Abstract
Glycogen, a branched polymer of glucose, forms an energy re-serve in numerous organisms. In mammals, the two largest glyco-gen stores are in skeletal muscle and liver, which express tissue-specific glycogen synthase isoforms. MGSKO mice, in which mGys1 (mouse glycogen synthase) is disrupted, are devoid of muscle glycogen [Pederson, Chen, Schroeder, Shou, DePaoli-Roach and Roach (2004) Mol. Cell. Biol. 24, 7179–7187]. The GSL30 mouse line hyper-accumulates glycogen in muscle [Manchester, Skurat, Roach, Hauschka and Lawrence (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10707–10711]. We performed a microarray analysis of mRNA from the anterior tibialis, medial gastrocnemius and liver of MGSKO mice, and from the gastroc-nemius of GSL30 mice. In MGSKO mice, transcripts of 79 genes varied in their expression in the same direction in both the anterior tibialis and gastrocnemius. These included several genes encoding proteins proximally involved in glycogen metabolism. The Ppp1r1a [protein phosphatase 1 regulatory (inhibitor) sub-unit 1A] gene underwent the greatest amount of downregulation. In muscle, the downregulation of Pfkfb1 and Pfkfb3, encoding isoforms of 6-phosphofructo-2-kinase/fructose-2,6-bisphospha-tase, is consistent with decreased glycolysis. Pathways for branched-chain amino acid, and ketone body utilization appear to be downregulated, as is the capacity to form the gluconeogenic precursors alanine, lactate and glutamine. Expression changes among several members of the Wnt signalling pathway were identified, suggesting an as yet unexplained role in glycogen meta-bolism. In liver, the upregulation of Pfkfb1 and Pfkfb3 expression is consistent with increased glycolysis, perhaps as an adaptation to altered muscle metabolism. By comparing changes in muscle expression between MGSKO and GSL30 mice, we found a subset of 44 genes, the expression of which varied as a function of muscle glycogen content. These genes are candidates for regulation by glycogen levels. Particularly interesting is the observation that 11 of these genes encode cardiac or slow-twitch isoforms of muscle contractile proteins, and are upregulated in muscle that has a greater oxidative capacity in MGSKO mice.
Keywords: branched-chain amino acid oxidation, cAMP, glyco-gen, glycogen synthase, phosphatase inhibitor 1 (I-1), Wnt
Abbreviations: ADRβ2, β2-adrenergic receptor; BCKDC, branched-chain α-ketoacid dehydrogenase complex; BDK, BCKD kinase; EST, expressed sequence tag; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GBE1, glycogen branching enzyme; GLUT, glucose transporter; GYS, glycogen synthase; HRP, horse radish peroxidase; I-1, phosphatase inhibitor-1; LRP5, low density lipoprotein receptor-related protein 5; MGSKO, muscle glycogen synthase knockout; mTOR, mammalian target of rapamycin; PFKFB, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase; Ppp1r1a, protein phosphatase 1 regulatory (inhibitor) subunit 1A; RT, reverse transcriptase; UGP2, UDP-glucose pyrophosphorylase 2
INTRODUCTION
Glycogen is a branched polymer of glucose and the principal reserve of carbohydrate in mammals, with the largest deposits being in liver and muscle [1]. Conversion of glucose to glycogen requires phosphorylation to glucose-6-phosphate, isomerization to glucose-1-phosphate and synthesis of UDP-glucose by UGP2 (UDP-glucose pyrophosphorylase2) (Figure 1). UDP-glucose is the immediate glucosyl donor for glycogen biosynthesis, at the stage of initiation by glycogenin, as well as bulk synthesis by glycogen synthase that forms the basic α-1,4-glycosidic linkages of glycogen. The branching enzyme introduces the α-1,6-glycosidic linkages that create the branched structure. Glycogen synthase is subject to complex regulation and is inactivated by phosphorylation at multiple sites. This inactivation can be overcome by the allosteric activator, glucose-6-phosphate.
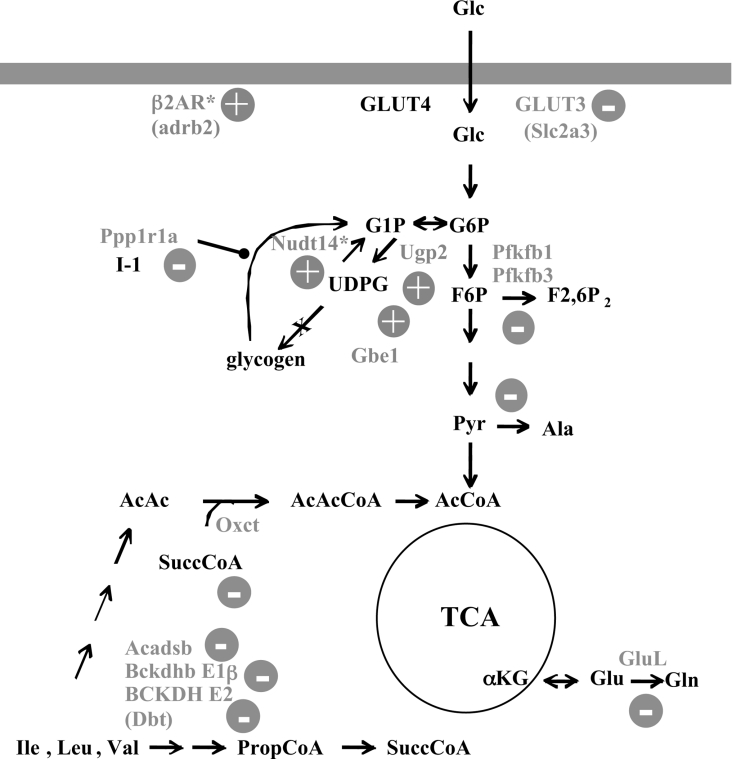
Figure 1. Partial diagram of intermediary metabolism.
Indicated are steps where there was a significant alteration in the corresponding transcript level in MGSKO muscle. (+) or (−) indicates the direction of the change in transcript level. Proteins are represented by their gene names. αKG, α-ketoglutarate; AcCoA, acetyl-CoA; AcAcCoA, acetoacetyl-CoA; AcAc, acetoacetate; F6P, fructose-6-phosphate; F2,6P2, fructose-2,6-bisphosphate; Glc, glucose; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; PropCoA, propionyl-CoA; Pyr, pyruvate; SuccCoA, succinyl-CoA; TCA, tricarboxylic acid; UDPG, uridine diphisphoglucose. The gene names are linked to Unigene citations in Supplementary Table 5 (http:www.BiochemJ.org/bj/395/bj3950137add.htm). *, Indicates a change only in gastrocnemius; all other changes are in both gastrocnemius and anterior tibialis.
Muscle and liver express different isoforms of several of the enzymes in glycogen metabolism, including glycogen synthase [2,3], glycogenin in humans [4], and the degradative enzyme glycogen phosphorylase [5]. In addition, the two organs utilize different GLUTs (glucose transporters) to mediate glucose entry into the cell, as well as different forms of hexokinase/glucokinase for the phosphorylation of glucose. There are two genes, GYS1 and GYS2, encoding glycogen synthase in both mouse and human genomes. To date, GYS2 is only known to be expressed in liver whereas GYS1 is expressed in skeletal muscle and a number of other tissues.
Glycogen is synthesized under conditions of nutritional sufficiency, acting as an energy reserve to be utilized over time. In liver, it functions primarily to supply glucose to the bloodstream. Muscle glycogen, by contrast, is used within muscle cells and depending on fibre type helps to fuel the contractile process. In humans, another role for muscle glycogen may be as a readily accessible repository for ingested glucose, to avoid excessive blood glucose accumulation. A large proportion, 40–90%, of post-prandial glucose is converted into muscle glycogen and one might postulate that defects in glycogen synthesis would be linked to impaired glucose disposal and diabetes [6–8]. There are indeed reports of an association between mutations in GYS1 and insulin resistance and diabetes in some human populations [9,10].
In an effort to determine how muscle glycogen affects whole-body glucose metabolism, we created 2 transgenic mouse models in which muscle glycogen levels were manipulated genetically. GSL30 mice overexpress hyperactive-phosphorylation site mu-tants of glycogen synthase, driven by the muscle-specific creatine kinase promoter, and overaccumulate glycogen in muscle [11]. MGSKO (muscle glycogen synthase knockout) mice have disrupted Gys1 and hence completely lack glycogen in muscle and several other tissues [12]. The MGSKO mice are 5–10% lighter, have less fat, a greater capacity to oxidize fat in their muscle tissue and an increased proportion of oxidative fibres in some muscles [13]. Fasted and fed blood-glucose levels were the same in wild-type and MGSKO animals. However, the MGSKO mice were significantly better at disposing of glucose in a glucose tolerance test [13] whereas GSL30 animals performed less well than wild-type littermates [13]. The relatively small muscle glycogen pool in mice possibly explains this somewhat paradoxical result, given the role of muscle glycogen in glucose disposal in humans. Neither of the genetically modified animals was significantly impaired by exhaustive exercise on a treadmill [14,15].
In the present study, we report that the expression of multiple genes is affected when glycogen synthase levels, and therefore the amount of glycogen accumulation, is genetically modified in murine skeletal muscle. Many of the genes encode proteins involved in metabolism and in the muscle's contractile apparatus. In addition, the comparison of transcript levels in the medial gastrocnemius muscle of wild-type, MGSKO and GSL30 mice defined a small number of transcripts that varied in expression either in direct or inverse proportion to glycogen levels, indicating that expression of the corresponding genes is linked to glycogen accumulation.
EXPERIMENTAL
Sample preparation
Generation of the GSL30 and MGSKO transgenic mouse lines has been described previously [11,12]. For the various biochem-ical determinations, unless otherwise indicated, fasted animals were killed in the early morning. Male mice (3-month-old) of comparable weight were used in these studies (wild-type/MGSKO, 29.9±1.3/28.5±1.5 g; WT/GSL30, 26.7±1.7/26.0±0.6 g; weights are the means±S.E.M. for 5 mice). All mice were maintained in temperature- and humidity-controlled conditions with a 12 h light/dark cycle and were allowed food and water ad libitum. Animals were maintained in the Association for Assessment of Accreditation of Laboratory Animal Care-approved animal facility at Indiana University. All procedures were approved by the Indiana University Animal Care and Use Committee. Total RNA was isolated from the anterior tibialis, medial gastrocnemius, or liver of 4 or 5 wild-type mice and their MGSKO or GSL30 littermates respectively using TRIzol™ Reagent (Molecular Research Center, Cincinnati, OH, U.S.A.). RNA was further purified using the RNeasy® kit (Qiagen Inc., Valencia, CA, U.S.A.). RNA integrity was confirmed by both electrophoresis on 1% agarose gels and from UV spectra. Samples were then labelled and hybridized to the Affymetrix MOE430A mouse expression GeneChips® using the standard Affymetrix protocol [16] at the Indiana University School of Medicine, Center for Medical Genomics. Briefly, 5–10 μg of total RNA was used to synthesize double-stranded cDNA using the SuperScript II kit (Invitrogen, Carlsbad, CA, U.S.A.) and the Affymetrix T7-(dT)24 primer, which contains a T7 promoter attached to a poly-dT sequence: 5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′. Labelled cRNA was produced by in vitro transcription using the T7 RNA polymerase with the Enzo BioArray, High Yield RNA Transcript Labeling Kit with biotinylated nucleoside 5′-triphosphates (ENZO Diagnostics Inc., Farmingdale, NY, U.S.A.). Labelled cRNA (15 μg) was used to make a 300 μl hybridization cocktail, of which 200 μl was injected into the GeneChip®, 1 sample per chip. Samples were hybridized for 17 h at 45 °C with constant rotation. The GeneChips® then were washed and stained with phycoerythrin-labelled streptavidin.
Data analysis
The GeneChips® were scanned using a Model 3000 scanner controlled by the MAS5 (Affymetrix Microarray Suite version 5) software [17]. Each sample was scaled to a target intensity of 1000 using the ‘all probe sets’ scaling option [17]. MAS5 ‘absolute’ expression analysis was performed for each sample [17]. These data were exported from Affymetrix MAS 5.0 and loaded into the MicroArray Data Portal [18] for further analysis. For each comparison between 2 groups of animals, only probe sets that were present in at least 50% of the samples in at least 1 of the 2 groups were analysed [19]. For analyses, we compared the log2 of the level of gene expression (signal) using a Welch's unpaired t test [20]. Genes of interest were selected, for which P≤0.05, and there was an increase or decrease in expression by a magnitude of 1.5-fold or more.
Quantitative RT (reverse transcriptase)-PCR
Double-stranded cDNA from each GeneChip experiment was used in quantitative RT-PCR reactions carried out using an ABI Prism 7700 Sequence Detection System [Applied Biosystems (ABI), Foster City, CA, U.S.A.] according to the manufacturer's instructions. The following primers were designed using Primer Express™ 1.0 software: rGys1 (5′-tgtcacagatcaccgccatc-3′, 5′-ccaacggcctgaatgtga-3′); mGys1 (5′-ccggctttggctgctttat-3′, 5′-ccgatccagaatgtaaatgcc-3′); Ppp1r1a (5′-acggaagaagatgacaaggacc-3′, 5′-ttgccctaggtgatgttcaacc-3′); Ugp2 (5′-acctgggatacctgccgtg-3′, 5′-cctgctcaccccttccttc-3′); Tnfrsf19 (5′-tggaagagcatggttgaccc-3′, 5′-tgccgtattacaaagccttgg-3′); Dyrk1b (5′-gtggacttcggcagttcctg-3′, 5′-agcggtagaagcggctctg-3′); Adrb2 (5′-tctgtgccttcgcaggtctt-3′, 5′-gtccgttctgccgttgcta-3′); Tnnt1 (5′-tcattgcactaaaagaccgca-3′, 5′-tctgaagcgctgttgctcag-3′); Bdnf (5′-gcaaacatgtctatgagggttcg-3′, 5′-ctcgctaatactgtcacacacgc-3′); GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (5′-gtcgtggatctgacgtgcc-3′, 5′-atgcctgcttcaccaccttc-3′); Dkk3 (5′-cctgtgtttcagctcctacctca-3′, 5′-cccctcctaggctgtggtg-3′); Gbe1 (5′-acaccagggaagttcaaaattgtac-3′, 5′-gtgttgtggtccagtctctgatg-3′); Glut3 (5′-ggatgctttcggtgatagtcct-3′, 5′-tgttctcggcagcaagtgtt-3′); MLP (5′-actggcgagcatcttggc-3′, 5′-agggttgcttgtggtggct-3′); Wnt4 (5′-ctccctccctgtctttggg-3′, 5′-gatggcgtatacaaaggccg-3′); Cpt1a (5′-tgcaaagatcaatcggaccc-3′, 5′-aggacgccactcacgatgtt-3′); Pfkfb1 (5′-acccgtgaacccgaggaa-3′, 5′-agacaatctgatcacttgaaaaggg-3′); Pfkfb3 (5′-agctgcccggacaaaacat-3′, 5′-ctcggctttagtgcttctggg-3′), and Igf1 (5′-gcttccggagctgtgatctg-3′, 5′-agcgggctgcttttgtagg-3′). Gene names are given in full in Tables 1 and 2. Reaction volumes were scaled to a 20 μl final volume and were composed of 10 μl of SYBR Green PCR Master Mix, 2 pmol of primer mix and approx. 10 ng of template cDNA. All reactions were repeated in triplicate. The expression levels of tested genes were normalized to an internal GAPDH control. Comparison of fold changes was determined to be significant if P<0.05 in a paired Student's t test.
Table 1. Confirmation of transcript levels by quantitative real-time PCR.
All fold-changes listed are P<0.05 compared with wild-type muscle. AT, anterior tibialis; Bdnf, brain derived neurotrophic factor; Cpt1a, carnitine palmitoyltransferase Ia; Dkk3, Dickkopf3; Dyrk1b, dual-specificity tyrosine-phosphorylation-regulated kinase; gast, gastrocnemius; Igf1, insulin-like growth factor 1; mGys1/Gys3, mouse glycogen synthase1/3; MLP, muscle LIM-domain protein; N/A, not applicable; rGys1, rabbit glycogen synthase 1; Tnfrsf19, tumour necrosis factor receptor superfamily, member 19; Tnnt1, troponin T1. Gys1 and Gys3 in the database correspond to the same gene.
| Transcript | UniGene Cluster | Genotype | Tissue | Microarray (fold-change) | qRT-PCR (fold-change) |
|---|---|---|---|---|---|
| Bdnf | Mm.1442 | MGSKO | AT | 2.6 | 2.44 |
| Dkk3 | Mm.55143 | MGSKO | AT | −4.3 | −5.06 |
| Gbe1 | Mm.29201 | MGSKO | AT | 2.26 | 3.46 |
| Glut3 | Mm.269857 | MGSKO | AT | −3.01 | −1.7 |
| MLP | Mm.17235 | MGSKO | AT | 2.3 | 3.02 |
| Ppp1r1a | Mm.143788 | MGSKO | AT | −9.98 | −14.28 |
| Ugp2 | Mm.28877 | MGSKO | AT | 1.87 | 1.75 |
| Wnt4 | Mm.20355 | MGSKO | AT | −5.6 | −2.27 |
| Adrb2 | Mm.5598 | MGSKO | Gast | 1.52 | 9.43 |
| Cpt1a | Mm.18522 | MGSKO | Gast | −2.15 | −2.47 |
| Igf1 | Mm.268521 | MGSKO | Gast | −1.62 | −5.45 |
| Ppp1r1a | Mm.143788 | MGSKO | Gast | −3.54 | −12.3 |
| Tnfrsf19 | Mm.281356 | MGSKO | Gast | −2.11 | −3.1 |
| Tnnt1 | Mm.258670 | MGSKO | Gast | 2.13 | 1.46 |
| Ugp2 | Mm.28877 | MGSKO | Gast | 1.6 | 7.46 |
| Pfkfb1 | Mm.249131 | MGSKO | Gast | −1.25 | −2.13 |
| Pfkfb3 | Mm.19669 | MGSKO | Gast | −2.64 | −5.76 |
| Glut3 | Mm.269857 | MGSKO | Gast | −2.04 | −7.93 |
| Pfkfb1 | Mm.249131 | MGSKO | Liver | 1.47 | 9.44 |
| Pfkfb3 | Mm.19669 | MGSKO | Liver | 2.04 | 4.72 |
| Dyrk1b | Mm.57249 | GSL30 | Gast | 1.75 | 2.28 |
| Adrb2 | Mm.5598 | GSL30 | Gast | −1.78 | −1.62 |
| mGys1/Gys3 | Mm.275654 | GSL30 | Gast | 2.9 | 6.02 |
| rGys1 | N/A | GSL30 | Gast | N/A | 14.1 |
| Ppp1r1a | Mm.143788 | GSL30 | Gast | 1.7 | 3.88 |
| Tnfrsf19 | Mm.281356 | GSL30 | Gast | 2.01 | 1.35 |
| Tnnt1 | Mm.258670 | GSL30 | Gast | −2.4 | −1.6 |
Table 2. Genes whose expression varies with glycogen levels in the gastrocnemius.
| Gastrocnemius microarray (fold-change) | ||||
|---|---|---|---|---|
| Gene symbol | Gene description | Unigene | MGSKO | GSL30 |
| Metabolism | ||||
| Gys1 | Glycogen synthase | Mm.275654 | −44.45* | 2.91 |
| Hsd17β7 | Hydroxysteroid (17-β) dehydrogenase 7 | Mm.12882 | −2.35 | 1.73 |
| Ugp2 | UDP-glucose pyrophosphorylase 2 | Mm.130747 | 1.59* | −1.59 |
| Car14 | Carbonic anhydrase 14 | Mm.224836 | 1.88 | −1.85 |
| Muscle/contractile proteins | ||||
| Mylk | Myosin, light polypeptide kinase | Mm.247544 | 1.52 | −1.79* |
| Utrn | Utrophin | Mm.331784 | 1.56 | −1.67 |
| Atp1β1 | ATPase, Na+/K+ transporting, β1 polypeptide | Mm.4550 | 1.57 | −1.61† |
| Atp2α2 | ATPase, Ca+2 transporting, cardiac muscle, slow-twitch 2 | Mm.227583 | 1.66 | −1.61‡ |
| Myl2 | Myosin, light polypeptide 2, regulatory, cardiac, slow-twitch | Mm.1529 | 1.75 | −1.91 |
| Tpm3 | Tropomyosin 3 | Mm.240839 | 1.84 | −2.39† |
| Myh11 | Myosin, heavy polypeptide 11, smooth muscle | Mm.250705 | 1.98 | −2.19 |
| Tncc | Troponin C, cardiac/slow-twitch skeletal | Mm.712 | 2.03 | −1.96 |
| Tnnt1 | Troponin T1, skeletal, slow-twitch | Mm.711 | 2.13 | −2.44 |
| Myl3 | Myosin, light polypeptide 3 | Mm.7353 | 2.10† | −1.73† |
| Myh6 | Myosin, heavy polypeptide 6, cardiac muscle, α | Mm.290003 | 2.30* | −2.75a |
| Receptors and signalling | ||||
| Ppp1r1a | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | Mm.143788 | −3.54 | 1.67 |
| Tnfrsf19 | Tumour necrosis factor receptor superfamily, member 19 | Mm.281356 | −2.00* | 1.95* |
| Map3k7 | Mitogen activated protein kinase kinase kinase 7 | Mm.258589 | −1.55 | 1.72* |
| Adrβ2 | Adrenergic receptor, β2 | Mm.5598 | 1.52 | −1.78 |
| C1qr1 | Complement component 1, q subcomponent, receptor 1 | Mm.681 | 1.54 | −1.58 |
| Snrk | SNF related kinase | Mm.257989 | 1.56 | −1.54 |
| Notch4 | Notch gene homologue 4 (Drosophila) | Mm.173813 | 1.56 | −1.75 |
| Trpm7 | Transient receptor potential cation channel | Mm.244705 | 1.61 | −2.01 |
| Bmp6 | Bone morphogenetic protein 6 | Mm.254978 | 1.62 | −1.79 |
| Cxcl14 | Chemokine (C-X-C motif) ligand 14 | Mm.30211 | 1.63 | −1.57 |
| Tie1 | Tyrosine kinase receptor 1 | Mm.4345 | 1.64 | −1.99 |
| Adam15 | Disintegrin and metalloproteinase domain 15, metargidin | Mm.274049 | 1.68 | −1.57 |
| Kitl | Kit ligand | Mm.45124 | 1.77 | −1.58 |
| VEC/Cdh5 | VE-cadherin/cadherin 5, calcium ion-binding | Mm.21767 | 2.31 | −1.71 |
| Nervous system | ||||
| Mal | Myelin and lymphocyte protein, T-cell differentiation | Mm.39040 | 1.77 | −1.79 |
| Mpz | Myelin protein zero | Mm.9986 | 3.51 | −1.71 |
| Nucleic acid associated proteins | ||||
| Dnase1 | Deoxyribonuclease I | Mm.239992 | −5.61 | 7.64 |
| 1700037B15Rik | RIKEN cDNA 1700037B15 gene | Mm.250841 | −2.52* | 1.68 |
| Gadd45α | Growth arrest and DNA-damage-inducible 45 α | Mm.1236 | −1.74 | 1.75 |
| 5830411E10Rik | RIKEN cDNA 5830411E10 gene | Mm.196290 | −1.61 | 1.84 |
| Eif4el3 | Eukaryotic translation initiation factor 4E like 3 | Mm.227183 | 1.57 | −2.08 |
| AI255170 | Expressed sequence AI255170 | Mm.214973 | 2.01 | −2.41 |
| Other | ||||
| Tekt1 | Tektin 1 | Mm.42257 | −2.02 | 2.73 |
| Sepr | Selenoprotein R | Mm.28212 | −1.53 | 1.58 |
| Timp4 | Tissue inhibitor of metalloproteinases 4 | Mm.255607 | 1.55 | −1.58 |
| Esam1 | Endothelial cell-specific adhesion molecule | Mm.41751 | 1.56 | −1.56 |
| 0610013D04Rik | RIKEN cDNA 0610013D04 gene | Mm.196330 | 1.76 | −1.64 |
| 1500005K14Rik | RIKEN cDNA 1500005K14 gene | Mm.34131 | 1.81 | −1.52 |
* Average of 2 probe sets detected in this experiment.
† Average of 3 probe sets detected in this experiment.
‡ Average of 4 probe sets detected in this experiment.
Western blot analysis
Skeletal muscle was excised quickly, frozen in liquid nitrogen, and stored at −80 °C. For Western blot analyses, frozen tissue samples were homogenized (w/v) in 10 vol. of a solution containing 50 mM Tris/HCl (pH 7.5), 0.5 mM EDTA, 2 mM EGTA, 1% Triton X-100, 0.1 mM N-p-tosyl-L-lysine chloromethyl ketone, 2 mM benzamidine, 0.5 mM PMSF and leupeptin 10 μg/ml using a Tissue Tearer model 285-370 (Biospec Products Inc., Bartlesville, OK, U.S.A.) at maximal setting for 20 s. Protein concentrations were determined by the method of Bradford [20a] using the protein assay dye reagent (Bio-Rad). Protein (50 μg) was separated by SDS/PAGE. Proteins were transferred on to nitrocellulose [I-1 (phosphatase inhibitor-1)] or PVDF membranes [BCKDC (branched-chain α-ketoacid dehydrogenase complex) and BDK (BCKD kinase)] (Millipore, Bedford, MA, U.S.A.) by the semi-dry method and then incubated with the appropriate antibodies. Antibodies to I-1 (a gift from A. Nairn, Department of Psychiatry, Yale University, CT, U.S.A.) were used at a dilution of 1:500. Antisera for each component of BCKDC (subunits E1 and E2) were generated against purified E1 and E2 components from isolated rat liver BCKDC and used at a dilution of 1:500 [21]. A monoclonal antibody against BDK was generously given by Y. Shimomura (Department of Materials Science and Engineering, Nagoya Institute of Technology, Japan) [22]. Membranes were blocked with 50 g/l skimmed milk in TBST [20 mM Tris/HCl (pH 7.5), 0.5 M NaCl and 0.1% (v/v) Tween-20]. The blots were then incubated with the primary antibody in blocking buffer, washed 3 times with TBST, and incubated with the secondary antibodies. The secondary antibody against I-1 was a rabbit anti-mouse HRP (horseradish peroxidase) conjugate (Sigma) used at a dilution of 1:5000. For BCKDC, HRP-conjugated secondary anti-(rabbit IgG) (Upstate, Lake Placid, NY, U.S.A.) was used at a dilution of 1:10000. For BDK, an anti-(mouse IgG) HRP-conjugate (Upstate) was used. Binding of the antibodies was detected by enhanced chemiluminescence (Amersham Pharmacia Biotech). Membranes were exposed to Kodak X-Omat LS Film and developed according to the manufacturer's recommendations. Levels of protein expression were quantified by densitometric scanning of the films.
RESULTS AND DISCUSSION
Overview
In the present study, microarray-chip analysis was used to investigate the effects of genetically altering the glycogen concentration on transcript profiles in skeletal muscles from the MGSKO and GSL30 mice that either lack or overaccumulate glycogen respectively. To assess whether loss of muscle glycogen caused any indirect metabolic repercussions in liver, we also analysed transcripts in liver from MGSKO mice. The complete microarray data sets have been deposited at the GEO (gene expression omnibus) of NCBI (National Center for Biotechnology Information) (http://www.ncbi.nlm.nih.gov/geo/), under accession number GSE2198. Some 22690 genes and ESTs (expressed sequence tags; hereafter referred to as genes) are represented on the Affymetrix MOE430A mouse expression GeneChip® arrays [16]. Our standard-significance filter included a detection call of 0.5, a P value of 0.05, and an increase or decrease in expression by a magnitude of 1.5-fold (Figure 2; for a complete list of these genes see Supplementary Tables 1–4 at http://www.BiochemJ.org/bj/395/bj3950137add.htm). We analysed changes in mRNA transcript expression levels in the medial gastrocnemius of GSL30 mouse lines and in the medial gastrocnemius and anterior tibialis of MGSKO mice. In the gastrocnemius of the GSL30 mouse line (see Supplementary Table 1 at http://www.BiochemJ.org/bj/395/bj3950137add.htm), the expression of 743 genes was altered (325 upregulated and 418 downregulated), whereas 467 genes were regulated in the same muscle of the MGSKO mouse line (see Supplementary Table 2 at http://www.BiochemJ.org/bj/395/bj3950137add.htm; 233 upregulated and 234 downregulated). We also examined the anterior tibialis of MGSKO mice (see Supplementary Table 3 at http://www.BiochemJ.org/bj/395/bj3950137add.htm) in which the expression of 435 genes was modified (193 upregulated and 242 downregulated). Analysis of liver from the MGSKO mouse line (see Supplementary Table 4 at http://www.BiochemJ.org/bj/395/bj3950137add.htm) revealed changes in the expression of 156 genes (71 upregulated and 85 downregulated). A number of genes were selected for confirmation by quantitative RT-PCR and, as seen in Table 1, there was perfect qualitative agreement with the results of microarray analysis and in most cases also a good quantitative correlation, giving faith in the overall performance of the gene chip analysis.
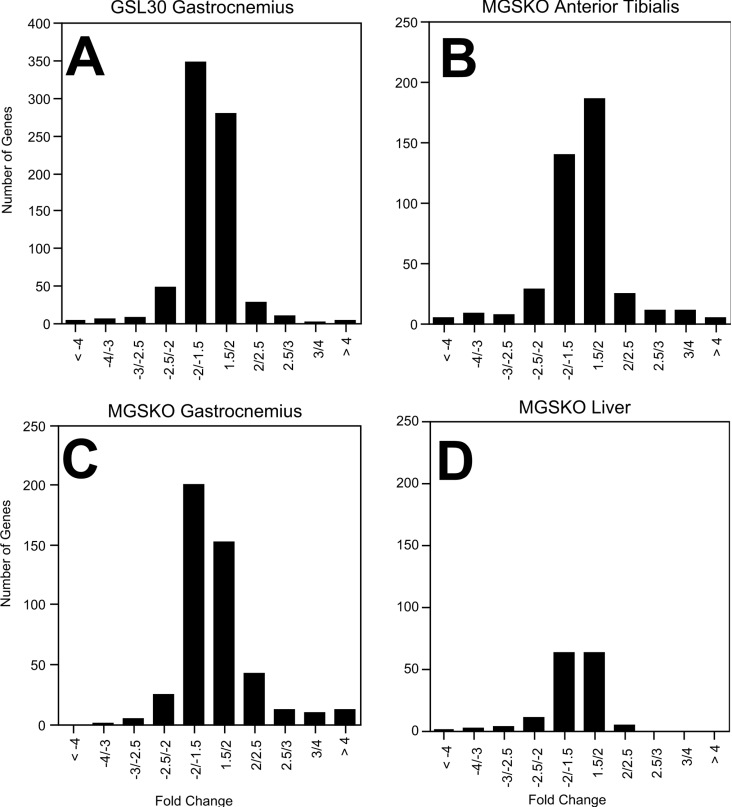
Figure 2. Distribution of the magnitude of changes in transcripts in different tissues.
The number of genes significantly up- and down-regulated in both the GSL30 (A) and MGSKO (B, C and D) mouse lines are plotted to show the distribution by the amount of fold-change (experimental/control).
Functional classification of affected genes
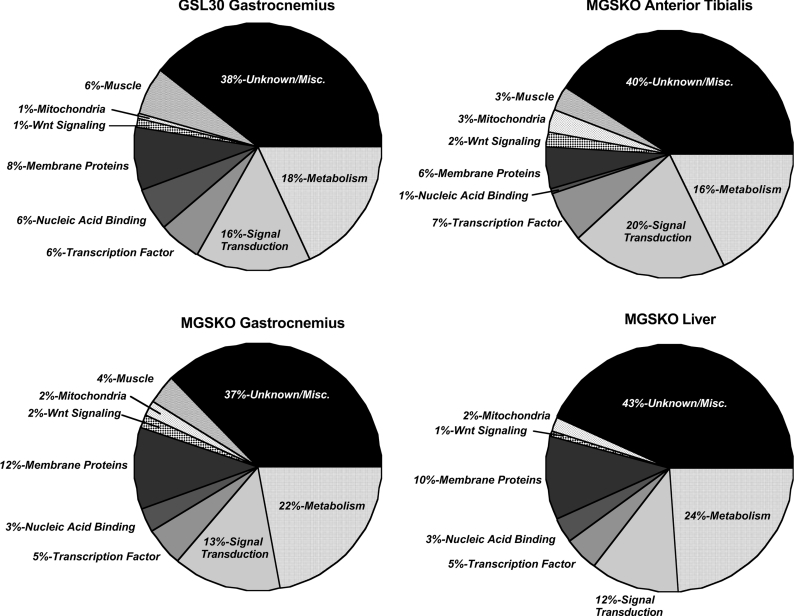
Implicated genes were annotated and assigned to functional groups. The largest group of genes with a significant change in expression level, 37–43%, consisted of ESTs and genes for which the protein products have unknown functions (Figure 3). The second largest functional group consisted of genes involved in metabolism in the GSL30 gastrocnemius (18%), and MGSKO anterior tibialis (18%), gastrocnemius (22%), and liver (24%). In the medial gastrocnemius of both mouse lines and in the anterior tibialis of the MGSKO mouse, genes involved in Wnt signalling and in muscle function were the other major functional categories. In MGSKO muscle, 79 genes were identified as being affected in both anterior tibialis and gastrocnemius, with all but 4 having an expression level altered in the same direction (see Supplementary Table 5 at http://www.BiochemJ.org/bj/395/bj3950137add.htm). When transcript profiles from the gastrocnemius of MGSKO and GSL30 mice were compared, the expression of a small subset of 44 genes varied either in direct or inverse proportion to muscle glycogen content (Table 2).
Figure 3. Functional groups of genes with altered expression.
Diagrams indicate the proportion of genes in different categories that passed the significance filters consisting of a detection call of 0.5, P≤0.05, and an increase or decrease in expression by a magnitude of 1.5-fold.
Muscle metabolism and its regulation in MGSKO mice
The expression of several genes encoding proteins intimately linked to glycogen metabolism was modified in MGSKO mice (Figure 1). The greatest downregulation in expression in both the gastrocnemius and anterior tibialis was of the targeted Gys1 gene, −45.4 and −144-fold respectively, as expected given the locus of disruption. This result provides an excellent validation of the specificity of the analysis. The genes for both UGP2 and GBE1 (glycogen branching enzyme) were upregulated, suggesting that they might normally be negatively controlled by glycogen levels. For Ugp2, this idea is strengthened by the observation that its transcript was downregulated in muscle in GSL30 mice. The UDP-glucose pyrophosphatase [Nudt14, where Nudt is (nucleoside diphosphate attached moiety ‘X’)-type motif] transcript, a recently characterized enzyme that converts UDP-glucose to uridine monophosphate and glucose-1-phosphate [23], was also increased in expression in the gastrocnemius of MGSKO mice. The physiological function of this enzyme is not well understood but the response of the corresponding gene to a loss of glycogen makes the case for its role in glycogen metabolism. The overall impact on UDP-glucose levels in muscle from MGSKO mice appears to be an elevation in expression levels. Initial measurements of UDP-glucose in whole hind-limb muscle from MGSKO mice indicate an approx. 5-fold increase over wild-type animals (results not shown). In previous studies, a decrease in UDP-glucose levels was observed in GSL30 mice [11,24], suggesting an inverse relationship between UDP-glucose concentration and glycogen. Therefore we cannot exclude the possibility that the impact of genetically modifying glycogen stores on gene expression is secondary to changes in the concentration of key metabolites, such as UDP-glucose.
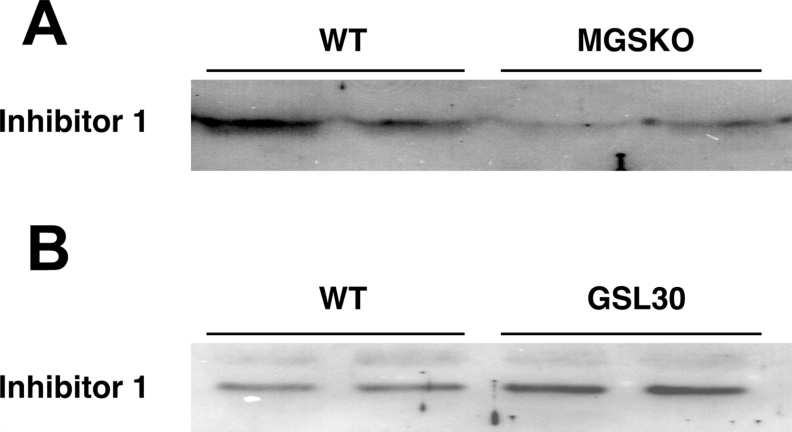
After Gys1, Ppp1r1a [protein phosphatase 1 regulatory (inhibitor) subunit 1A] showed the greatest downregulation in muscle from MGSKO mice (Table 2; [25]). Corresponding changes in I-1 protein levels were also confirmed by Western blotting (Figure 4). This protein acts as a potent phosphatase inhibitor only after phosphorylation by cAMP-dependent protein kinase [25,26] and hence has been proposed to be involved in mediating β-adrenergic stimulation of skeletal muscle [27]. Glycogen-associated forms of type 1 phosphatase are thought to dephosphorylate glycogen synthase and phosphorylase [28] and so a decrease in I-1 levels might have been expected to correlate with a greater activation of phosphorylase phosphatase and hence less activation of phosphorylase. In fact, the opposite is true and phosphorylase in muscle from MGSKO mice is in a more activated state [12]. However, this interpretation is complicated by the fact that I-1 is an effective inhibitor only after phosphorylation. A decrease in protein levels does mean that the maximal achievable inhibition would be decreased. The lack of a correlation between I-1 levels and phosphorylase activation has already been reported by Scrimgeour et al. [29] in their studies of Ppp1r1a knockout mice. The absence of I-1 had no effect on basal muscle glycogen synthase levels or phosphorylase activities, or the response of these enzymes to insulin or adrenaline (epinephrine). The role of I-1 in skeletal muscle glycogen metabolism is thus obscure but the results of the present study reaffirm a genetic link with glycogen, even more so since I-1 is elevated in the GSL30 glycogen overaccumulating mice, at both the transcript (Table 2) and protein levels (Figure 4).
Figure 4. I-1 protein levels are changed in the skeletal muscle of glycogen synthase altered animals.
Homogenates were prepared from bulk hind-limb skeletal muscle of wild-type (WT), MGSKO (A) and GSL30 (B) mice. Extracts were separated by SDS/PAGE and proteins were transferred to membranes. Membranes were probed with an antibody to I-1 and proteins detected by chemiluminescence. The migration of the I-1 protein is shown.
A possible explanation for the activation of phosphorylase in MGSKO mice is the observed upregulation of the Adrb2 (β2-adrenergic receptor) gene that could correlate with enhanced signalling through cAMP. This, in turn, would be consistent with the activation of phosphorylase. The Adrb2 transcript level is inversely proportional to muscle glycogen content and is decreased in muscle from GSL30 mice. We observed that the Pfkfb3 (6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase3) isoform of the bifunctional enzyme was downregulated in our screen. We confirmed this by real-time PCR (Table 1) and also examined the transcript for the major Pfkfb1 isoform found in liver and skeletal muscle. Pfkfb1 was downregulated but by a magnitude that fell below our filter in the array experiment; however, by quantitative PCR it was shown to be downregulated 2.13-fold (Table 1). Pfkfb1 is phosphorylated by cAMP [30]. Increased cAMP signalling would reinforce decreased kinase activity of PFKFB, since phosphorylation of the enzyme by cAMP-dependent protein kinase enhances the phosphatase activity [31]. Decreased fructose-2,6-bisphosphate levels would suppress glycolysis. These results are consistent with a reduced flux from glucose to pyruvate. Pfkfb3 encodes an isoform of the bifunctional enzyme that is ubiquitous, has a relatively high kinase to phosphatase activity ratio [32] and is induced by hypoxia [33]. Downregulation of this gene would be likely to decrease the level of fructose-2,6-bisphosphate, a potent activator of glycolysis [34]. Consistent with this idea, Minchenko et al. [35] suggested that the induction of Pfkfb3 by hypoxia would be correlated with elevated glycolysis. The same case has been made in a recent report demonstrating that increased Pfkfb3 expression in cultured cells caused increased glycolysis [36]. Therefore it is reasonable to expect the opposite to occur after downregulation of this gene. Thus the expression changes for both Pfkfb1 and Pfkfb3 would mitigate changes towards decreased glycolysis. The MGSKO mice have muscle with a greater oxidative capacity [13] and so pyruvate is possibly diverted more towards the tricarboxylic acid cycle, and the greater energy return from the oxidation of glucose may explain why the glycolytic flux is decreased.
Another downregulated gene potentially involved in glucose utilization is Slc2a3, which encodes GLUT3. This transporter is generally associated with glucose transport into neurons [37] and its role in skeletal muscle has not been clearly established. However, knock-down of GLUT3 in L6 cells has been reported to substantially decrease basal levels of glucose uptake [38]. It is intriguing that both the Slc2a3 and Pfkfb3 genes are induced by hypoxia under the control of HIF-1 (hypoxia-inducible transcription factor-1) [35,39,40].
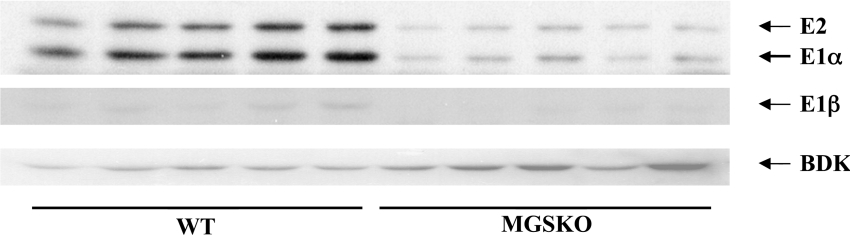
Branched-chain amino acid catabolism appears to be downregulated in MGSKO animals, with the expression of Bckdhb1 (BCKD, beta polypeptide) and Dbt (dihydrolipoamide branched chain transacylase), encoding subunits E1β and E2 of the BCKDC, being decreased, as well as the expression of Acadsb (branched/short-chain acyl-CoA dehydrogenase). Parallel changes in protein levels of BCKD subunits E1α, E1β and E2 were confirmed by Western blotting (Figure 5). The protein level of BDK (BCKD kinase) was also increased in muscle from MGSKO mice. Thus the absence of glycogen has resulted in a co-ordinated response to shut down this regulatory step in branched chain amino acid catabolism [41], both decreasing enzyme levels and ensuring that any remaining enzyme is inactivated by phosphorylation. Also diminished was the transcript for the SuccCoA (succinyl CoA):AcAcCoA (acetoacetyl-CoA) transferase (Oxct1), an enzyme important for ketone body utilization and branched chain amino acid oxidation. There is some evidence for a link between branched chain amino acids and glycogen metabolism. Nishitani et al. [42] reported that leucine activated glycogen synthase in the liver of cirrhotic rats. Leucine is also implicated in activating the mTOR (mammalian target of rapamycin) pathway [43] that is potentially involved in the activation of glycogen synthase [44]. In fact, the mTOR transcript was upregulated 1.72-fold in anterior tibialis but not gastrocnemius muscle (see Supplementary Tables 2 and 3 at http://www.BiochemJ.org/bj/395/bj3950137add.htm). One could speculate that a sufficiency of glycogen signals an increase in branched-chain amino acid oxidation, diverting intermediates away from conversion to glycogen. Gpt2 (alanine aminotrans-ferase 2) and Glul (glutamine synthase) were also downregulated. Combined with decreased lactate formation from diminished glycolysis, the production of 3 key gluconeogenic substrates, lactate, alanine and glutamine, is likely to be impaired. The overall pattern of the changes in metabolic gene expression is consistent with decreased utilization of glucose, ketone bodies and amino acids to fuel muscle; the difference is likely to be made up by a greater oxidative capacity of muscle in MGSKO animals.
Figure 5. BCKDC and BDK protein levels are altered in the skeletal muscle of MGSKO animals.
Homogenates were prepared from bulk hind-limb skeletal muscle of MGSKO and wild-type (WT) mice. Extracts were separated by SDS/PAGE and proteins were transferred to membranes. Membranes were probed with appropriate antibodies and proteins detected by chemiluminescence. The migration of the BDCK components E1 and E2, and BDK are shown.
Muscle proteins
A group of 11 genes encoding muscle proteins was upregulated in MGSKO gastrocnemius and downregulated in the GSL30 mouse line (Table 2). Essentially, these genes encode cardiac or slow-twitch isoforms of the corresponding proteins, consistent with the observation that in MGSKO mice there was an increase in the relative proportion of oxidative fibres [13]. The genes include those encoding myosin light (Myl2 and Myl3) and heavy (Myh6 and Myh11) chains, troponins (Tnnt1 and Tncc), and ATPases (Atp1b1 and Atp2a2). The potential role of glycogen in this adaptation is interesting since glycogen, as a fuel for contraction, is associated with fast-twitch fibres. Does the level of glycogen accumulation have a direct influence on fibre type determination or does fibre type commitment dictate the extent of glycogen accumulation? From these data, it is impossible to distinguish between these two hypotheses. But, for the MGSKO mice, it is tempting to consider that the lack of glycogen has influenced fibre type determination.
Wnt signalling
Wnt signalling events are critical in development and are implicated in a growing number of cellular processes [45–47]. There is a report linking the pathway to glycogen metabolism [48] but this connection is not well established even though insulin and Wnt signalling pathways share GSK-3 (GYS kinase-3) as a key element. Multiple components and targets of the Wnt pathway were modulated in the gastrocnemius of GSL30 and MGSKO mouse lines (see Supplementary Tables 1 and 2 at http://www.BiochemJ.org/bj/395/bj3950137add.htm). These include Dickkopf 3 (4.82-fold decrease), Wnt4 (1.84-fold decrease) and cadherin 5 (2.3-fold increase) in the MGSKO line, and Dickkopf 2 (2.31-fold increase) and Wnt6 (1.59-fold decrease) in the GSL30 line. Also, the expression of Lrp5 (low density lipoprotein receptor-related protein 5), a Wnt coreceptor, is upregulated in the anterior tibialis of MGSKO mice (1.71-fold increase, see Supplementary Table 3 at http://www.BiochemJ.org/bj/395/bj3950137add.htm). It has been reported that LRP5 is essential for normal cholesterol metabolism and glucose-induced insulin secretion [49]. Additionally, the expression of several genes regulated by Wnt signalling events was altered. These include Herpud1 (homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1; a 1.52-fold decrease in the MGSKO line), and Pkd1 (polycystic kidney disease 1 homologue; a 1.85-fold increase in the GSL30 line).
Liver
In the course of attempting to understand why MGSKO mice performed better than wild-type animals in a glucose tolerance test, we considered the hypothesis that there might be indirect effects on liver and hence included the liver in the present study. There is a precedent for such organ–organ communication in that decreased muscle glycogen levels have been reported to increase interleukin-6 production with subsequent metabolic consequences in the liver [50]. As might have been expected, many fewer genes were affected in the liver (156) compared with muscle (435–467) from MGSKO mice (See Supplementary Tables 2–4 at http://www.BiochemJ.org/bj/395/bj3950137add.htm). Of particular interest was the identification of Pfkfb3 expression in the array analysis, which was upregulated in liver and downregulated in skeletal muscle. The expression of Pfkfb1, considered to encode the major isoform of the bifunctional enzyme found in liver [51], was significantly increased 1.47-fold in the array data but fell below the magnitude set in our filter (Table 1). However, by quantitative PCR, a 9.44-fold upregulation was recorded (Table 1). It is possible, then, that the liver of knockout mice increases glycolysis to compensate for altered skeletal muscle metabolism, and contributes to enhanced glucose disposal [13].
In summary, we have shown that alterations in glycogen synthase, and ultimately glycogen levels, result in significant changes in gene expression in skeletal muscle. Glycogen levels affected several genes that encode proteins proximally involved in glycogen metabolism. Pathways for glucose, branched chain amino acid and ketone body utilization appear to be downregulated, consistent with a greater reliance of muscle on oxidative metabolism. Unexpectedly, the expression of several members of the Wnt signalling pathway emerged as being modified, suggesting an as yet unexplained role in glycogen metabolism. By comparing gene expression changes in MGSKO and GSL30 mice, we found that the expression of a subset of 44 genes varied as a function of muscle glycogen content. Particularly interesting is the fact that 11 of these genes encode cardiac or slow-twitch isoforms of muscle contractile proteins and are upregulated in MGSKO muscle which has a greater oxidative capacity. This group also includes a number of genes encoding proteins potentially involved in signalling, including I-1 and the ADRβ2, as discussed above, as well as 2 poorly characterized protein kinases, Snrk (Snf1 related kinase) and the MAPK (mitogen-activated protein kinase) pathway enzyme Map3k7. The former is interesting because its relatives, Snf1p in yeast [52] or AMP-activated kinase in mammals [53,54], are known to bind glycogen [55,56]. These 44 genes are all candidates for regulation by cellular glycogen levels.
Online data
Acknowledgments
We are grateful to A. DePaoli-Roach, H. Edenberg, R. Jerome, J. McClintick, A. Nairn, Y. Shimomura, M. Stephens, H. Tian, W. Wilson and P. Wu for reagents and helpful advice. We are indebted to Jean M. Francois and Marie-Odile Loret of the Laboratoire de Biotechnologie et Bioprocédés, Toulouse, France, for performing UDP-glucose measurements. This study was supported by grants to P.J.R. (DK 27221), R.A.H. (R01DK 047844), and G.E.P. (1 F32 DK66983) from the National Institutes of Health. B.A.P. was supported by a Mentor-based postdoctoral research award from the American Diabetes Association (to P.J.R.).
References
- 1.Roach P. J. Glycogen and its metabolism. Cur. Mol. Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 2.Roach P. J., Skurat A. V., Harris R. A. Regulation of glycogen metabolism. In: Jefferson L. S., Cherrington A. D., editors. Handbook of Physiology. Oxford: Oxford University Press; 2001. pp. 609–647. [Google Scholar]
- 3.Skurat A. V., Roach P. J. Multiple mechanisms for the phosphorylation of C-terminal regulatory sites in rabbit muscle glycogen synthase expressed in COS cells. Biochem. J. 1996;313:45–50. doi: 10.1042/bj3130045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mu J., Skurat A. V., Roach P. J. Glycogenin-2, a novel self-glucosylating protein involved in liver glycogen biosynthesis. J. Biol. Chem. 1997;272:27589–27597. doi: 10.1074/jbc.272.44.27589. [DOI] [PubMed] [Google Scholar]
- 5.Newgard C. B., Littman D. R., van Genderen C., Smith M., Fletterick R. J. Human brain glycogen phosphorylase. Cloning, sequence analysis, chromosomal mapping, tissue expression, and comparison with the human liver and muscle isozymes. J. Biol. Chem. 1988;263:3850–3857. [PubMed] [Google Scholar]
- 6.Meyer C., Dostou J. M., Welle S. L., Gerich J. E. Role of human liver, kidney, and skeletal muscle in postprandial glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 2002;282:E419–E427. doi: 10.1152/ajpendo.00032.2001. [DOI] [PubMed] [Google Scholar]
- 7.Shulman G. I., Rothman D. L., Jue T., Stein P., DeFronzo R. A., Shulman R. G. Quantitation of muscle glycogen synthesis in normal subjects and subjects with non-insulin-dependent diabetes by 13C nuclear magnetic resonance spectroscopy. N. Engl. J. Med. 1990;322:223–228. doi: 10.1056/NEJM199001253220403. [DOI] [PubMed] [Google Scholar]
- 8.Jue T., Rothman D. L., Shulman G. I., Tavitian B. A., DeFronzo R. A., Shulman R. G. Direct observation of glycogen synthesis in human muscle with 13C NMR. Proc. Natl. Acad. Sci. U.S.A. 1989;86:4489–4491. doi: 10.1073/pnas.86.12.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orho-Melander M., Shimomura H., Sanke T., Rasmussen S. K., Nanjo K., Pedersen O., Groop L. C. Expression of naturally occurring variants in the muscle glycogen synthase gene. Diabetes. 1999;48:918–920. doi: 10.2337/diabetes.48.4.918. [DOI] [PubMed] [Google Scholar]
- 10.Orho-Melander M., Almgren P., Kanninen T., Forsblom C., Groop L. C. A paired-sibling analysis of the XbaI polymorphism in the muscle glycogen synthase gene. Diabetologia. 1999;42:1138–1145. doi: 10.1007/s001250051282. [DOI] [PubMed] [Google Scholar]
- 11.Manchester J., Skurat A. V., Roach P., Hauschka S. D., Lawrence J. C., Jr Increased glycogen accumulation in transgenic mice overexpressing glycogen synthase in skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10707–10711. doi: 10.1073/pnas.93.20.10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pederson B. A., Chen H., Schroeder J. M., Shou W., DePaoli-Roach A. A., Roach P. J. Abnormal cardiac development in the absence of heart glycogen. Mol. Cell. Biol. 2004;24:7179–7187. doi: 10.1128/MCB.24.16.7179-7187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pederson B. A., Schroeder J. M., Parker G. E., Smith M. W., Depaoli-Roach A. A., Roach P. J. Glucose metabolism in mice lacking muscle glycogen synthase. Diabetes. 2005;54:3466–3473. doi: 10.2337/diabetes.54.12.3466. [DOI] [PubMed] [Google Scholar]
- 14.Pederson B. A., Cope C. R., Schroeder J. M., Smith M. W., Irimia J. M., Thurberg B. L., DePaoli-Roach A. A., Roach P. J. Exercise capacity of mice genetically lacking muscle glycogen synthase: in mice, muscle glycogen is not essential for exercise. J. Biol. Chem. 2005;280:17260–17265. doi: 10.1074/jbc.M410448200. [DOI] [PubMed] [Google Scholar]
- 15.Pederson B. A., Cope C. R., Irimia J. M., Schroeder J. M., Thurberg B. L., Depaoli-Roach A. A., Roach P. J. Mice with elevated muscle glycogen stores do not have improved exercise performance. Biochem. Biophys. Res. Commun. 2005;331:491–496. doi: 10.1016/j.bbrc.2005.03.206. [DOI] [PubMed] [Google Scholar]
- 16.Affymetrix. Santa Clara: 2001. Affymetrix: GeneChip Expression Analysis Technical Manual. [Google Scholar]
- 17.Affymetrix. version 5.0. Santa Clara: 2001. Affymetrix: Microarray Suite User's Guide. [Google Scholar]
- 18.Breese M. R., Stephens M. J., McClintick J. N., Grow M. W., Edenberg H. J. Proceedings of the 18th ACM Symposium on Applied Computing. Melbourne, Florida: 2003. Labrat LIMS: an extensible framework for developing laboratory information management, analysis, and bioinformatics solutions for microarrays. [Google Scholar]
- 19.McClintick J. N., Jerome R. E., Nicholson C. R., Crabb D. W., Edenberg H. J. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:4. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zar J. H. Upper Saddle River: Prentice Hall; 1999. Biostatistical Analysis. [Google Scholar]
- 20a.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 21.Obayashi M., Shimomura Y., Nakai N., Jeoung N. H., Nagasaki M., Murakami T., Sato Y., Harris R. A. Estrogen controls branched-chain amino acid catabolism in female rats. J. Nutr. 2004;134:2628–2633. doi: 10.1093/jn/134.10.2628. [DOI] [PubMed] [Google Scholar]
- 22.Shimomura Y., Paxton R., Ozawa T., Harris R. A. Purification of branched chain alpha-ketoacid dehydrogenase complex from rat liver. Anal. Biochem. 1987;163:74–78. doi: 10.1016/0003-2697(87)90094-7. [DOI] [PubMed] [Google Scholar]
- 23.Yagi T., Baroja-Fernandez E., Yamamoto R., Munoz F. J., Akazawa T., Hong K. S., Pozueta-Romero J. Cloning, expression and characterization of a mammalian Nudix hydrolase-like enzyme that cleaves the pyrophosphate bond of UDP-glucose. Biochem. J. 2003;370:409–415. doi: 10.1042/BJ20021140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azpiazu I., Manchester J., Skurat A. V., Roach P. J., Lawrence J. C., Jr Control of glycogen synthesis is shared between glucose transport and glycogen synthase in skeletal muscle fibers. Am. J. Physiol. Endocrinol. Metab. 2000;278:E234–E243. doi: 10.1152/ajpendo.2000.278.2.E234. [DOI] [PubMed] [Google Scholar]
- 25.Huang F. L., Glinsmann W. H. Separation and characterization of two phosphorylase phosphatase inhibitors from rabbit skeletal muscle. Eur. J. Biochem. 1976;70:419–426. doi: 10.1111/j.1432-1033.1976.tb11032.x. [DOI] [PubMed] [Google Scholar]
- 26.Huang F. L., Glinsmann W. A second heat-stable protein inhibitor of phosphorylase phosphatase from rabbit muscle. FEBS. Lett. 1976;62:326–329. doi: 10.1016/0014-5793(76)80086-5. [DOI] [PubMed] [Google Scholar]
- 27.Foulkes J. G., Cohen P., Strada S. J., Everson W. V., Jefferson L. S. Antagonistic effects of insulin and beta-adrenergic agonists on the activity of protein phosphatase inhibitor-1 in skeletal muscle of the perfused rat hemicorpus. J. Biol. Chem. 1982;257:12493–12496. [PubMed] [Google Scholar]
- 28.Cohen P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 29.Scrimgeour A. G., Allen P. B., Fienberg A. A., Greengard P., Lawrence J. C., Jr Inhibitor-1 is not required for the activation of glycogen synthase by insulin in skeletal muscle. J. Biol. Chem. 1999;274:20949–20952. doi: 10.1074/jbc.274.30.20949. [DOI] [PubMed] [Google Scholar]
- 30.Pilkis S. J., Claus T. H., Kurland I. J., Lange A. J. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase: a metabolic signaling enzyme. Annu. Rev. Biochem. 1995;64:799–835. doi: 10.1146/annurev.bi.64.070195.004055. [DOI] [PubMed] [Google Scholar]
- 31.el-Maghrabi M. R., Claus T. H., Pilkis J., Pilkis S. J. Regulation of 6-phosphofructo-2-kinase activity by cyclic AMP-dependent phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 1982;79:315–319. doi: 10.1073/pnas.79.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Maghrabi M. R., Noto F., Wu N., Manes N. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: suiting structure to need, in a family of tissue-specific enzymes. Curr. Opin. Clin. Nutr. Metab. Care. 2001;4:411–418. doi: 10.1097/00075197-200109000-00012. [DOI] [PubMed] [Google Scholar]
- 33.Marsin A. S., Bouzin C., Bertrand L., Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J. Biol. Chem. 2002;277:30778–30783. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- 34.Okar D. A., Manzano A., Navarro-Sabate A., Riera L., Bartrons R., Lange A. J. PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem. Sci. 2001;26:30–35. doi: 10.1016/s0968-0004(00)01699-6. [DOI] [PubMed] [Google Scholar]
- 35.Minchenko A., Leshchinsky I., Opentanova I., Sang N., Srinivas V., Armstead V., Caro J. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 2002;277:6183–6187. doi: 10.1074/jbc.M110978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atsumi T., Nishio T., Niwa H., Takeuchi J., Bando H., Shimizu C., Yoshioka N., Bucala R., Koike T. Expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase/PFKFB3 isoforms in adipocytes and their potential role in glycolytic regulation. Diabetes. 2005;54:3349–3357. doi: 10.2337/diabetes.54.12.3349. [DOI] [PubMed] [Google Scholar]
- 37.Maher F., Vannucci S. J., Simpson I. A. Glucose transporter proteins in brain. FASEB J. 1994;8:1003–1011. doi: 10.1096/fasebj.8.13.7926364. [DOI] [PubMed] [Google Scholar]
- 38.Stuart C. A., Deling Y., Green A. American Diabetes Association Annual Meeting. San Diego, CA: 2005. GLUT3 siRNA inhibits glucose uptake and induces apoptosis in L6 myocytes. [Google Scholar]
- 39.O'Rourke J. F., Pugh C. W., Bartlett S. M., Ratcliffe P. J. Identification of hypoxically inducible mRNAs in HeLa cells using differential-display PCR. Role of hypoxia-inducible factor-1. Eur. J. Biochem. 1996;241:403–410. doi: 10.1111/j.1432-1033.1996.00403.x. [DOI] [PubMed] [Google Scholar]
- 40.Maxwell P. H., Dachs G. U., Gleadle J. M., Nicholls L. G., Harris A. L., Stratford I. J., Hankinson O., Pugh C. W., Ratcliffe P. J. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris R. A., Joshi M., Jeoung N. H. Mechanisms responsible for regulation of branched-chain amino acid catabolism. Biochem. Biophys. Res. Commun. 2004;313:391–396. doi: 10.1016/j.bbrc.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Nishitani S., Takehana K., Fujitani S., Sonaka I. Branched-chain amino acids improve glucose metabolism in rats with liver cirrhosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G1292–G1300. doi: 10.1152/ajpgi.00510.2003. [DOI] [PubMed] [Google Scholar]
- 43.Kimball S. R., Jefferson L. S. Molecular mechanisms through which amino acids mediate signaling through the mammalian target of rapamycin. Curr. Opin. Clin. Nutr. Metab. Care. 2004;7:39–44. doi: 10.1097/00075197-200401000-00008. [DOI] [PubMed] [Google Scholar]
- 44.Azpiazu I., Saltiel A. R., DePaoli-Roach A. A., Lawrence J. C. Regulation of both glycogen synthase and PHAS-I by insulin in rat skeletal muscle involves mitogen-activated protein kinase-independent and rapamycin-sensitive pathways. J. Biol. Chem. 1996;271:5033–5039. doi: 10.1074/jbc.271.9.5033. [DOI] [PubMed] [Google Scholar]
- 45.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 46.Nusse R. The Wnt gene family in tumorigenesis and in normal development. J. Steroid Biochem. Mol. Biol. 1992;43:9–12. doi: 10.1016/0960-0760(92)90181-h. [DOI] [PubMed] [Google Scholar]
- 47.Derksen P. W., Tjin E., Meijer H. P., Klok M. D., MacGillavry H. D., van Oers M. H., Lokhorst H. M., Bloem A. C., Clevers H., Nusse R., et al. Illegitimate WNT signaling promotes proliferation of multiple myeloma cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6122–6127. doi: 10.1073/pnas.0305855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamazaki H., Yanagawa S. Axin and the axin/arrow-binding protein DCAP mediate glucose-glycogen metabolism. Biochem. Biophys. Res. Commun. 2003;304:229–235. doi: 10.1016/s0006-291x(03)00582-5. [DOI] [PubMed] [Google Scholar]
- 49.Fujino T., Asaba H., Kang M. J., Ikeda Y., Sone H., Takada S., Kim D. H., Ioka R. X., Ono M., Tomoyori H., et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 2003;100:229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keller C., Steensberg A., Pilegaard H., Osada T., Saltin B., Pedersen B. K., Neufer P. D. Transcriptional activation of the IL-6 gene in human contracting skeletal muscle: influence of muscle glycogen content. FASEB J. 2001;15:2748–2750. doi: 10.1096/fj.01-0507fje. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y. H., Li Y., Uyeda K., Hasemann C. A. Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. J. Biol. Chem. 2003;278:523–530. doi: 10.1074/jbc.M209105200. [DOI] [PubMed] [Google Scholar]
- 52.Wiatrowski H. A., Van Denderen B. J., Berkey C. D., Kemp B. E., Stapleton D., Carlson M. Mutations in the gal83 glycogen-binding domain activate the snf1/gal83 kinase pathway by a glycogen-independent mechanism. Mol. Cell. Biol. 2004;24:352–361. doi: 10.1128/MCB.24.1.352-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardie D. G., Carling D., Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 54.Chen Z., Heierhorst J., Mann R. J., Mitchelhill K. I., Michell B. J., Witters L. A., Lynch G. S., Kemp B. E., Stapleton D. Expression of the AMP-activated protein kinase β1 and β2 subunits in skeletal muscle. FEBS. Lett. 1999;460:343–348. doi: 10.1016/s0014-5793(99)01371-x. [DOI] [PubMed] [Google Scholar]
- 55.Polekhina G., Gupta A., Michell B. J., van Denderen B., Murthy S., Feil S. C., Jennings I. G., Campbell D. J., Witters L. A., Parker M. W., et al. AMPK beta subunit targets metabolic stress sensing to glycogen. Curr. Biol. 2003;13:867–871. doi: 10.1016/s0960-9822(03)00292-6. [DOI] [PubMed] [Google Scholar]
- 56.Hudson E. R., Pan D. A., James J., Lucocq J. M., Hawley S. A., Green K. A., Baba O., Terashima T., Hardie D. G. A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Curr. Biol. 2003;13:861–866. doi: 10.1016/s0960-9822(03)00249-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.