Abstract
Two sPPases (soluble inorganic pyrophosphatases, EC 3.6.1.1) have been isolated from the microalga Chlamydomonas reinhardtii. Both are monomeric proteins of organellar localization, the chloroplastic sPPase I [Cr (Ch. reinhardtii)-sPPase I, 30 kDa] is a major isoform and slightly larger protein than the mitochondrial sPPase II (Cr-sPPase II, 24 kDa). They are members of sPPase family I and are encoded by two different cDNAs, as demonstrated by peptide mass fingerprint analysis. Molecular phylogenetic analyses indicated that Cr-sPPase I is closely related to other eukaryotic sPPases, whereas Cr-sPPase II resembles its prokaryotic counterparts. Chloroplastic sPPase I may have replaced a cyanobacterial ancestor very early during plastid evolution. Cr-sPPase II orthologues are found in members of the green photosynthetic lineage, but not in animals or fungi. These two sPPases from photosynthetic eukaryotes are novel monomeric family I sPPases with different molecular phylogenies and cellular localizations.
Keywords: Chlamydomonas reinhardtii, chloroplast, cyanelle, Cyanophora paradoxa, mitochondrion, soluble inorganic pyrophosphatase (sPPase)
Abbreviations: DTT, dithiothreitol; EST, expressed sequence tag; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; ORF, open reading frame; sPPase, soluble inorganic pyrophosphatase; Cp-sPPase, Cyanophora paradoxa sPPase; Cr-sPPase, Chlamydomonas reinhardtii sPPase; Y-sPPase, Saccharomyces cerevisiae sPPase
INTRODUCTION
sPPases (soluble inorganic pyrophosphatases; also known as pyrophosphate phosphohydrolases; EC 3.6.1.1) are essential enzymes for cellular anabolism that catalyse the hydrolysis of PPi to Pi, providing a thermodynamic advantage for many biosynthetic reactions [1]. Both eubacteria and archaea possess Mg2+-dependent sPPases [2], which, together with their eukaryotic homologues in fungi and metazoa, constitute family I of the sPPases. They differ significantly from family II of the Mn2+-dependent sPPases that are found only in a limited number of bacteria and archaea [3,4]. The two best-studied examples are the hexameric sPPase of Escherichia coli, and the dimeric enzyme of Saccharomyces cerevisiae, which are the prototypes of the prokaryotic and eukaryotic essential family I sPPases respectively [5–7].
The presence of sPPases in higher plants has been documented [8,9], but the enzymes remain poorly characterized. Although all of these sPPases are quite active and generally comprise as much as 0.1–0.5% of total cell protein, cytosolic PPi concentrations in photosynthetic plant tissues are maintained at millimolar levels (0.2–0.5 mM), clearly higher than those in yeast (2–5 μM), suggesting that these enzymes may be either compartmentalized or strongly regulated. This also suggests that PPi could have an important role as an energy donor in plant metabolism [10,11].
We performed molecular studies of the plastidic sPPases of the green alga Chlamydomonas reinhardtii, the primitive photosynthetic protist Cyanophora paradoxa and other microalgae and plants. The plastidic sPPases described here are the first natural monomeric sPPases reported. Ch. reinhardtii possesses two Mg2+-dependent sPPases, Cr (Ch. reinhardtii)-sPPase I and Cr-sPPase II, which are located in the chloroplast and mitochondrion respectively. Although different genes of widely divergent phylogeny encode them, both enzymes possess the conserved functional residues seen in all members of family I sPPases [2,5,6], but show changes in residues involved in oligomer stabilization [12]. Chloroplastic Cr-sPPase I is related to a eukaryotic sPPase, and we suggest that it replaced the ancestral cyanobacterial plastid sPPase at a very early stage of plastid evolution. Cr-sPPase II is phylogenetically related to the prokaryotic enzymes and has no orthologues in animals or fungi. The sPPases I and II described in the present study constitute a new class of monomeric family I sPPases with distinct molecular phylogenies and specific subcellular localizations.
MATERIALS AND METHODS
Organisms and growth conditions
The unicellular green algae (Chlorophyceae) Ch. reinhardtii 21 gr, Chlorella fusca 211-8b and Monoraphidium braunii 202-7b were grown under photoautotrophic conditions in Sueoka liquid mineral medium with NH4Cl as the nitrogen source [13]; when indicated Ch. reinhardtii cultures were supplemented with 12 mM sodium acetate. The photosynthetic flagellated protist (Glaucocystophyceae) Cy. paradoxa LB55 was cultured as described previously [14]. The unicellular red algae (Rhodophyceae) Porphyridium purpureum 1380-1a and Cyanidium caldarium 16/91 were grown as described previously [15]. The Chrysophycean microalga Ochromonas danica 933-7 and the Euglenophycean protists Euglena gracilis 1224-5/15 and Astasia longa 1204-17a were cultured in SAG 9 liquid medium. Diatoms Phaeodactylum tricornutum 1090-1a and Navicula pelliculosa 1050-3 were grown as described previously [16]. Arabidopsis thaliana Columbia plants were grown in soil under 12 h light–12 h dark cycles in a phytotron. Commercial spinach plants were obtained from a local greenhouse. The cyanobacteria Pseudanabaena sp. PCC 6903 and Synechocystis sp. PCC 6803 were cultured in BG11 medium [17]. Chlorobium tepidum cells were kindly provided by Professor M. T. Madigan (Deparment of Microbiology, Southern Illinois University, Carbondale, IL, U.S.A.). The E. coli strain MC1061YPPA, which expresses the yeast YPPA gene instead of the native bacterial ppa gene [12], was kindly provided by Professor R. Lahti (Department of Biochemistry and Food Chemistry, University of Turku, Vatselankatu 2, FIN-20014 Turku, Finland).
Reagents
Restriction enzymes, T4 DNA ligase, Taq polymerase, PMSF and leupeptin were purchased from Boehringer Manheim. Aprotinin, trypsin inhibitor, DTT (dithiothreitol), S. cerevisiae sPPase (catalogue number 83205) and other analytical grade chemicals were purchased from Sigma–Aldrich. Oligonucleotides were obtained from Amersham Biosciences.
Monospecific polyclonal antibodies against the Cr-sPPases I and II were obtained by subcutaneous immunization of rabbits with samples of purified proteins (500 μg of Cr-sPPase I; 250 μg of Cr-sPPase II) dissolved in 0.5 ml of 50 mM Tris/HCl (pH 7.5) buffer plus identical volumes of incomplete Freund's adjuvant.
Protein techniques
Enzyme assays
PPi activity was assayed by the colorimetric determination of Pi produced from the enzymatic hydrolysis of PPi at room temperature [18] with Mg2-PPi as a substrate under the conditions described previously [19]. For optimum pH determinations, a mixture of 50 mM Mes, Tris and glycine was used as a buffer. The reaction rates are expressed as μmoles of Pi produced per min. sPPase activity assay following standard non-denaturing PAGE was performed as described by Sugino and Miyoshi [20], and the gels were stained according to the method of Baykov and Volk [21]. Protein bands were visualized with Coomassie Brilliant Blue R-250 after non-denaturing PAGE or SDS/PAGE. Protein concentrations were determined by the method of Bradford [22].
The electrophoretic mobilities of standard proteins of known molecular mass [aldolase (154 kDa), BSA (66 kDa), ovoalbumin (47 kDa) and cytochrome c (12 kDa)] were used to estimate the native molecular masses of plastidic algal sPPases on a series of non-denaturing (native) PAGE gels with different polyacrylamide percentages, as described by Hedrick and Smith [23].
Western blot assays
After SDS/PAGE the proteins were transferred on to nitrocellulose membranes using a transblot apparatus (Bio-Rad Laboratories), blocked with 5% (v/v) non-fat milk in PBS and incubated overnight at 4 °C. The anti-Cr-sPPase sera were diluted (1:1000) in blocking buffer and incubated with blots at room temperature for 60 min. The nitrocellulose membrane was washed three times for 20 min each with PBS containing 0.1% (v/v) Tween 20 before the addition of a 1:10000 dilution of goat anti-rabbit IgG peroxidase antibody conjugate in blocking buffer for 30 min.
Protein purification
Unless otherwise specified for biochemical and immunochemical studies, Ch. reinhardtii cells and other microalgae and plant tissues were prepared by resuspension in 25 mM Tris/HCl (pH 7.5)/2 mM MgCl2/2 mM DTT/1 mM EDTA, supplemented with a cocktail of protease inhibitors (0.1 mM PMSF, 1 mM benzamidine and 2 mM ϵ-aminohexanoic acid) at 0.1 g of wet mass/ml. Algal cells were then disrupted at 4 °C with a Branson model B-12 sonifier, for 120 s (30 s pulses) at 70 W. Cy. paradoxa cells and cyanelles were disrupted by osmotic shock and ultrasonic treatment respectively, as described previously [14]. Plant leaves were sliced, suspended in buffer, as described above, and mechanically disrupted with a Waring blender. Cell-free extracts were obtained by centrifugation of homogenates at 40000 g for 20 min.
Purification of two sPPases from Ch. reinhardtii
Cells at late logarithmic growth phase were harvested at 10000 g in a Sorval RC5C centrifuge at 4 °C, washed twice with 50 mM Tris/HCl (pH 7.5) and stored at −20 °C. Frozen cells were resuspended in buffer A [50 mM Tris/HCl (pH 7.5), 10% (v/v) glycerol, 1 mM MgCl2, 10 mM 2-mercaptoethanol, 1 mM DTT and 0.5 mM EDTA] supplemented with protease inhibitors (1 mM PMSF, 2 mM ϵ-aminohexanoic acid and 0.5 mM benzamidine) at 3 ml/g of cells (wet mass). The cells were disrupted by ultrasonic treatment in a water bath at 4 °C with a Branson 25U Sonifier (six 30 s pulses with intermediate 30 s of cooling intervals). The suspension was then supplemented with MgCl2 at a final concentration of 10 mM and centrifuged at 40000 g for 30 min. The greenish supernatant was treated with 10 mM streptomycin sulphate (final concentration) for 1 h at 4 °C with continuous stirring and centrifuged as described above. The crude extract was subjected to ammonium sulphate fractionation (40–80% saturation), resuspended in 10 ml of buffer A and dialysed overnight against 5 litres of 20 mM Tris/HCl buffer (pH 7.5) with 0.1 mM DTT. The resulting extract was applied at 20 ml/h to a DEAE-cellulose DE-52 column (3 cm×12 cm) (Whatman) previously equilibrated with buffer A. Proteins were eluted with a linear gradient of NaCl, (300 ml, 0–300 mM) in the same buffer. Solid ammonium sulphate was added up to 10% (w/v) to the pooled fraction with enzymatic activity, and the sample was applied to a phenyl-Sepharose HP column (1.6 cm×18 cm; Amersham Biosciences) equilibrated in buffer A containing 10% (w/v) ammonium sulphate. The column was washed and developed in a linear gradient (150 ml) of decreasing ammonium sulphate (20%–0%) and increasing glycerol [10%–20% (v/v)] concentrations in buffer A. Collected fractions with sPPase activity were pooled, dialysed against buffer A and then loaded on to a hydroxyapatite column (1 cm×3 cm) (Macro-Prep Ceramic HP, Bio-Rad Laboratories) previously equilibrated with the same buffer. sPPase proteins were eluted with a linear gradient of ammonium sulphate (0–10 mM). Purified Cr-sPPases I and II appeared to co-elute, as verified by SDS/PAGE. The sample was eventually subjected to high-performance gel-filtration chromatography on a Protein Pak 300SW column (Waters) equilibrated with buffer A plus 0.1 M NaCl to separate the two sPPase isoforms.
Analytical FPLC (Amersham Biosciences) gel filtration of the resolved sPPases was performed under non-denaturing conditions by isocratic elution on a Superose 12HR 10/30 column (1 cm×30 cm, Amersham Biosciences) using an automated Pharmacia FPLC system. The physicochemical parameters reported are the average values of three independent experiments. The column was equilibrated with 50 mM Tris/HCl buffer (pH 7.5) supplemented with 2 mM MgCl2, 1 mM DTT, 0.5 mM EDTA and 0.1 M NaCl. The following proteins were used as molecular markers: ferritin (420 kDa), catalase (232 kDa), aldolase (154 kDa), lipoamide dehydrogenase (113 kDa), BSA (66 kDa), ovoalbumin (47 kDa) and cytochrome c (12 kDa). The chromatographic procedure was carried out at 4 °C. Protein samples were either filtered or centrifuged (12000 g, 5 min) before being applied on to the column. Elution was performed with the same buffer at a flow rate of 0.4 ml/min. The A280 was continuously monitored.
Protein sequencing and MS analyses
N-terminal sequences of purified sPPases from Cy. paradoxa (XAITAEPVGTPETLEYRVFIQKDGK), O. danica (XYAFGINGXRI) and Eu. gracilis (XDTKLGLVEEEAAN) were obtained by Edman degradation with an automatic Applied Biosystem 460A sequencer at the protein analysis facilities of the Vienna Biocenter (University of Vienna, Vienna, Austria). These protein sequences were submitted to the SwissProt database with accession numbers P80887, P81988 and P81987, respectively. No results were obtained for Cn. caldarium and Cr-sPPases, which was probably due to the blocked N-termini. Molecular mass determinations by MALDI-TOF (matrix-assisted laser desorption ionization–time-of-flight) MS of some of the purified sPPases were performed on a Bruker apparatus by Dr G. Allmaier (Institute of Chemical Technologies and Analytics, University of Vienna, Vienna, Austria). Purified Cr-sPPase I and II were digested in situ with trypsin after SDS/PAGE for peptide mass fingerprint analysis by MALDI–TOF MS (AutoFlex; Bruker-Daltonics) at the proteomic facilities of the Instituto de Bioquímica Vegetal y Fotosíntesis (CSIC-Universidad de Sevilla). Only peptides with masses in the range 500–3000 Da were used for analyses. Protein identification was carried out by database searches using the Mascot search engine (http://www.matrixscience.com).
Cr-sPPases subcellular localization
The Ch. reinhardtii cw15 mutant, which is deficient in cell wall synthesis, was used for cellular fractionation and isolation of organelles. Cultures at exponential phase were harvested by centrifugation at 5000 g for 20 min. The fractionation was carried out by centrifugation on discontinuous Percoll gradients as described previously [24–26]. After centrifugation, gradients were collected in 1 ml fractions which were analysed for enzyme activity, total protein and chlorophyll content (at A700). Two pigmented bands were resolved, a dark-green band containing chloroplasts (fractions 8–11) and a denser brown band including mitochondria (fractions 14–16). Western blots (approx. 20 μg of protein loaded per lane) with anti-(Cr-sPPase I) and anti-sPPase II antibodies showed that the 30 kDa Cr-sPPase I was associated with the green-coloured band, and the 24 kDa Cr-sPPase II appears linked with the denser brown band. Chloroplasts from Spinacea oleracea and cyanelles from Cy. paradoxa were isolated as described previously [14,27].
DNA manipulation and sequencing
cDNA preparations from A. thaliana leaves and Ch. reinhardtii were obtained using the TimeSaver™ cDNA synthesis kit (Amersham Biosciences) with random primer oligonucleotides and M-MLV (Moloney murine leukaemia virus) retrotranscriptase.
Cloning of a cDNA encoding a chloroplastic sPPase precursor from A. thaliana
The AT5G09650 gene identified by BLAST analysis [28] of EST (expressed sequence tag) databases was amplified by PCR using the following oligonucleotides: F1, 5′-ccatggcggctactagagtgtt-aact-3′ and F4 5′-ccatggttcaagaagaaggtccggcc-3′, with NcoI sites added; and R, 5′-ggagacctttcactttactgagaattc-3′, with an EcoRI site added (restriction sites indicated in bold). These primers were used to amplify cDNA fragments corresponding to the sPPase I precursor (F1 and R) and the putative mature protein (F4 and R) as predicted by the ChloroP and TargetP web-based programs [29]. The fragments were cloned into pTrc99A (Amersham Biosciences) and the sequence was submitted to EMBL/GenBank® with accession number AJ252210.
PCR amplification and sequencing of two Ch. reinhardtii cDNAs encoding putative sPPases
BLAST analysis [28] of EST databases identified two Ch. reinhardtii cDNAs designated PPAI and PPAII, encoding two different putative sPPases. Both cDNAs were amplified using algal cDNA preparations as templates with the two following oligonucleotide pairs: PPAIF (forward) 5′-gtcgcccacacaccagatc-3′ and PPAIR (reverse) 5′-ttttttacgtctggcgggg-3′ for the 1.2 kb cDNA encoding a putative chloroplastic precursor [PPAI gene, with an 843 bp ORF (open reading frame) encoding an sPPase I-like protein], and PPAIIF (forward) 5′-acaagcacaccacatccttg-3′ and PPAIIR (reverse) 5′-cggtccaagagacatggctg-3′ for a 0.8 kb cDNA encoding a putative bacterial-like sPPase (PPAII gene, with a 579 bp ORF encoding a sPPase II-like protein). The cDNA sequences corresponding to the Ch. reinhardtii PPAI and PPAII genes were submitted to the EMBL/GenBank® database under accession numbers AJ298231 and AJ298232 respectively.
Protein sequence comparisons and phylogenetic analyses
A multiple amino acid sequence alignment of the sPPases from photosynthetic eukaryotes and other selected eukaryotic and prokaryotic family I sPPases was performed using ClustalX version 1.8 [30]. This alignment was used to construct a phylogenetic distance tree (neighbour-joining method, BLOSUM matrix) with the same program. Sequence data from public databases and unfinished microbial genome projects were obtained by similarity searches using BLAST algorithms [28] against websites of the NCBI (National Center of Biotechnology Information) (http://www.ncbi.nih.gov/), JGI (Joint Genomic Institute) (http://genome.jgi-psf.org/euk_home.html), the Sanger Institute (http://www.sanger.ac.uk/Projects/) or Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/mdb/mdbcomplete.html). Deduced eukaryotic sPPase sequences were analysed for protein sorting motifs, N-terminal targeting sequences and protein motifs for specific subcellular localizations using the web-based programs SignalP version 2.0.b2, TargetP version 1.0 and ChloroP version 1.0 [29,31].
RESULTS
Identification of sPPase activity in photosynthetic eukaryotes: isolation of two sPPases from the green microalga Ch. reinhardtii
Cell-free extracts from all photosynthetic eukaryotes tested, both microalgae and plants, contain substantial levels (0.1–0.5 units/mg of protein) of an alkaline sPPase activity. The activity was strictly dependent on bivalent metal cations, of which Mg2+ is by far the most effective. Most sPPase activity appears to be localized in the photosynthetic plastids (4.0–5.5 units/mg of protein, approx. 10-fold higher than in whole-cell extracts, results not shown). It is undetectable in the cytosol, as was demonstrated by subcellular fractionation and enzyme localization experiments performed with Ch. reinhardtii (see below), the cyanelle-bearing photosynthetic protist Cy. paradoxa and spinach leaves. A purification procedure involving cell mechanical disruption, ammonium sulphate fractionation of soluble proteins and three column-chromatography steps (ionic exchange, hydrophobic and hydroxyapatite chromatographies) was used to isolate, to electrophoretic homogeneity, the sPPases from the above-mentioned sources and from Cn. caldarium (a rhodophycean microalga), O. danica (a chromophycean microalga) and Eu. gracilis (a photosynthetic euglenoid) (see Figures 1–4). The purification methods yielded electrophoretically pure sPPases with specific activities in the range 110–140 units/mg of protein and recoveries of 30–50%. Since analyses of purified preparations of Ch. reinhardtii always showed two different sPPase proteins that co-purified using this procedure (Figure 1A), these isoforms were studied in more detail. Native PAGE and SDS/PAGE (Figure 1) and analytical FPLC gel-filtration chromatography (Figure 2) revealed two active monomeric sPPases. Cr-sPPase I (isoform I), accounting for 80–85% of the total activity and purified protein, was a polypeptide of 33–37 kDa, and Cr-sPPase II (isoform II) was a smaller polypeptide of 28–32 kDa (Figures 1 and 2). This is in contrast with S. cerevisiae where the cytosolic and mitochondrial sPPases are stable dimers (Figure 2B and [5–7]). Sequencing of Cr-sPPases by Edman degradation yielded no results, suggesting covalent N-terminal blocking of these proteins. Molecular masses of 29850 Da and 24350 Da were identified by MALDI–TOF MS for Cr-sPPase I and Cr-sPPase II respectively, slightly lower than, but in fairly good agreement with, values of apparent molecular masses estimated by indirect methods (FPLC, native and denaturing PAGE).
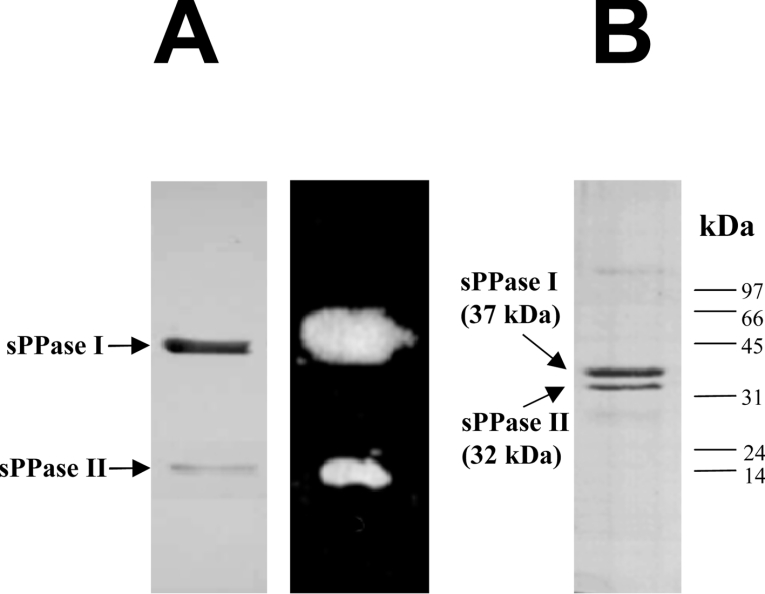
Figure 1. Native and SDS/PAGE analysis of Cr-sPPases.
(A) Non-denaturing PAGE analysis of purified Cr-sPPases (hydroxyapatite eluate) separated by a 7% polyacrylamide gel. A Coomassie-Blue-stained electrophoretogram showed two protein bands with different intensities (left-hand panel), both with sPPase activity in situ (right-hand panel): a major Cr-sPPase I and a minor Cr-sPPase II. Approx. 6 μg of protein was loaded per lane. (B) SDS/PAGE (12% gel) analysis of the same preparation. The Coomassie-Blue-stained electrophoretogram also showed two protein bands with different intensities. Apparent molecular masses of 37 and 32 kDa were estimated for the major and minor Cr-sPPases respectively. The positions and molecular masses of protein standards are indicated on the right. Approx. 5 μg of protein was loaded.
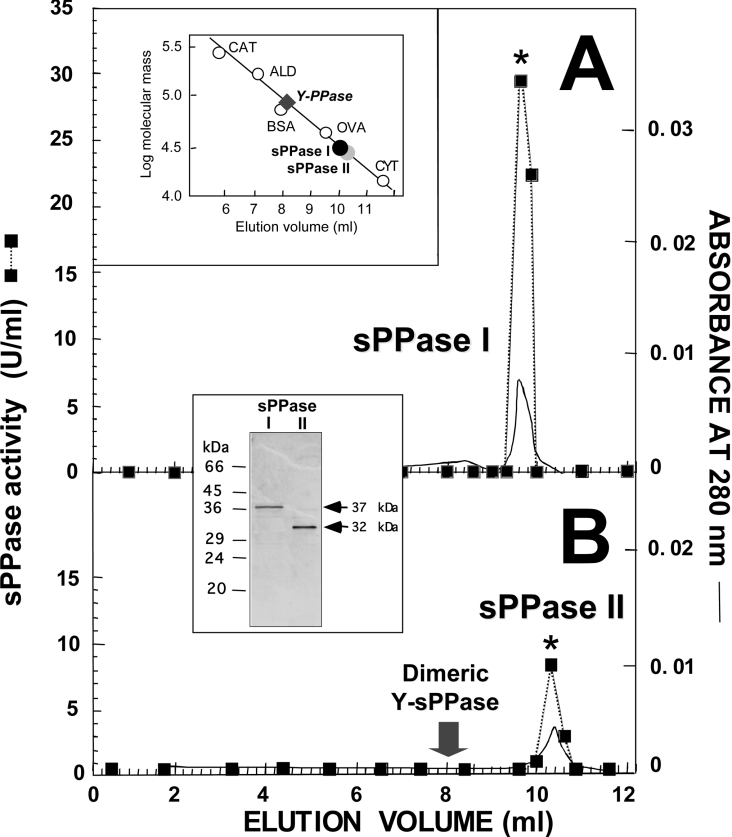
Figure 2. FPLC gel-filtration chromatography on Superose 12HR column of the sPPase isoforms co-purified from Ch. reinhardtii.
Aliquots (0.5 ml) of Cr-sPPase I (approx. 70 μg) (A) and Cr-sPPase II (approx. 35 μg) (B) purified preparations, previously resolved using this technique, were applied on to a Superose 12HR 10/30 column. Isocratic elution was achieved at a flow rate of 1 ml/min and 0.2 ml fractions were collected. The upper inset shows the calibration of the column with protein standards (CAT, catalase; ALD, aldolase; OVA, ovoalbumin; CYT, cytochrome c) and the positions of the sPPase peaks, which correspond to native molecular masses of approx. 33 kDa (black circle, Cr-sPPase I) and approx. 28 kDa (grey circle, Cr-sPPase II). The Coomassie-Blue-stained SDS/PAGE gel (lower inset; approx. 4 μg protein per lane) shows the single 37 kDa and 32 kDa protein bands present in the fractions with highest activity (*), indicating that the proteins are monomers. The positions and molecular masses of protein standards are indicated on the left-hand side of the SDS/PAGE gel. The elution position of a commercial cytosolic Y-sPPase is consistent with a dimeric structure (approx. 70 kDa), indicated for comparison by an arrow in (B) and by a diamond in the upper inset.
Cr-sPPases were absolutely dependent on bivalent cations. As in crude extracts, Mg2+ was the most effective, activity levels with Zn2+, Fe2+, Cu2+ or Mn2+ being only 20–30% of that with Mg2+, and no activity was obtained with Ca2+ (results not shown). The substrate for both enzymes was Mg2-PPi with Km values of 10 and 65 μM for Cr-sPPase I and II respectively, and inorganic polyphosphates could not replace PPi. Both isoforms exhibit an optimum pH of 7.5.
The purification of Cp-sPPase (Cy. paradoxa sPPase) was carried out with the same procedure from whole-cell extracts and cyanelles obtained from this protist. A single sPPase (Cp-sPPase), purified from whole cells or isolated cyanelles, was also separated by gel-filtration chromatography (Figure 3) and behaved on native PAGE gels as a monomer of approx. 32 kDa (results not shown). A lower molecular mass of 26560 Da was estimated by MALDI–TOF MS. When the N-terminal sequence of Cp-sPPase, determined by Edman degradation (XAITAEPVGTPETLEYRVFIQKDGK), was aligned with other sequences obtained from databases it showed a high degree of sequence identity with cytosolic sPPases from non-photosynthetic eukaryotes and mitochondrial sPPase from yeast (see below). These results suggest that Cp-sPPase from cyanelles should be encoded in the nuclear genome as a precursor endowed with a transit peptide, and are in agreement with the monomeric state found for other plastidic sPPases described in the present study, all of which are encoded as protein precursors (see below). Indeed, the sPPases purified from the phylogenetic diverse microalgae Cn. caldarium, O. danica and Eu. gracilis, as well as the sPPase purified from spinach chloroplasts, were all clearly larger than their cyanobacterial counterparts, but also monomers of approx. 35 kDa, as estimated by SDS/PAGE (Figure 4A) and FPLC gel filtration (results not shown). However, the sPPase from the thermophilic microalga Cn. caldarium exhibited a slightly larger apparent molecular mass of approx. 40 kDa, estimated by FPLC gel filtration and SDS/PAGE (Figure 4A), consistent with MALDI–TOF MS that yielded a value of 37230 Da for this protein. Attempts to sequence these algal sPPases by Edman degradation yielded no results for the Cn. caldarium protein, suggesting N-terminal blocking of this protein, and only short sequences with O. danica (XYAFGINGXRI) and Eu. gracilis (XDTKLGLVEEEAAN) (proteins with only limited similarity to other sPPases), as was expected due to the hypervariable character of the N-terminal protein regions.
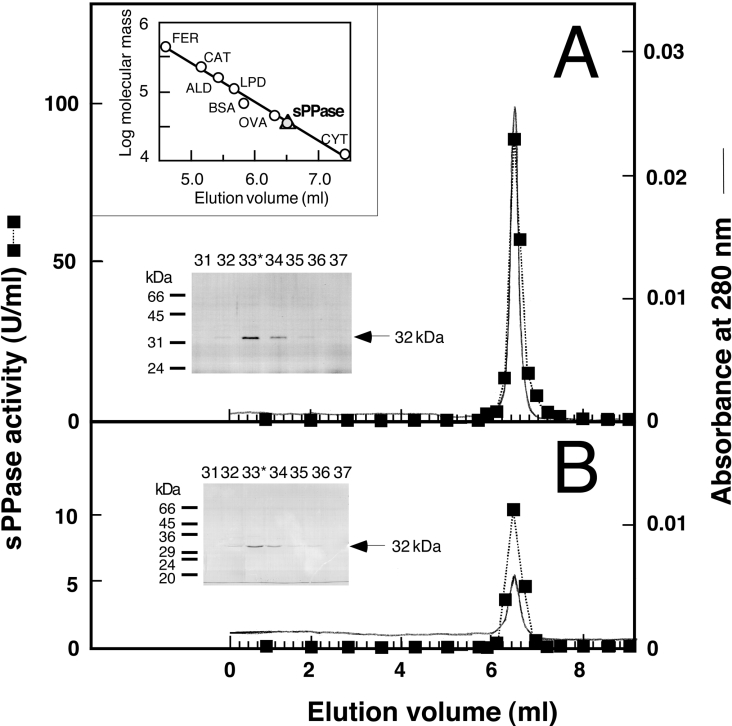
Figure 3. FPLC gel-filtration chromatography on Superose 12HR column of the sPPase purified from whole cells and cyanelles of Cy. paradoxa.
Aliquots (0.1 ml) of sPPase preparations (hydroxyapatite eluate) purified from whole cells (approx. 150 μg) (A) or isolated cyanelles (approx. 50 μg) (B) were applied on to a Superose 12HR 10/30 column. Elution (0.4 ml/min) and fractions were collected as described in Figure 2. The upper inset shows the calibration of the column with protein standards (FER, ferritine; CAT, catalase; ALD, aldolase; LPD, lipoamide dehydrogenase; OVA, ovoalbumin; CYT, cytochrome c) and the position of sPPase peaks (triangle and grey circle) that corresponds to a native molecular mass of approx. 33 kDa. The Coomassie-Blue-stained SDS/PAGE gels of the indicated fractions around the activity peak (*, highest activity fraction) show a single 32 kDa protein (arrows) that co-eluted with sPPase activity in both cases. The positions and molecular masses of protein standards are indicated on the left-hand side of the SDS/PAGE gels.
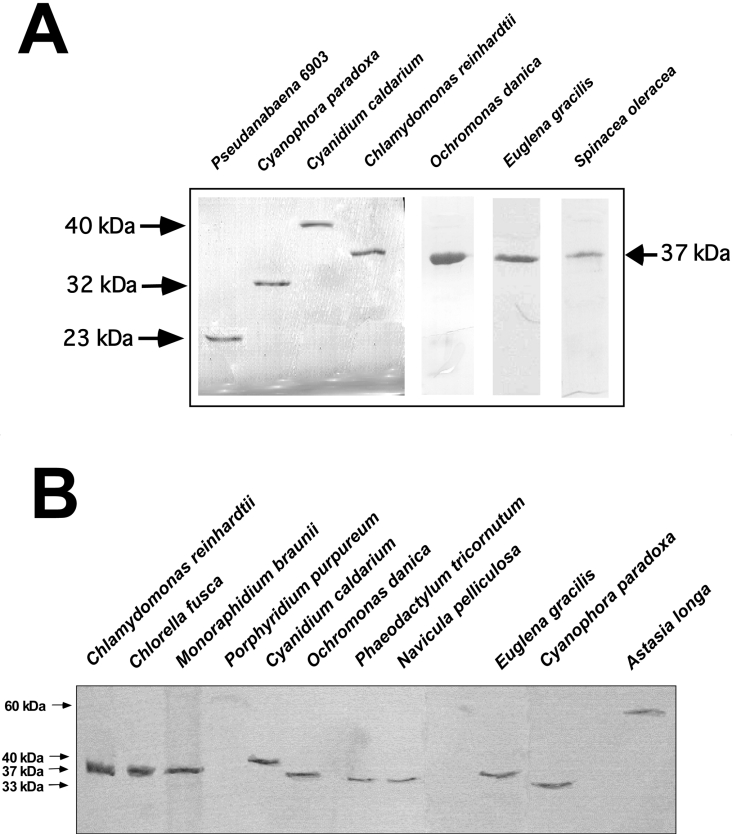
Figure 4. SDS/PAGE and Western blot analyses of chloroplastic sPPases from microalgae and plants.
(A) Coomassie-Blue-stained SDS/PAGE of sPPases purified from different photosynthetic organisms: cyanobacterium Pseudanabaena sp. PCC 6903, microalgae Cy. paradoxa, Cn. caldarium, Ch. reinhardtii (chloroplastic Cr-sPPase I), O. danica, Eu. gracilis and spinach chloroplasts. Molecular masses are indicated by arrows. Approx. 3 μg of purified proteins were applied per lane. (B) Western blot probed with the monospecific anti-(Cr-sPPase I) antibody showing Cr-sPPase I orthologues in diverse photosynthetic protists. Approx. 30 μg of protein extracts were loaded per lane for Chlorophyceae (Ch. reinhardtii, Ch. fusca and M. braunii) and 80 μg per lane for the other microalgae. A single protein band of 33–40 kDa was immnunodetected, except in the secondary non-photosynthetic euglenoid As. Longa, which exhibited a 60 kDa band, and the rhodophycean alga P. purpureum, where no protein was immnunodetected.
The catalytic properties of algal and plant sPPases studied in this work are shown in Table 1. These enzymes exhibit a high affinity for the substrate with Km values (10–30 μM) and catalytic efficiencies (estimated as kcat/Km ratios) similar to those found for their homologues in photosynthetic prokaryotes (M. R. Gómez-García and A. Serrano, unpublished work). However, catalytic efficiencies were clearly lower than that of the dimeric yeast sPPase [32], mainly due to the comparatively higher (approx. 10-fold) Km values of the algal proteins.
Table 1. Kinetics parameters of sPPases from photosynthetic eukaryotes.
*Data for comparison, from [32].
| Values | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Organism… | Ch. reinhardtii | O. danica | E. gracilis | Cy. paradoxa | Cn. caldarium | Sp. oleracea | S. cerevisiae* | ||
| Kinetic parameter | sPPaseI | sPPaseII | |||||||
| Km (μM) | 10.5 | 65.2 | 29.2 | 32.4 | 32.4 | 10.5 | 29.4 | 1.5 | |
| kcat (s−1) | 314.6 | 334.6 | 204.5 | 280.0 | 208.5 | 184.5 | 102.5 | 197.0 | |
| kcat/Km | 30.0 | 5.1 | 7.1 | 8.6 | 6.4 | 17.6 | 3.5 | 131.3 | |
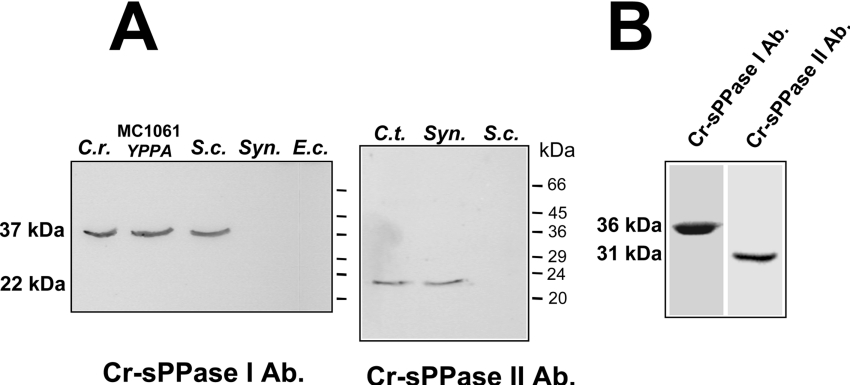
Western blot analysis, performed using monospecific polyclonal antibodies against Cr-sPPase I and II with soluble protein extracts from microalgae belonging to different taxonomic groups (Chlorophyceae, Rhodophyceae, Chromophyceae, Bacillariophyceae, Euglenophyceae and Glaucocystophyceae), is shown in Figure 4(B). These microalgae exhibit Mg2+-dependent sPPase activity and, consistent with this finding, possess 30–40 kDa polypeptides which cross-reacted with the anti-(Cr-sPPase I) antibody, suggesting they possess eukaryotic-type family I sPPase. Since this antibody recognized the sPPases purified from Cy. paradoxa cyanelles and Ch. reinhardtii and spinach chloroplasts (results not shown), the immunodetected proteins in the algal crude extracts should probably correspond to plastidic sPPases. The antibody against Cr-sPPase I also recognized sPPase I-like polypeptides in Western blots of crude extracts from S. cerevisiae and E. coli strain MC1061, expressing the cytosolic Y-sPPase (yeast sPPase), but no protein bands were immunodetected in cyanobacterial or E. coli extracts (Figure 5A). Moreover, the anti-(Cr-sPPase II) antibody recognized prokaryotic sPPases (24 kDa), such as those from the photosynthetic sulphur bacterium Chl. tepidum and the unicellular cyanobacterium Synechocystis sp. PCC 6803, but no protein bands were immunodetected in yeast or chloroplast extracts (Figure 5A). Interestingly, these antibodies recognized a protein band for sPPase I-like and sPPase II-like polypeptides, of 35 and 30 kDa (estimated by SDS/PAGE) respectively, in crude extracts from A. thaliana leaves, suggesting the presence of orthologues of both algal proteins in plant photosynthetic tissues (Figure 5B).
Figure 5. Immunoreaction patterns of the anti-(Cr-sPPase I) and II antibodies.
(A) Cell-free extracts of different micro-organisms (C.r., Ch. reinhardtii, 10 μg of protein; MC1061 YPPA, E. coli strain MC1061 expressing Y-sPPase, 30 μg of protein; S.c., S. cerevisiae, 30 μg of protein; Syn, Synechocystis PCC 6803, 90 μg of protein; E.c. E. coli DH5α, 90 μg of protein). (B) A soluble extract of A. thaliana leaves (40 μg of protein per lane). Molecular mass markers positions are indicated. Note the different capacity of both antibodies to recognize eukaryotic- and prokaryotic-type enzymes, and that both types of sPPases are detected in plant photosynthetic tissue. Ab, antibody.
Subcellular localization of the two Cr-sPPase paralogues
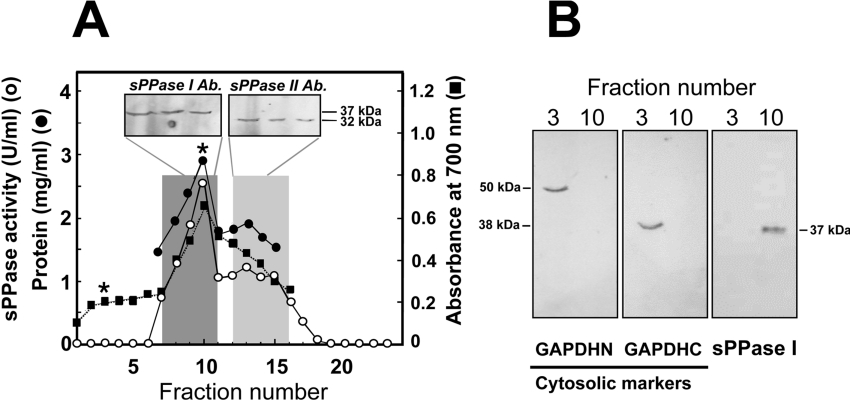
Percoll gradients [24–26] show that Cr-sPPase I (30 kDa protein) is mostly in the chloroplastic fraction from Ch. reinhardtii, whereas the 24 kDa Cr-sPPase II was related to the mitochondria (Figure 6A). In parallel, Western blots were performed as controls using antibodies that detect two cytosolic marker proteins in algae, GAPDHN (non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase) and GAPDHC (NAD-dependent glyceraldehyde-3-phosphate dehydrogenase), as reported previously [33], both of which are involved in glycolytic carbon metabolism in photosynthetic eukaryotes (Figure 6B). This analysis confirmed that the cytosol of Ch. reinhardtii lacks sPPase and that there is no significant contamination with the chloroplastic fraction. Similar results were obtained with Cy. paradoxa where virtually all sPPase is detected in the cyanelles.
Figure 6. Cr-sPPases subcellular localization by Percoll density gradients.
(A) Western blots (approx. 20 μg of protein loaded per lane) with anti-(Cr-sPPase I) and anti-(Cr-sPPase II) antibodies shows that the 37 kDa Cr-sPPase I was associated with the green-coloured band (dark grey bar) and the 32 kDa Cr-sPPase II appears to be associated with a denser band, including mitochondria (light grey bar). (B) Western blot analyses of fractions 3 (cytosol) and 10 (chlorophyll peak) of the gradient with antibodies against two cytosolic protein markers in photosynthetic eukaryotes, GAPDHN (non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase; left-hand panel) and GAPDHC (NAD-dependent glyceraldehyde-3-phosphate dehydrogenase; middle panel), as well as with the anti-(Cr-sPPase I) antibody (right-hand panel). Approx. 30 μg of protein was loaded per lane.
Cloning of a PPA gene encoding the chloroplastic sPPase from A. thaliana and expression in E. coli under trc promoter
A BLAST search in the Arabidopsis Genome Displayer (http://kazusa.or.jp/kaos/cgi) using the yeast YPPA gene as a query sequence (eukaryotic type sPPase) identified a genomic sequence encoding the putative chloroplastic sPPase (precursor polypeptide) from A. thaliana. It corresponds to the AT5G09650 gene, which is well represented in the EST databases.
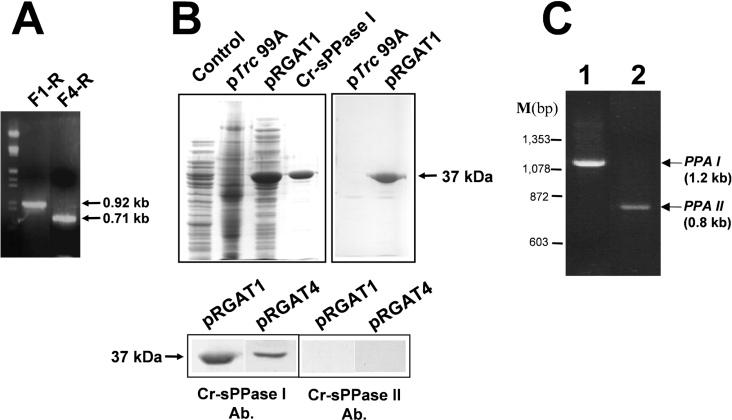
PCR amplifications performed using a total cDNA preparation from A. thaliana leaves as a template and specific primers designed for the precursor of the chloroplastic sPPase and the probable mature protein, as predicted by computer programs, produced the predicted amplicons (0.92 kb, precursor; 0.71 kb, predicted mature protein) (Figure 7A). They were cloned in E. coli expression plasmid pTrc99A to obtain the constructs pRGAT1 and pRGAT4 respectively.
Figure 7. Cloning of two A. thaliana cDNA amplicons encoding a chloroplastic sPPase putative precursor and its predicted mature protein, and immunochemical characterization of the corresponding recombinant proteins produced in E. coli.
(A) Electrophoresis on 0.7% agarose gel of the single PCR bands amplified with specific primer pairs F1-R (0.92 kb, complete sPPase I precursor ORF) and F2-R (0.71 kb, predicted sPPase I mature protein ORF). Both ORFs correspond to the A. thaliana gene AT5G09650. Lambda phage DNA cleaved with HindIII and EcoRI was used as a bp marker. (B) Upper panel: Coomassie-Blue-stained SDS/PAGE (left-hand panel) of cell-free extracts (50 μg of protein per lane) from E. coli XL1blue transformed with empty pTrc99A (control), transformed with pRGAT1 (F1-R cDNA construct) expressing a 37 kDa protein and 5 μg of purified Cr-sPPase I, and immunodetection (right-hand panel) of the recombinant plant sPPase I-like protein by the anti-(Cr-sPPase I) antibody. Lower panel: immunodetection of over-expressed plant 37 kDa protein with the anti-(Cr-sPPase I) antibody in cell-free extracts of E. coli XL1blue clones (40 μg of protein per lane) transformed with pRGAT1 (F1-R cDNA construct, sPPase I chloroplast precursor ORF) and pRGAT4 (F4-R cDNA construct, predicted sPPase I mature protein ORF). Note that no equivalent protein band was recognized by the anti-(Cr-sPPase II) antibody. (C) Electrophoretic analysis (0.7% agarose gel) of PCR-amplified Ch. reinhardtii cDNAs corresponding to the two different PPA genes encoding putative sPPases identified in EST databases. Single cDNA bands of the expected size for PPAI (1.2 kb, lane 1) and PPAII (0.8 kb, lane 2) cDNAs were obtained using a total cDNA preparation from Ch. reinhardtii cells as a template. ΦX174 phage DNA cleaved with HaeIII was used as a bp marker (M).
The expression of the largest cDNA fragment in E. coli (Figures 7A and 7B) yielded a 37 kDa protein with sPPase activity recognized by the antibody against Cr-sPPase I, whereas the anti-(Cr-sPPase II) antibody did not recognize any protein in the bacterial extracts (Figure 7B). Moreover, apparent sizes of the immunodetected protein bands were the same for the pRGAT1 and pRGAT4 clones (Figure 7B), suggesting that plant chloroplastic sPPase precursor was processed by the bacterial host cell. This is also in agreement with the fact that sPPase specific activity levels measured in extracts from E. coli transformed with pRGAT1 and pRGAT4 (Table 2) were approx. 15-fold that of the native sPPase activity level in the bacterium. In the case of pRGAT1 almost 50% of sPPase activity was detected in the pellet fraction, probably because part of the heterologous plastidic sPPase could be anchored to the bacterial plasma membrane.
Table 2. E. coli pRGAT1 and pRGAT4 sPPase specific activities in soluble and membrane fractions.
| sPPase activity (units/mg of protein) | ||||
|---|---|---|---|---|
| Fraction | E. coli… | pRGAT1 | pRGAT4 | pTrc99A |
| Membrane | 24.0±0.12 | 11.5±0.12 | 0.8±0.04 | |
| Soluble | 59.0±0.12 | 49.0±0.15 | 4.5±0.10 | |
Cloning of two cDNAs encoding sPPases from Ch. reinhardtii
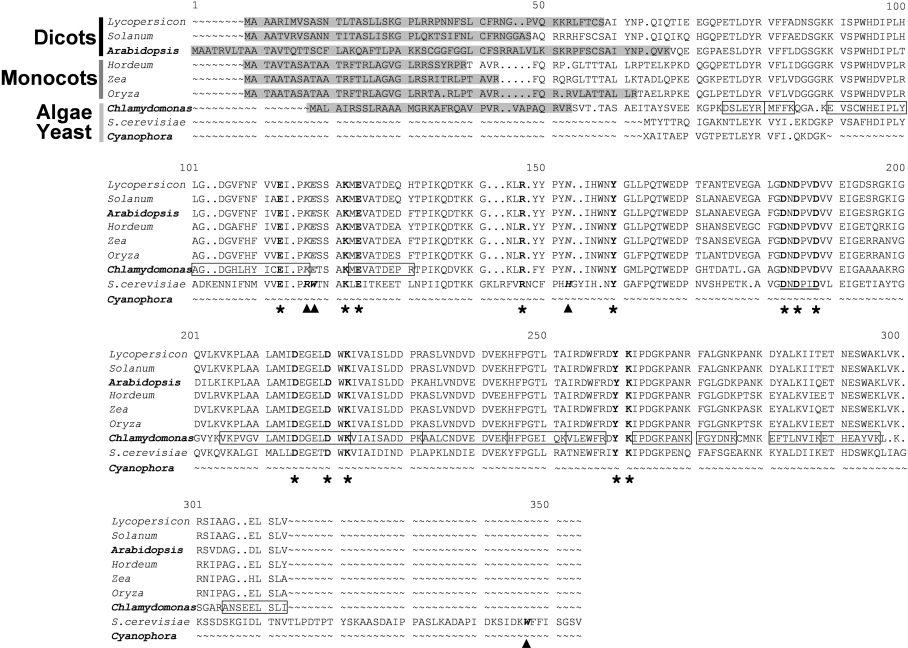
BLAST similarity searches carried out with the EST public databases (ChlamyEST, http://www.kazusa.or.jp/eh/plant/chlamy/EST/, and ‘dbest Chlamydomonas’ on the NCBI BLAST server), using the protein sequences deduced from the chloroplastic PPA gene described above and from another sPPase-encoding cDNA from A. thaliana previously reported in [34] as queries, allowed us to identify two different Ch. reinhardtii sPPase-encoding cDNAs. Both of these cDNAs were PCR amplified from phototrophic algal cells (Figure 7C) and sequenced. The longest one (approx. 1.2 kb) contained a 843 bp ORF (PPAI gene) encoding an sPPase I-like protein precursor with a N-terminal chloroplastic transit peptide of 34 amino acids, identified by the ChloroP and TargetP programs (Figure 8). The shortest cDNA (approx. 0.8 kb) contained a 579 bp ORF (PPAII gene) and encodes a putative prokaryotic-type sPPase II-like protein with no clear N-terminal organelle transit peptide (results not shown). Shorter transit peptides were predicted for algal PPA gene products when compared with orthologues from plants, in agreement with previous reports on transit peptides for organelles in Ch. reinhardtii [35]. Both cDNAs have the PROSITE motif and the conserved catalytic residues of the sPPase family I [5] (Figure 8). MALDI–TOF peptide mass fingerprint identified the two purified sPPases from Ch. reinhardtii as products of the two cloned algal cDNAs (Figure 8 and results not shown). These cDNAs should correspond to the Ch. reinhardtii genes C_260136 (PPAI gene) and C_80147 (PPAII gene) annotated in the Chlamydomonas genome version 2.0 database (http://genome.jgi-psf.org/chlre2/chlre2.home.html). A third putative sPPase gene in the Chlamydomonas genome database (C_700081, suggested to be mitochondrial) was, in agreement with our results, probably not expressed, since no ESTs were found in databases; moreover, its predicted product should be inactive, because it exhibits deleterious mutations in four active-site residues that are reported to be essential for catalytic activity (A. Serrano, unpublished work).
Figure 8. ClustalX sequences alignment of several putative chloroplastic sPPase orthologues of plants (monocots and dicots) and the photosynthetic protists Ch. reinhardtii (PPAI-encoded Cr-sPPase I) and Cy. paradoxa, with the homodimeric sPPase of S. cerevisiae.
Full protein sequences derived from complete ORFs are shown except for Cp-sPPase, for which only the N-terminal region is known so far. The 13 active-site residues functionally important for sPPase activity [48,49] are in bold and indicated by asterisks. The PROSITE motif of active-site residues, characteristic of family I sPPases, are underlined. N-terminal extensions predicted to be chloroplastic transit peptides are indicated in grey boxes. All available chloroplastic sPPase sequences show substitutions (italicized and further indicated by arrowheads) of residues Arg51, Trp52 and His87 and lack of Trp279 (yeast sPPase numbering, italics and bold) that are involved in yeast sPPase homodimer stabilization [12]. Peptides of purified Cr-sPPase I identified by MALDI–TOF fingerprint MS analysis are boxed in the protein sequence deduced from PPAI cDNA; they cover approx. 60% of the predicted mature protein sequence. Most plant protein sequences were identified by BLAST searches in non-redundant and EST databases and obtained by gene reconstruction by EST overlapping. The sequences used are (accession numbers in parentheses): S. cerevisiae cytosolic sPPase (CAA31629), Ch. reinhardtii PPAI putative chloroplastic precursor (AJ298231); Cy. paradoxa cyanellar sPPase N-terminal sequence (P80887); A. thaliana chloroplastic precursor (AJ252210); Lycopersicon esculentum (BF623033 and BG126436); Solanum tuberosum (BI432577 and BI434604); Hordeum vulgare (BF623033 and BJ464714); Zea mais (BI596040 and CA404176); Oryza sativa (CB681947 and CB681948, AK059725).
DISCUSSION
Eukaryotes have specific cellular compartments, such as chloroplasts and mitochondria, where biosynthetic reactions take place with the concomitant release of PPi hydrolysed by sPPases. Family I sPPases are the most widespread and probably the most ancestral ones [5,6]. The sPPases described so far are oligomeric proteins present in the cytosol of archaea, bacteria, fungi and metazoa, as well as in some organelles (mitochondria). Molecular phylogenetic studies indicate the presence of two divergent evolutionary lineages in this sPPase group: the ‘eukaryotic’ (fungi, plants, metazoa and many protists) and the ‘prokaryotic’ (bacteria, archaea and photosynthetic eukaryotes), as shown in the present study (Figure 9). The family I sPPases are, therefore, an ancient conserved family of orthologues from evolutionarily very distant organisms.
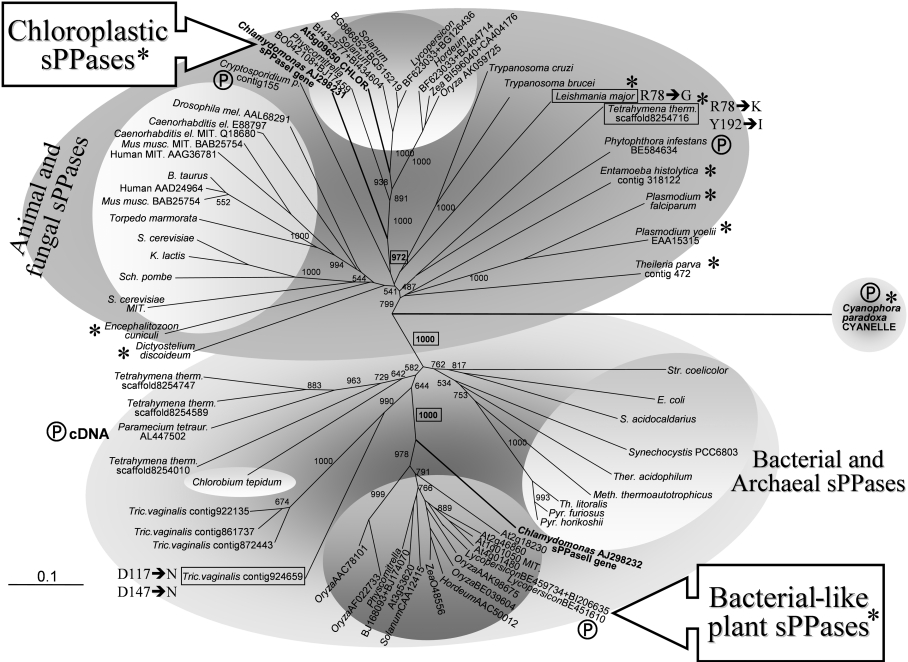
Figure 9. Molecular phylogenetic analysis of the two paralogous family I sPPases identified in photosynthetic eukaryotes and their evolutionary relationships with sPPases of prokaryotes (bacteria and archaea), protists (parasitic and free-living), fungi and metazoa.
Amino acid sequences deduced from algal and plant genes were aligned with sequences of selected family I sPPases using ClustalX. Most sequences have all the amino acid residues reported to be functionally important for sPPase activity [48,49] and the PROSITE motif of family I sPPases. A few sPPase sequences from primitive protists (boxed) exhibit changes in some of these functionally important residues (indicated by Y-sPPase numbering). These sequences are found among natural sPPase mutants, some of them with unusual catalytic features [19,32]. Eukaryotic sequences showing substitutions in residues involved in yeast sPPase homodimer stabilization (Arg51, Trp52 and His87, Y-sPPase numbering; [12]) are marked with asterisks (all chloroplastic sPPases and many orthologous sequences from protists). The few partial sequences used are marked with a Ⓟ symbol. Proteins clearly identified directly, or inferred by computer programs, to have organellar (mitochondrial or chloroplastic) localization, and therefore synthesized as precursors, are labelled accordingly. Numbers at nodes indicate the statistical support (bootstrap values from 1000 replicates) of selected associated groups. The 0.1 bar represents amino acid substitutions per site. Two main clearly defined sPPase assemblies are well supported (bootstrap value 1000, boxed in bold). They cluster on one side as eukaryotic proteins (cytosolic and organellar) from protists, plants, fungi and animals (dark grey upper area), and on the other side prokaryotic (bacterial and archaeal) proteins, as well as bacterial-like sPPases from photosynthetic eukaryotes and some non-photosynthetic protists (ciliates and diplomonads) (light grey lower area). The two clusters of sPPases from photosynthetic eukaryotes (plants and microalgae) tentatively defined in this report, namely chloroplastic sPPase I and bacterial-like sPPase II proteins, are well supported (bootstrap values 972 and 1000 respectively, boxed in bold) and arrange with the eukaryotic (animal and fungal) and prokaryotic sPPases respectively. Accession numbers (in parentheses) for other sequences used are: S. cerevisiae cytosolic sPPase (CAA31629) and mitochondrial sPPase precursor (NP_013994.1); human cytosolic sPPase (AAD24964) and mitochondrial sPPase precursor (AAG36781); Mus musculus cytosolic sPPase (BAB25754) and mitochondrial sPPase precursor (BAB22922); Kluyveromyces lactis cyotosolic sPPase (CAA32466); Encephalitozoon cuniculi (AL590449); Plasmodium falciparum (NP_473275.1); Entamoeba histolytica (contig 318122, TIGR); Dictyostelium discoideum (AU061000 and AU034313); Trypanosoma brucei (contigs 90C7TF and 81B5TF, TIGR); Trypanosoma cruzi (contig TCKGB96TR, TIGR); E. coli (P17288); Synechocystis PCC6803 (AJ252207); Streptomyces coelicolor (O9×819); E. coli (P17288); Sulfolobus acidocaldarius (P50308); Thermoplasma acidophilum (P37981); Methanobacterium thermoautotrophicus (O26363); Thermococcus litoralis (P77992); Pyrococcus horikoshii (O59570). Other putative sPPase sequences obtained with BLAST searches are identified by accession numbers and contig codes indicated in the Figure.
The two paralogous sPPases of S. cerevisiae are encoded by the nuclear genes YPIRI and YPIR2, encoding cytosolic [36] and mitochondrial [7] sPPases respectively, both of which carry out similar functions in these compartments. These enzymes are highly homologous eukaryotic family I sPPases. In contrast with fungi and animals, photosynthetic plant cells lack cytosolic sPPase [11], but possess an integral membrane H+-translocating PPase, unrelated to sPPases, that uses chemical energy from PPi to generate an electrochemical gradient [37]. sPPase levels similar to those obtained in yeast and other non-photosynthetic organisms were detected in extracts of microalgae and plants, but no sPPase activity was located in the cytosol of any of the photosynthetic eukaryotes tested in the present study.
Two distinct sPPases from the green microalga Ch. reinhardtii, Cr-sPPase I and Cr-sPPase II, with molecular masses of 30 and 24 kDa respectively, were isolated and characterized. The monomeric state determined is ununsual for eukaryotic family I enzymes. Subcellular fractionation indicated a chloroplastic localization of Cr-sPPase I, whereas Cr-sPPase II was associated with mitochondrial fractions.
sPPases isolated from other phylogenetic diverse microalgae, Cn. caldarium, O. danica and Eu. gracilis, and from Sp. oleracea chloroplasts, are also monomeric proteins, presumably similar to Cr-sPPase I. Chloroplastic sPPases have structural features more similar to eukaryotic (animal and fungal) sPPases than to cyanobacterial sPPases, although cyanobacterial-like ancestral endosymbionts (structurally similar to the extant cyanelles) are considered the evolutionary ancestors of chloroplasts [38]. Cyanelles are peptidoglycan-surrounded plastids that phylogenetic analysis has placed on the earliest branch of phototrophic eukaryotes after the primary endosymbiotic event [39–41]. Interestingly, cyanelles, despite close similarity to unicellular cyanobacteria and to ancestral primitive plastids, contain a momomeric sPPase that resembles its homologous chloroplastic enzymes. Its N-terminal sequence is similar to algal and plant chloroplastic sPPases (Figure 8) and also to cytosolic sPPases from fungi and animals (Figures 8 and 9). The presence of a non-cyanobacterial eukaryotic type enzyme, in this extant primitive plastid-like organelle, would suggest that the ancestral cyanobacterial endosymbiont sPPase was lost quite early during the evolutionary development of plastids and was replaced by the host cell sPPase [42,43]. As sPPases may have been essential for the evolution of cellular anabolism [7,44], a functional substitution of the cyanobacterial enzyme probably helped to stabilize the primary endosymbiotic event that gave rise to current chloroplasts. This may be a remarkable example of endosymbiotic orthologous gene displacement during organelle evolution. A similar process may have taken place with the mitochondrial sPPase in fungi, where the ancestral proteobacterial sPPase was not transferred into the nuclear genome and was lost; the current mitochondrial enzyme is a nuclear-encoded eukaryotic sPPase very similar to the cytosolic isoform [7].
The two organellar sPPases of Ch. reinhardtii, Cr-sPPase I (major chloroplastic isoform) and II (mitochondrial isoform), are active monomers. Monomeric Cr-sPPase I orthologues seem to be ubiquitous among photosynthetic eukaryotes (from protists with primitive cyanelles to plants) and cross-reacted with an anti-(Cr-sPPase I) antibody. All algal and plant sPPase I-like proteins identified to date should have N-terminal extensions predicted to be chloroplastic transit peptides.
The two PPA genes of Ch. reinhardtii found in EST databases encode Cr-sPPase I and II. PPAI codes for a plastidic eukaryotic-type sPPase endowed with a chloroplastic transit peptide for this organelle. PPAII encodes a smaller prokaryotic-type protein with no clear transit peptide. Both have the PROSITE motif of family I sPPases. PCR amplification revealed two genes obtained from photoautotrophic preparations of the microalga, indicating that both are expressed in these conditions, in agreement with the results obtained from Percoll experiments. Although further expression studies are needed to confirm these results, purified Cr-sPPases I and II were identified as the predicted PPAI and PPAII gene products respectively by MALDI–TOF MS analyses.
Y-sPPases are dimeric enzymes with an oligomerization state completely different than those from prokaryotes, involving two kinds of highly conserved subunit interactions: hydrophobic between aromatic residues (Trp52 and His87) and hydrogen bridges between basic residues (Arg51) [6,12]. This interface network is highly conserved among fungal and animal sPPases [45,46] (Figure 9). Y-sPPase variants with mutations in these residues generate monomeric proteins in solution [12] with increased Km and decreased kcat values for PPi, which reduce the catalytic efficiency to values similar to those of plastidic monomeric sPPases (Table 1). Sequence alignment of eukaryotic sPPases shows that Trp52 and His87, involved in dimeric structure stabilization in fungal cytosolic sPPases, identified the substitutions W136E and H175N in Cr-sPPase I, as well as in its plant orthologues (Figure 8). A C-terminal extension containing the fourth residue involved in yeast sPPase homodimer stabilization (Trp279) was also absent in plastidic sPPases (Figure 8). Changes in these residues are presumably responsible for the unusual monomeric structure of these proteins, which are natural variants of eukaryotic family I-sPPases, and are in accordance with the catalytic features of plastidic sPPases [12].
The putative precursor of the A. thaliana chloroplastic sPPase and the possible mature protein were expressed in E. coli and immunodetected with antibodies against Cr-sPPase I, suggesting that the plant plastidic sPPase is processed to produce a protein of the expected molecular mass; however, a significant proportion appears to be anchored to the bacterial membrane. These results indicate that the natural plant sequence encodes a protein with a chloroplastic transit peptide.
A. thaliana possess five prokaryotic-type Cr-sPPase II orthologues (Figure 9) that are expressed in diverse plant photo- and hetero-trophic tissues (A. Serrano, unpublished work). In agreement with the monomeric state of the bacterial-like Cr-sPPase II, the sequences of all plant and algal sPPase II identified so far show substitutions in six residues corresponding to E. coli sPPase, Asn24, Tyr77, Gln80, His136, His140 and Asp143 (results not shown), which form contact clusters at the subunit interfaces involved in hexamer stabilization [6,46]. Some of them have N-terminal extensions suggested to be mitochondrial transit peptides. In summary, all sPPases from photosynthetic eukaryotes are monomeric family I proteins with unusual structural and catalytic features.
Molecular phylogenetic analyses of the sPPases from photosynthetic eukaryotes and their evolutionary relationships with sPPases of prokaryotes, protists, fungi and metazoa confirmed the distant evolutionary positions of the two paralogous family I sPPase classes (plastidic and prokaryotic-type) found in microalgae and plants (Figure 9). The two main, clearly defined, sPPase assemblies are well supported. On one side they cluster with eukaryotic (cytosolic and organellar) proteins from animals, fungi, protists and plants, and on the other side, with prokaryotic (bacterial and archaeal) proteins, and prokaryotic-type sPPases from photosynthetic eukaryotes and some non-photosynthetic protists (ciliates and diplomonads). The two compact sPPase clusters from photosynthetic eukaryotes, chloroplastic sPPase I and prokayotic-type sPPase II proteins, are well supported and arrange with the eukaryotic (animal and fungal) and prokaryotic (bacterial and archaeal) sPPases respectively. Remarkably, a few sPPases from primitive protists (trypanosomes, diplomonads and ciliates) exhibit changes in some catalytic important residues, probably being natural eukaryotic sPPase functional mutants (Figure 9); they could be either pseudo-genes or sPPases with unusual catalytic features (see below). Chloroplastic sPPases and some orthologous sequences from protists in the eukayotic assembly show substitutions in residues involved in Y-sPPase homodimer stabilization. As discussed above, they are expected to be monomeric enzymes, an sPPase class that seems widely distributed among lower eukaryotes (Figure 9). Some of these eukaryotic proteins were synthesized as precursors and identified to have organellar (mitochondrial or plastidic) localization. Fungal and animal sPPases group as a compact cluster in the eukaryotic assembly, showing closely related cytosolic and mitochondrial paralogues (Figure 9). Some heterotrophic protists (diplomonads and ciliates) have, similar to photosynthetic eukaryotes, sPPase paralogues of very distant phylogenetic assignments (eukaryotic and bacterial), suggesting photosynthetic ancestries in agreement with recent studies [47]. Natural sPPase with mutations of catalytically important residues may have unexpected functional features, as described recently in the parasitic protist Leishmania major [19]. This divergent and probably ancient family I sPPase belongs to the eukaryotic assembly, in agreement with its unusual catalytic properties. This family clusters with other trypanosomal sPPases in an evolutionarily diverse-branched group. This group is closely related to orthologues of other ancient protists (apicomplexans and amoebae) and is more distantly related to the fungi–metazoa assemblies.
The presence of prokaryotic- and eukaryotic-type sPPases in Ch. reinhardtii, its close relative the colonial microalga Volvox carteri (A. Serrano, unpublished work), the moss Physcomitrella patens and many higher plants (Figure 9) shows photosynthetic eukaryotes as a remarkable example of apparent redundancy of paralogous sPPases (all of them nucleus encoded) that are arranged in two distant phylogenetic groups, a situation that was maintained during the evolution of the green photosynthetic lineage. Overall, the results of the present study reveal that photosynthetic eukaryote sPPases represent a remarkable and so far unique example of endosymbiotic orthologous gene displacement during the early stages of organelle evolution.
Acknowledgments
We thank Dr C. D. Fraley and Dr J. Josse for their comments on the manuscript prior to submission, and Professor W. Löffelhardt and Dr G. Allmaier (University of Vienna, Austria) for their assistance in the work with Cy. paradoxa and N-terminal protein sequencing and MALDI–TOF analyses. This work was supported by research grants from the Spanish (BMC2001-563 and BFU2004-00843, MEC) and Andalusian Regional (PAI group CVI-261, CEC) Administrations.
References
- 1.Kornberg A. On the metabolic significance of phosphorolytic and pyrophosphorolytic reactions. In: Kasha M., Pullman D., editors. Horizons in Biochemistry. New York: Academic Press; 1962. pp. 251–254. [Google Scholar]
- 2.Lahti R., Kolakowski L. F., Vihinem M., Pohjanoksa K., Cooperman B. S. Conservation of functional residues between yeast and E. coli inorganic pyrophosphatases. Biochim. Biophys. Acta. 1990;1038:338–345. doi: 10.1016/0167-4838(90)90246-c. [DOI] [PubMed] [Google Scholar]
- 3.Young T. W., Kuhn N. J., Wadeson A., Ward S., Burges D., Cooke G. D. Bacillus subtilis ORF yybQ encodes a manganese-dependent inorganic pyrophosphatase with distinctive properties: the first of a new class of soluble pyrophosphatase? Microbiology. 1998;144:2563–2571. doi: 10.1099/00221287-144-9-2563. [DOI] [PubMed] [Google Scholar]
- 4.Shintani T., Uchiumi T., Yonezawa T., Salminen A., Baykov A. A., Lahti R., Hachimori A. Cloning and expression of a unique inorganic pyrophosphatase from Bacillus subtilis: evidence for a new family of enzymes. FEBS Lett. 1998;439:263–266. doi: 10.1016/s0014-5793(98)01381-7. [DOI] [PubMed] [Google Scholar]
- 5.Cooperman B. S., Baykov A. A., Lahti R. Evolutionary conservation of the active site of soluble inorganic pyrophosphatase. Trends Biochem. Sci. 1992;17:262–266. doi: 10.1016/0968-0004(92)90406-y. [DOI] [PubMed] [Google Scholar]
- 6.Baykov A. A., Cooperman B. S., Goldman A., Lahti R. Cytoplasmic inorganic pyrophosphatase. Prog. Mol. Subcell. Biol. 1999;23:127–150. doi: 10.1007/978-3-642-58444-2_7. [DOI] [PubMed] [Google Scholar]
- 7.Lundin M., Baltscheffsky H., Ronne H. Yeast PPA2 gene encodes a mitochondrial inorganic pyrophosphatase that is essential for mitochondrial function. J. Biol. Chem. 1991;266:12168–12172. [PubMed] [Google Scholar]
- 8.Simmon S., Butler L. G. Alkaline inorganic pyrophosphatase of maize leaves. Biochim. Biophys. Acta. 1969;172:150–157. doi: 10.1016/0005-2728(69)90100-5. [DOI] [PubMed] [Google Scholar]
- 9.Klemme B., Jacobi G. Separation and characterization of two inorganic pyrophosphatases from spinach leaves. Planta. 1974;120:147–153. doi: 10.1007/BF00384924. [DOI] [PubMed] [Google Scholar]
- 10.Bennet V. L., Ristrophe D. L., Hamming J. J., Butler L. G. Maize leaf inorganic pyrophosphatase: isoenzymes, specificity for substrates, inhibitors, and divalent metal ions, and pH optima. Biochim. Biophys. Acta. 1973;293:232–241. doi: 10.1016/0005-2744(73)90396-3. [DOI] [PubMed] [Google Scholar]
- 11.Weiner H., Stitt M., Heldt H. W. Subcellular compartmentation of pyrophosphate and alkaline pyrophosphatase in leaves. Biophys. Biochem. Acta. 1987;893:13–21. [Google Scholar]
- 12.Salminen A., Parfenyev A. N., Salli K., Efimova I. S., Magretova N. N., Goldman A., Baykov A. A., Lahti R. Modulation of dimer stability in yeast pyrophosphatase by mutations at the subunit interface and ligand binding to the active site. J. Biol. Chem. 2002;277:15465–15471. doi: 10.1074/jbc.M200101200. [DOI] [PubMed] [Google Scholar]
- 13.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U.S.A. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano A., Löffelhardt W. Identification of two different glyceraldehyde-3-phosphate dehydrogenases (phosphorylating) in the photosynthetic protist Cyanophora paradoxa. Arch. Microbiol. 1994;162:14–19. [Google Scholar]
- 15.Rigano C. Studies on nitrate reductase from Cyanidium caldarium. Arch. Microbiol. 1971;76:265–276. doi: 10.1007/BF00409121. [DOI] [PubMed] [Google Scholar]
- 16.Darley W. M., Volcani B. E. Synchronized cultures: diatoms. Methods Enzymol. 1971;23:85–96. [Google Scholar]
- 17.Rippka R., Deruelles J., Waterbury J. B., Hermann M., Stainer R. Y. Generic assignment, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979;111:1–16. [Google Scholar]
- 18.Fiske C. H., Subarow Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- 19.Gomez-Garcia M. R., Ruiz-Perez L. M., Gonzalez-Pacanowska D., Serrano A. A novel calcium-dependent soluble inorganic pyrophosphatase from the trypanosomatid Leishmania major. FEBS Lett. 2004;560:158–166. doi: 10.1016/S0014-5793(04)00097-3. [DOI] [PubMed] [Google Scholar]
- 20.Sugino Y., Miyoshi Y. The specific precipitation of orthophosphate and some biochemical applications. J. Biol. Chem. 1964;239:2360–2364. [PubMed] [Google Scholar]
- 21.Baykov A., Volk S. Quantitative detection of orthophosphate in polyacrylamide gels. Anal. Biochem. 1985;151:13–15. doi: 10.1016/0003-2697(85)90045-4. [DOI] [PubMed] [Google Scholar]
- 22.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 23.Hedrick I. L., Smith A. J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch. Biochem. Biophys. 1968;126:155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- 24.Klein U., Chen C., Gibbs M., Platt-Aloia K. A. Cellular fractionation of Chlamydomonas reinhardtii with emphasis on the isolation of chloroplasts. Plant Physiol. 1983;72:481–487. doi: 10.1104/pp.72.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris E. H. San Diego: Academic Press; 1988. The Chlamydomonas Source Book: A comprehensive Guide to Biology and Laboratory Use; pp. 25–31. [Google Scholar]
- 26.Price C. A., Reardon E. M. Isolation of chloroplasts for protein synthesis from spinach and Euglena gracilis by centrifugation in silica sols. In: Edelman M., Hallick R. B., Chua N. H., editors. Methods in Chloroplasts Molecular Biology. Amsterdam: Elsevier Biomedical Press; 1982. pp. 189–209. [Google Scholar]
- 27.Kuwavara T., Murata N. Purification and characterization of 33 kDa protein of spinach chloroplasts. Biochim. Biophys. Acta. 1979;581:228–236. doi: 10.1016/0005-2795(79)90242-3. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S. F. M. T., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–33402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emanuelsson O., von Heijne G. Prediction of organellar targeting signals. Biochim. Biophys. Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J. D., Gibson T. J., Plewnisk F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emanuelsson O., Nielsen H., Von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohjanjoki P., Lahti R., Goldman A., Cooperman B. S. Evolutionary conservation of enzymatic catalysis: quantitative comparison of the effects of mutation of aligned residues in Saccharomyces cerevisiae and Escherichia coli inorganic pyrophosphatases on enzymatic activity. Biochemistry. 1998;37:1754–1761. doi: 10.1021/bi971771r. [DOI] [PubMed] [Google Scholar]
- 33.Valverde F., Ortega J. M., Losada M., Serrano A. Sugar-mediated transcriptional regulation of the Gap gene system and concerted photosystem II functional modulation in the microalga Scenedesmus vacuolatus. Planta. 2005;221:937–952. doi: 10.1007/s00425-005-1501-0. [DOI] [PubMed] [Google Scholar]
- 34.Kieber J. K., Singer E. R. Cloning and characterization of an inorganic pyrophosphatase gene from Arabidopsis thaliana. Plant Mol. Biol. 1991;16:345–348. doi: 10.1007/BF00020567. [DOI] [PubMed] [Google Scholar]
- 35.Franzen L. G., Rochaix J. D., von Heijne G. Chloroplasts transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plants chloroplasts presequences. FEBS Lett. 1990;260:165–168. doi: 10.1016/0014-5793(90)80094-y. [DOI] [PubMed] [Google Scholar]
- 36.Cooperman B. S., Chiu N., Bruckmann R. H., Bunick G. J., McKenna G. P. Yeast inorganic pyrophosphatase. I: new methods of purification, assay, and crystallization. Biochemistry. 1973;12:1665–1669. doi: 10.1021/bi00733a001. [DOI] [PubMed] [Google Scholar]
- 37.Heldt H. W. Oxford: Oxford University Press; 1997. Plant Biochemistry and Molecular Biology. [Google Scholar]
- 38.Brown J., Doolitle F. Archaea and the prokaryote-to eukaryote transition. Microbiol. Mol. Biol. Rev. 1997;61:466–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janssen I., Jakowitsch J., Michalowski C. B., Bohnert H. J., Löffelhardt W. Evolutionary relationship of psbA genes from cyanobacteria, cyanelles and plastids. Curr. Genet. 1989;15:335–340. doi: 10.1007/BF00419913. [DOI] [PubMed] [Google Scholar]
- 40.Löffelhardt W., Bohnert H. J., Bryant D. A. The cyanelles of Cyanophora paradoxa. Crit. Rev. Plant. Sci. 1997;16:393–413. [Google Scholar]
- 41.Steiner J. M., Löffelhardt W. Protein import into cyanelles. Trends Plant Sci. 2002;7:72–77. doi: 10.1016/s1360-1385(01)02179-3. [DOI] [PubMed] [Google Scholar]
- 42.Schwartzbach S. D., Osafune T., Löffelhardt W. Protein import into cyanelles and complex chloroplasts. Plant Mol. Biol. 1998;38:247–263. [PubMed] [Google Scholar]
- 43.Steiner J. M., Kocher T., Nagy C., Löffelhardt W. Chloroplast SecE: evidence for spontaneous insertion into the thylakoid membrane. Biochem. Biophys. Res. Commun. 2002;293:747–752. doi: 10.1016/S0006-291X(02)00285-1. [DOI] [PubMed] [Google Scholar]
- 44.Chen J. B. A., Fromant M., Leveque F., Schmitter J. M., Blanquet S., Plateau P. Pyrophosphatase is essential for growth of Escherichia coli. J. Bacteriol. 1990;172:5686–5689. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baykov A. A., Shestakov A. S. Two pathways of pyrophosphate hydrolysis and synthesis by yeast inorganic pyrophosphatase. Eur. J. Biochem. 1992;206:463–470. doi: 10.1111/j.1432-1033.1992.tb16947.x. [DOI] [PubMed] [Google Scholar]
- 46.Siluva T., Salminen A., Parkenyev A. N., Pohjanjoki P., Goldman A., Cooperman B. S., Baykov A. A., Lahti R. Evolutionary aspects of inorganic pyrophosphatase. FEBS Letts. 1999;454:75–80. doi: 10.1016/s0014-5793(99)00779-6. [DOI] [PubMed] [Google Scholar]
- 47.Waller R. F., McConville M. J., McFadden G. I. More plastids in human parasites? Trends Parasitol. 2004;20:54–57. doi: 10.1016/j.pt.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 48.Kankare J., Neal G. S., Salminen T., Glumoff T., Cooperman B. S., Lahti R., Goldman A. The structure of E. coli soluble inorganic pyrophosphatase at 2.7 Å resolution. Protein Eng. 1994;7:823–830. doi: 10.1093/protein/7.7.823. [DOI] [PubMed] [Google Scholar]
- 49.Kankare J., Salminen T., Lahti R., Cooperman B. S., Baykov A. A., Goldman A. Crystallographic identification of metal-binding sites in Escherichia coli inorganic pyrophosphatase. Biochemistry. 1996;35:4670–4677. doi: 10.1021/bi952637e. [DOI] [PubMed] [Google Scholar]