and cation-exchange HPLC yielded a single material with potent activity in the oocyte assay (Figures 4A–4E). The observed mass number, 713.34 [M+2H]2+ in the nano ESI-TOF-MS analysis was in good agreement with the calculated mass number (713.28 [M+2H]2+) for oct-GnRH. Moreover, the fragmentation pattern of the isolated peptide in the TOF-MS/MS analysis was in accord with that of oct-GnRH. Co-injection of native and sythetic oct-GnRH showed a single peak on both a reversed-phase and a cation-exchange HPLC (results not shown). However, other fractions failed to activate the oct-GnRHR. These results confirmed that Octopus possesses a single oct-GnRH species.
Figure 4. Purification of the endogenous ligand of oct-GnRHR from Octopus brain.
(A) HPLC fractionation on a Capcell Pak UG80 column. Fraction 20 showed a Cl− current in the oocyte assay. (B) Fraction 20 in (A) was condensed in vacuo, dissolved in 20 mM phosphate buffer and injected into a cation-exchange HPLC TSKgel SP-5PW column. Fractions 6 and 7 were active in the oocyte assay. (C) Fractions 6 and 7 in (B) were separated by a reversed-phase HPLC with a Capcell Pak C18 UG80 column and an 18–30% linear gradient of acetonitrile. Fractions 11 and 12 both exhibited Cl− currents. (D and E): Fractions 11 and 12 in (C) were separated by a reversed-phase HPLC with a Capcell Pak C18 UG80 column and a 15–27% linear gradient of acetonitrile (D) and a 16.8% isocratic elution of acetonitrile (E). A single material was eluted at the peak. Bars indicate active fractions showing a Cl− current in the oocyte assay.
The response of oct-GnRHR to other GnRH analogues was investigated using the same assay. mGnRH and cGnRH-I and -II showed no effects at concentrations higher than 10−5 M. More-over, des-NY-oct-GnRH failed to trigger the current at concen-trations higher than 10−5 M, whereas NY-cGnRH-II evoked a slight current at 10−5 M (Figure 3C). AKH I, AKH II and cor-azonin were devoid of any effects at concentrations higher than 10−5 M (results not shown), although oct-GnRHR shows high amino acid sequence similarity to receptors for those peptides (Table 2). Taken together, these results provided evidence that oct-GnRHR is an authentic receptor for oct-GnRH.
Localization of oct-GnRHR mRNA in Octopus
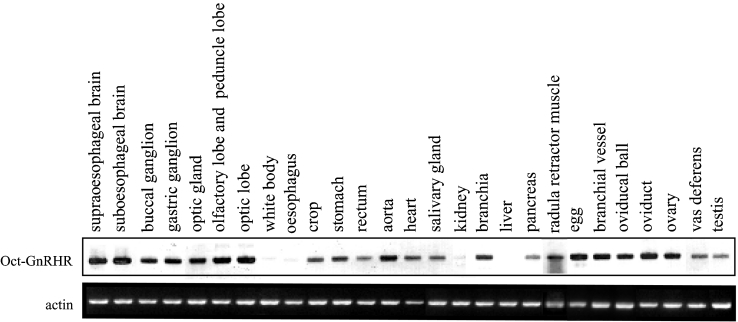
Southern-blot analysis of RT-PCR products for oct-GnRHR verified a wide distribution of oct-GnRHR mRNAs in the CNS and peripheral nervous systems, and in several peripheral tissues of Octopus (Figure 5), whereas β-actin genes were shown to be expressed to a similar degree in all tissues.
Figure 5. Southern-blot analysis of RT-PCR products for oct-GnRHR and β-actin transcripts isolated from Octopus tissues.
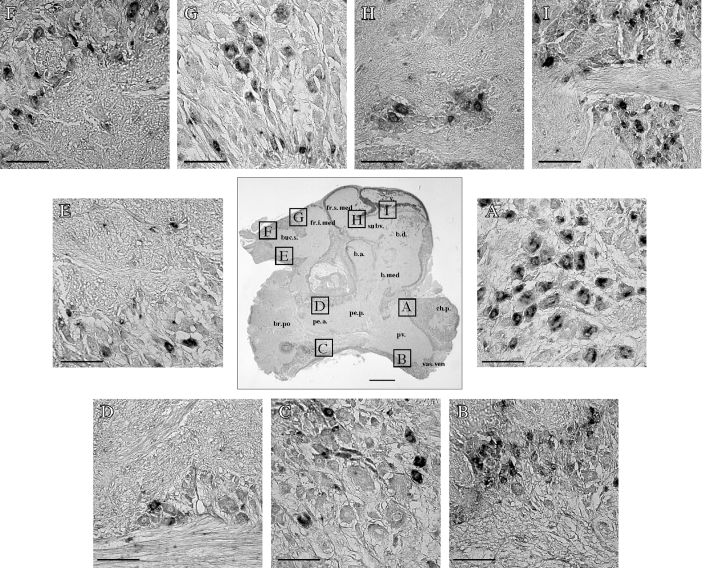
Furthermore, the localization of oct-GnRHR mRNA in the CNS was in detail observed by in situ hybridization to 5 μm serial sections of the brain using an antisense DIG-labelled oct-GnRHR cRNA probe. The oct-GnRHR mRNA was highly expressed in the palliovisceral lobe and superior buccal lobe, whereas low expression was seen in the subvertical lobe, superior and inferior frontal lobe, posterior brachial lobe and pedal lobe (Figures 6A–6I). Such positive staining was only detected when the antisense probe was used, but was not observed with the sense probe (results not shown). These results suggest that oct-GnRH has multiple functions via oct-GnRHR in both the Octopus nervous systems and peripheral tissues.
Figure 6. Localization of oct-GnRHR mRNA in the Octopus brain.
The centre sagittal section was stained with Nissl reagent. The scale bar represents 1 mm. (A–I) In situ hybridization with the DIG-labelled oct-GnRHR cRNA probe to fixed 5 μm-thick sections of Octopus brain. The scale bars represent 100 μm. Abbreviations: v., vertical lobe; fr.s.med., median superior frontal lobe; fr.i.med., median inferior frontal lobe; buc.s., superior buccal lobe; subv., subvertical lobe; b.d., dorsal basal lobe; b.a., anterior basal lobe; b.med., median basal lobe; br.po., posterior brachial lobe; pe.a., anterior pedal lobe; pe.p., posterior pedal lobe; pv., palliovisceral lobe; ch.p., posterior chromatophore lobe; vas.ven., ventral vasomotor lobe.
Biological effects of oct-GnRH
Oct-GnRH mRNA-expressing cell bodies and immunoreactive fibres were observed in the superior buccal lobe [7,8], which controls the movement of the buccal mass [22], and oct-GnRHR was expressed in the radula retractor muscle (Figure 5). These findings indicated the possibility that oct-GnRH regulates the action of the radula retractor muscle. To determine the function of oct-GnRH in the buccal mass, a biological assay using radula retractor muscle was used. The application of oct-GnRH evoked contraction to the radula retractor muscle at the concentration of 10−7 M (Figure 7). The effect was also concentration-dependent over the range 10−10–10−7 M (results not shown).
Figure 7. Contractile and modulatory effects of oct-GnRH on the radula retractor muscle of Octopus.
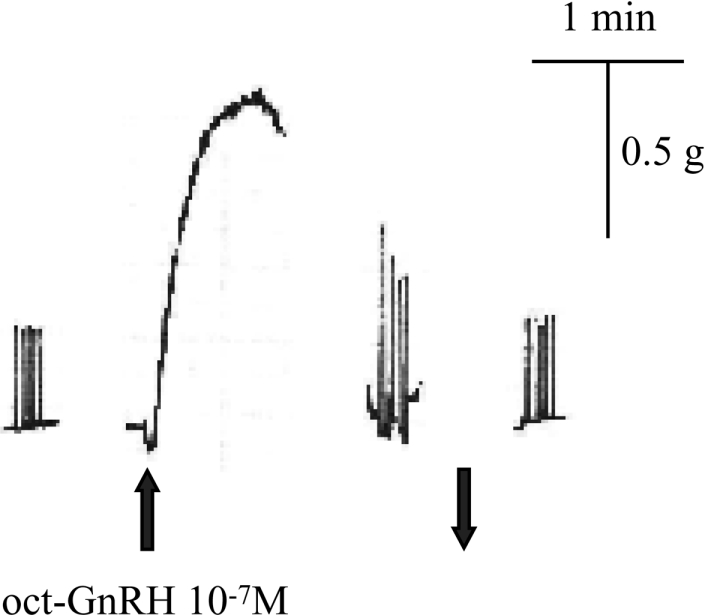
Twitch contractions were produced by a train of electrical pulses (20 V, 1 ms, 0.2 Hz, five pulses). The upward arrow indicates application of the peptide to the muscle. The downward arrow indicates washing-out of the peptide.
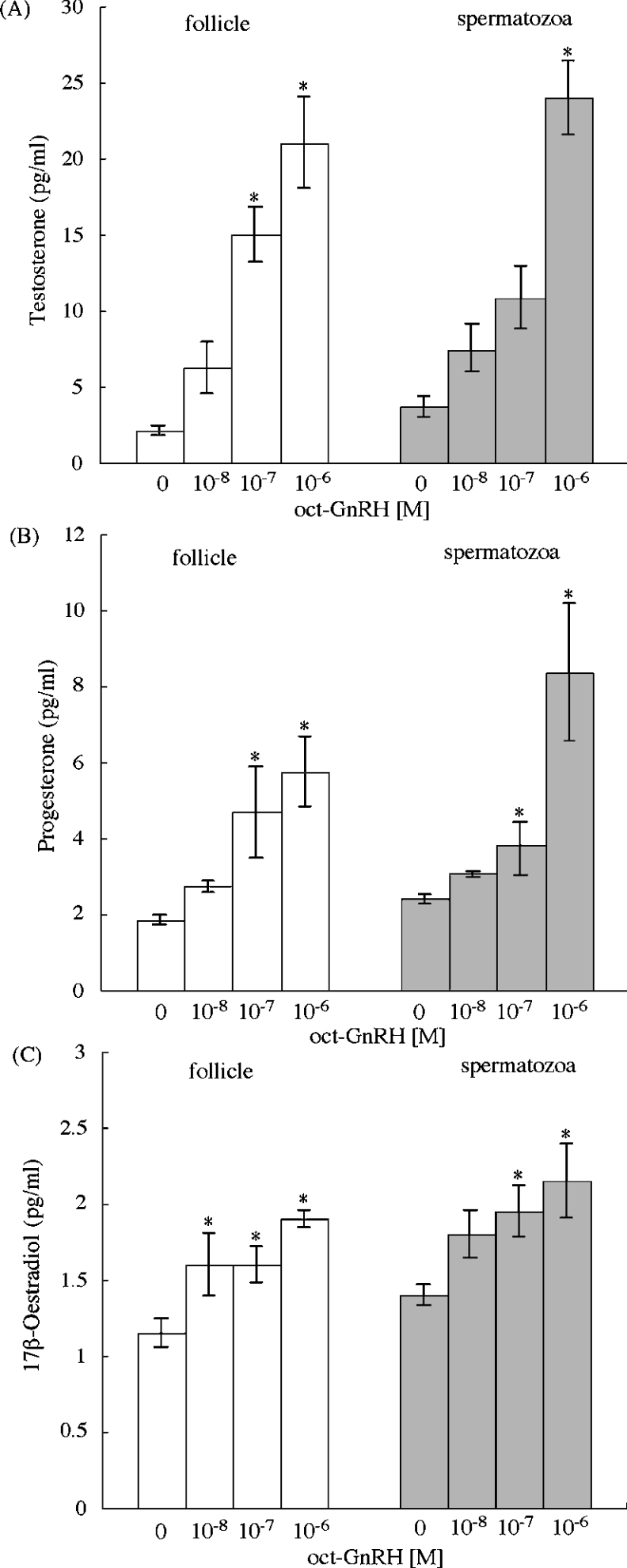
In vertebrates, GnRH is known to play a critical role in synthesis and release of sex steroids in reproductive tissues [3,23]. Therefore we examined the effect of oct-GnRH on steroidogenesis of the male and female reproductive tissues such as the oct-GnRHR-expressing ovary and testis. The treatment of follicle and spermatozoa with media containing oct-GnRH led to an elevation of basal steroidogenesis of testosterone, progesterone and 17β-oestradiol, in a concentration-dependent manner (10−8 M–10−6 M) in both Octopus follicle and spermatozoa (Figures 8A–8C), whereas the medium alone and cGnRH-I at 10−6 M showed no effect (results not shown). In addition, the short-time incubation (1 and 3 h) showed no significant effects (results not shown). These results supported the view that oct-GnRH participates in steroidogenesis via oct-GnRHR in the male and female repro-ductive tissues.
Figure 8. In vitro induction of releasing sex steroid by oct-GnRH on the Octopus follicle and spermatozoa.
(A) Testosterone; (B) progesterone; (C) 17β-oestradiol. Gonads were incubated for 16 h with various concentrations of oct-GnRH. White bars indicate follicle and grey bars indicate spermatozoa. Each point represents the mean±S.E.M. for three preparations. *P<0.05 compared with sex steroid in the absence of oct-GnRH.
DISCUSSION
Among protostomes, octopuses are endowed with several excep-tional properties. The dioecism and highly advanced nervous and endocrine systems [6] which they display are due to mole-cular and functional evolution of neuropeptides and hormones. For instance, two oxytocin/vasopressin superfamily peptides (octo-pressin and cephalotocin) and their three receptors were charac-terized from Octopus in our previous study, whereas other protostomes have been shown to possess only one oxytocin/vasopressin superfamily peptide [9,24]. The ligand–receptor sel-ectivity were established through different evolutionary lineages from those of their vertebrate counterparts although those of receptors are highly conserved among known receptors of the oxytocin/vasopressin superfamily in vertebrates as well as invert-ebrates [9]. Therefore, structural and functional identification of octopus neuropeptides and hormones is expected to contribute a great deal to an investigation of the biological mechanism underlying the advanced behaviour of octopuses and evolutionary aspects of neuropeptides and hormones.
GnRH is a hypothalamic decapeptide that plays important roles in regulating reproduction in vertebrates, but molecular forms and activities of protostome GnRHs have yet to be investigated. Previously, we showed that Octopus has an oct-GnRH with sequence similarity to that of chordate GnRH [7]. In the present study, we have cloned a novel receptor from Octopus brain. The amino acid sequence similarities with the other GnRHRs, analysis of intron/exon structures, and the functional analysis in Xenopus oocyte assay, led to the conclusion that the cloned receptor is specific for oct-GnRH (oct-GnRHR) and that oct-GnRHR shares a common ancestor with chordate GnRHRs. In addition, oct-GnRHR conserves the cytoplasmic tail, which is presumed to be truncated during divergence into non-mammalian and mammal lineages in the evolutionary process of vertebrates [25,26]. These findings suggest that oct-GnRHR has the characteristics of an ancestral GnRHR.
Although Di Cosmo and co-workers [27,28] implied that Octopus possesses cGnRH-I-like peptide because cGnRH-I-like immunoreactivities were detected in the CNS lobe and the reproductive ducts of the female and male of Octopus, Xenopus oocytes that express oct-GnRHR responded to only native and synthetic oct-GnRH, but not to any other GnRH species (Figures 3 and 4). Furthermore, we could not characterize any other homo-logous GnRHRs. These results suggest that Octopus, unlike chordates, has a single GnRH and GnRHR.
Homology modelling of the human GnRHR based on the X-ray structure of rhodopsin accommodates the experimentally determined or putative interactions of GnRH-I and receptor residues, such as Asp98, Trp101, Asn102, Lys121, Asn212, Trp280, Tyr290, and Asp302 residues in the human GnRHR I [1]. These amino acid residues are also highly conserved in GnRHR II and non-mammalian GnRHR I, suggesting that these residues serve the same function. In oct-GnRHR, Asn102, Lys121, Asn212, Tyr290, and Asp302 residues are replaced by serine, threonine, serine, phenylalanine and serine residues respectively (Figure 1A). mGnRH, cGnRH-I and -II were devoid of any activity in Xenopus oocytes expressing oct-GnRHR (Figure 3). The difference in the GnRH family peptide residues at position 5, 7, and 8 is believed to enable GnRHs to interact selectively with their respective receptors [1]. Although almost all GnRHs consist of ten amino acid residues, oct-GnRH is composed of 12 amino acids, in which asparagine and tyrosine have been inserted after the leading pyroglutamate residue in the other GnRHs (Table 1). The asparagine and tyrosine residues at positions 2 and 3 of oct-GnRH are requisite for activity, because oct-GnRHR did not respond to des-NY-oct-GnRH and cGnRH-II, but NY-cGnRH-II exhibited weak activation of oct-GnRHR (Figure 3). Similar reactivities were observed in cardioactivities of oct-GnRH, NY-cGnRH-II, des-NY-oct-GnRH, and cGnRH-II in the systemic heart [8], where oct-GnRHR was shown to be expressed. Consequently, it is sug-gested that the replacements of amino acid residues in oct-GnRHR are primarily responsible for specific interaction with oct-GnRH, and the Octopus unique ligand–receptor binding for oct-GnRHR is specific to 12-residue peptides, including oct-GnRH.
Oct-GnRHR was widely distributed in both the nervous systems and peripheral tissues, including reproductive tissues (Figure 5). The distribution of oct-GnRHR in various tissues is in agreement with our previous study, given that oct-GnRH mRNA-expressing cell bodies and oct-GnRH-immunoreactive cell bodies and fibres were distributed in the CNS [8] and oct-GnRH-immunoreactive fibres were detected in peripheral organs, including the heart, oviduct and oviducal gland [7,8]. The contraction of the heart by oct-GnRH was demonstrated in our previous study [8]. In the present study, oct-GnRH was found to have a contractile effect on the radula retractor muscle of Octopus (Figure 7), which expressed oct-GnRHR (Figure 5). This is compatible with the fact that oct-GnRH mRNA-expressing cell bodies and immunoreactive fibres were observed in the superior buccal lobe [8]. In combination, these results and findings strongly suggest that oct-GnRH is involved in feeding behaviour generated by contractions of the muscles of the buccal mass. Moreover, the expressions of oct-GnRHR mRNA in the brain were distributed in the same lobes (Figure 6) that are histochemically correlated with localizations of oct-GnRH-immunoreactive cells [8]. The palliovisceral lobe is the control nervous region for cardiac control, the superior buccal lobe is the central region for feeding behaviour, the superior and inferior frontal lobe play an important part in regulating exploratory and learning behaviour, and the pedal lobe is related to the usage of the arms [6,22]. These findings support the notion that oct-GnRH not only regulates the contraction of muscles in peripheral tissues, but acts as a neuromodulator and/or a neurotransmitter in brain functions for autonomic functions, feeding, memory and movement. In addition, oct-GnRHR was expressed in both peripheral tissues and the CNS lobes which control the functions of those tissues, implying the possibility that oct-GnRH affects feedback regulation for the contraction of muscles and other neuronal functions in the CNS. Detailed functions of oct-GnRH in the brain are now being examined.
Oct-GnRH potentiated sex steroidogenesis in the Octopus follicle and spermatozoa, both of which expressed oct-GnRHR (Figure 8). These results are compatible with the finding that Octopus has sex steroids, sex steroid-binding proteins and 3β-hydroxysteroid dehydrogenase which is a steroid-metabolizing enzyme in the male and female reproductive systems [31–33]. Furthermore, progesterone induces activation of the Octopus spermatozoa, and 17β-oestradiol and progesterone fluctuations are accompanied with the morphological changes in the ovary, oviduct and oviducal gland during the reproductive cycle, sug-gesting that the reproductive system is under hormonal control [34,35]. The pacific oyster (Crassostrea gigas), GnRHR mRNA was expressed in both male and female gonads during the repro-ductive cycle [36]. The Octopus ovary and testis did not express oct-GnRH mRNA (A. Kanda and H. Minakata, unpublished data), and the optic gland in Octopus, which has oct-GnRH-immuno-reactive cells [7], plays an important role in the regulation of gonadal maturation [37], suggesting that oct-GnRH in the CNS, such as the optic gland, is released into the bloodstream and acts on the reproductive tissues. Taken together, these results and findings suggested that oct-GnRH induces sexual maturation and spawning by regulating the sex steroids via oct-GnRHR in a hormonal fashion and shares reproductive functions with chordate GnRHs, and that only the single pair of oct-GnRH and oct-GnRHR are responsible for multifunction in Octopus, although several combinations of GnRHs and those of receptors have respective specific physiological roles in vertebrates.
In conclusion, we have presented the primary sequence, gene organization, reactivity, localization and physiological roles of an octopus GnRH receptor, oct-GnRHR. Our data provide fruitful insights into both the structural and functional evolution of GnRH systems between octopuses and chordates.
Acknowledgments
A part of this work was funded by Grants-in-Aid for Scientific Research (12640669 and 15207007) to H.M. from the Japan Society for the Promotion of Science.
References
- 1.Millar R. P., Lu Z. L., Pawson A. J., Flanagan C. A., Morgan K., Maudsley S. R. Gonadotropin-releasing hormone receptors. Endocr. Rev. 2004;2:235–275. doi: 10.1210/er.2003-0002. [DOI] [PubMed] [Google Scholar]
- 2.Fink G. Gonadotropin secretion and its control. In: Knobil E., Neill J., editors. The Physiology of Reproduction. New York: Raven Press; 1998. pp. 1349–1377. [Google Scholar]
- 3.Cheng C. K., Leung P. C. Molecular biology of gonadotropin-releasing hormone (GnRH)-I, GnRH-II, and their receptors in humans. Endocr. Rev. 2005;26:283–306. doi: 10.1210/er.2003-0039. [DOI] [PubMed] [Google Scholar]
- 4.Leung P. C., Peng C. Gonadotropin-releasing hormone receptor: gene structure, expression and regulation. Biol. Signals. 1996;5:63–69. doi: 10.1159/000109175. [DOI] [PubMed] [Google Scholar]
- 5.Morgan K., Millar R. P. Evolution of GnRH ligand precursors and GnRH receptors in protochordate and vertebrate species. Gen. Comp. Endocrinol. 2004;139:191–197. doi: 10.1016/j.ygcen.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Young J. Z. Multiple matrices in the memory system of Octopus. In: Abbott N. J., Williamson R., Maddock L., editors. Cephalopod Neurobiology. Oxford: Oxford University Press; 1995. [Google Scholar]
- 7.Iwakoshi E., Takuwa-Kuroda K., Fujisawa Y., Hisada M., Ukena K., Tsutsui K., Minakata H. Isolation and characterization of a GnRH-like peptide from Octopus vulgaris. Biochem. Biophys. Res. Commun. 2002;291:1187–1193. doi: 10.1006/bbrc.2002.6594. [DOI] [PubMed] [Google Scholar]
- 8.Iwakoshi-Ukena E., Ukena K., Takuwa-Kuroda K., Kanda A., Tsutsui K., Minakata H. Expression and distribution of octopus gonadotropin-releasing hormone in the central nervous system and peripheral organs of the octopus (Octopus vulgaris) by in situ hybridization and immunohistochemistry. J. Comp. Neurol. 2004;477:310–323. doi: 10.1002/cne.20260. [DOI] [PubMed] [Google Scholar]
- 9.Kanda A., Satake H., Kawada T., Minakata H. Novel evolutionary lineages of the invertebrate Oxytocin/Vasopressin superfamily peptides and their receptors in octopus. Biochem. J. 2005;387:85–91. doi: 10.1042/BJ20041230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Kimura M. Cambridge: Cambridge University Press; 1983. The Neutral Theory of Molecular Evolution. [Google Scholar]
- 10.Kanda A., Takuwa-Kuroda K., Iwakoshi-Ukena E., Furukawa Y., Matsushima O., Minakata H. Cloning of Octopus cephalotocin receptor, a member of the oxytocin/vasopressin superfamily. J. Endocrinol. 2003;17:281–291. doi: 10.1677/joe.0.1790281. [DOI] [PubMed] [Google Scholar]
- 11.Satake H., Takuwa K., Minakata H., Matsushima O. Evidence for conservation of the vasopressin/oxytocin superfamily in Annelida. J. Biol. Chem. 1999;274:5605–5611. doi: 10.1074/jbc.274.9.5605. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi M., Muneoka Y. Modulatory actions of octopamine and serotonin on the contraction of buccal muscles in Rapana thomasiana – I. Enhancement of contraction in radula protractor. Comp. Biochem. Physiol. C. 1980;65:73–79. doi: 10.1016/0306-4492(80)90024-6. [DOI] [PubMed] [Google Scholar]
- 13.Muneoka Y., Kobayashi M. Modulatory actions of octopamine and serotonin on the contraction of buccal muscles in Rapana thomasiana – II. Inhibition of contraction in radula retractor. Comp. Biochem. Physiol. C. 1980;65:81–86. doi: 10.1016/0306-4492(80)90024-6. [DOI] [PubMed] [Google Scholar]
- 14.Bockaert J., Pin J. P. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Bogerd J., Choi H. S., Seong J. Y., Soh J. M., Chun S. Y., Blomenrohr M., Troskie B. E., Millar R. P., Yu W. H., et al. Three distinct types of GnRH receptor characterized in the bullfrog. Proc. Natl. Acad. Sci. U.S.A. 2001;98:361–366. doi: 10.1073/pnas.011508498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora K. K., Cheng Z., Catt K. J. Dependence of agonist activation on an aromatic moiety in the DPLIY motif of the gonadotropin-releasing hormone receptor. Mol. Endocrinol. 1996;10:979–986. doi: 10.1210/mend.10.8.8843414. [DOI] [PubMed] [Google Scholar]
- 17.Ballesteros J., Kitanovic S., Guarnieri F., Davies P., Fromme B. J., Konvicka K., Chi L., Millar R. P., Davidson J. S., Weinstein H., Sealfon S. C. Functional microdomains in G-protein-coupled receptors. The conserved arginine-cage motif in the gonadotropin-releasing hormone receptor. J. Biol. Chem. 1998;273:10445–10453. doi: 10.1074/jbc.273.17.10445. [DOI] [PubMed] [Google Scholar]
- 18.Kusakabe T., Mishima S., Shimada I., Kitajima Y., Tsuda M. Structure, expression, and cluster organization of genes encoding gonadotropin-releasing hormone receptors found in the neural complex of the ascidian Ciona intestinalis. Gene. 2003;322:77–84. doi: 10.1016/j.gene.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Okubo K., Nagata S., Ko R., Kataoka H., Yoshiura Y., Mitani H., Kondo M., Naruse K., Shima A., Aida K. Identification and characterization of two distinct GnRH receptor subtypes in a teleost, the medaka Oryzias latipes. Endocrinology (Baltimore) 2001;142:4729–4739. doi: 10.1210/endo.142.11.8475. [DOI] [PubMed] [Google Scholar]
- 20.Kakar S. S., Musgrove L. C., Devor D. C., Sellers J. C., Neill J. D. Cloning, sequencing, and expression of human gonadotropin releasing hormone (GnRH) receptor. Biochem. Biophys. Res. Commun. 1992;189:289–295. doi: 10.1016/0006-291x(92)91556-6. [DOI] [PubMed] [Google Scholar]
- 21.Staubli F., Jorgensen T. J., Cazzamali G., Williamson M., Lenz C., Sondergaard L., Roepstorff P., Grimmelikhuijzen C. J. Molecular identification of the insect adipokinetic hormone receptors. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3446–3451. doi: 10.1073/pnas.052556499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young J. Z. London and New York: Oxford University Press; 1971. The anatomy of the nervous system of Octopus vulgaris. [Google Scholar]
- 23.Leung P. C., Steele G. L. Intracellular signalling in the gonads. Endocr. Rev. 1992;13:476–498. doi: 10.1210/edrv-13-3-476. [DOI] [PubMed] [Google Scholar]
- 24.Takuwa-Kuroda K., Iwakoshi-Ukena E., Kanda A., Minakata H. Octopus, which owns the most advanced brain in invertebrates, has two members of vasopressin/oxytocin superfamily as in vertebrates. Regul. Pept. 2003;115:139–419. doi: 10.1016/s0167-0115(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 25.Cui J., Smith R. G., Mount G. R., Lo J. L., Yu J., Walsh T. F., Singh S. B., DeVita R. J., Goulet M. T., Schaeffer J. M., Cheng K. Identification of Phe313 of the gonadotropin-releasing hormone (GnRH) receptor as a site critical for the binding of nonpeptide GnRH antagonists. Mol. Endocrinol. 2000;14:671–681. doi: 10.1210/mend.14.5.0464. [DOI] [PubMed] [Google Scholar]
- 26.Troskie B. E., Hapgood J. P., Millar R. P., Illing N. Complementary deoxyribonucleic acid cloning, gene expression, and ligand selectivity of a novel gonadotropin-releasing hormone receptor expressed in the pituitary and midbrain of Xenopus laevis. Endocrinology (Baltimore) 2000;141:1764–1771. doi: 10.1210/endo.141.5.7453. [DOI] [PubMed] [Google Scholar]
- 27.Di Cosmo A., Di Cristo C. Neuropeptidergic control of the optic gland of Octopus vulgaris: FMRF-amide and GnRH immunoreactivity. J. Comp. Neurol. 1998;398:1–12. doi: 10.1002/(sici)1096-9861(19980817)398:1<1::aid-cne1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Di Cristo C., Paolucci M., Iglesias J., Sanchez J., Di Cosmo A. Presence of two neuropeptides in the fusiform ganglion and reproductive ducts of Octopus vulgaris: FMRFamide and gonadotropin-releasing hormone (GnRH) J. Exp. Zool. 2002;292:267–276. doi: 10.1002/jez.90000. [DOI] [PubMed] [Google Scholar]
- 29. Reference deleted.
- 30. Reference deleted.
- 31.Di Cosmo A., Paolucci M., Di Cristo C., Botte V., Ciarcia G. Progesterone receptor in the reproductive system of the female of Octopus vulgaris: characterization and immunolocalization. Mol. Reprod. Dev. 1998;50:451–460. doi: 10.1002/(SICI)1098-2795(199808)50:4<451::AID-MRD9>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 32.Di Cosmo A., Di Cristo C., Paolucci M. A estradiol-17β receptor in the reproductive system of the female of Octopus vulgaris: characterization and immunolocalization. Mol. Reprod. Dev. 2002;61:367–375. doi: 10.1002/mrd.10014. [DOI] [PubMed] [Google Scholar]
- 33.D'Aniello A., Di Cosmo A., Di Cristo C., Assisi L., Botte V., Di Fiore M. M. Occurrence of sex steroid hormones and their binding proteins in Octopus vulgaris Lam. Biochem. Biophys. Res. Commun. 1996;227:782–788. doi: 10.1006/bbrc.1996.1585. [DOI] [PubMed] [Google Scholar]
- 34.Tosti E., Di Cosmo A., Cuomo A., Di Cristo C., Gragnaniello G. Progesterone induces activation in Octopus vulgaris spermatozoa. Mol. Reprod. Dev. 2001;59:97–105. doi: 10.1002/mrd.1011. [DOI] [PubMed] [Google Scholar]
- 35.Di Cosmo A., Di Cristo C., Paolucci M. Sex steroid hormone fluctuations and morphological changes of the reproductive system of the female of Octopus vulgaris throughout the annual cycle. J. Exp. Zool. 2001;289:33–47. doi: 10.1002/1097-010x(20010101/31)289:1<33::aid-jez4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 36.Rodet F., Lelong C., Dubos M. P., Costil K., Favrel P. Molecular cloning of a molluscan gonadotropin-releasing hormone receptor orthologue specifically expressed in the gonad. Biochim. Biophys. Acta. 2005;1730:187–195. doi: 10.1016/j.bbaexp.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Wells M. J., Wells J. Hormonal control of sexual maturity in Octopus. J. Exp. Biol. 1959;36:1–33. [Google Scholar]