Abstract
Invasive tumour cells, such as gliomas, frequently express EGF (epidermal growth factor) receptor at a high level and they exhibit enhanced cell migration in response to EGF. We reported previously that tumour cell migration is associated with ectodomain cleavage of CD44, the major adhesion molecule that is implicated in tumour invasion and metastasis, and that the cleavage is enhanced by ligation of CD44. In the present study, we show that EGF promotes CD44 cleavage and CD44-dependent cell migration. Introduction of a dominant-negative mutant of the small GTPase Rac1 or depletion of Rac1 by RNAi (RNA interference) abrogated CD44 cleavage induced by EGF. Treatment with PD98059, an inhibitor for MEK (mitogen-activated protein kinase/extracellular-signal-regulated kinase kinase), also suppressed the CD44 cleavage. Furthermore, RNAi studies showed that EGF induced ADAM10 (a disintegrin and metalloproteinase 10)-dependent CD44 cleavage and cell migration. These results indicate that EGF induces ADAM10-mediated CD44 cleavage through Rac1 and mitogen-activated protein kinase activation, and thereby promotes tumour cell migration and invasion.
Keywords: a disintegrin and metalloproteinase (ADAM), CD44, cell-surface adhesion molecule, epidermal growth factor (EGF), Rac GTPase, shedding
Abbreviations: ADAM, a disintegrin and metalloproteinase; DMEM, Dulbecco's modified Eagle's medium; EGF, epidermal growth factor; EGFR, EGF receptor; ERK, extracellular-signal-regulated kinase; FRET, fluorescence resonance energy transfer; GST, glutathione S-transferase; HB-EGF, heparin-binding EGF-like growth factor; HRP, horseradish peroxidase; mAb, monoclonal antibody; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; pAb, polyclonal antibody; PBD, p21-binding domain; RNAi, RNA interference; siRNA, small interfering RNA; TIMP, tissue inhibitor of metalloproteinases; YFP, yellow fluorescent protein
INTRODUCTION
CD44 is a major cell-surface adhesion molecule for hyaluronan, a component of the extracellular matrix, and is implicated in leucocyte rolling [1,2], tumour invasion and metastasis [3,4]. We reported previously that hyaluronan fragments enhance ectodomain cleavage of CD44 [5], and that the cross-linking effect on CD44 elicits intracellular signals via Rac GTPase leading to ADAM (a disintegrin and metalloproteinase)-mediated CD44 cleavage and tumour cell migration [6].
The migratory properties of invasive tumour cells were affected by growth factor receptor signalling in addition to the interaction between adhesion molecules on tumour cells and the surrounding extracellular matrices [7]. One of the most prominent abnormalities observed in tumour cells, such as gliomas, involves overexpression of the EGFR [EGF (epidermal growth factor) receptor], a transmembrane tyrosine kinase receptor, and such receptor defects may be relevant to tumorigenesis [8–13]. EGFR overexpression contributes to migratory properties of tumour cells in addition to tumour cell growth [7,14–17]. In certain glioma cells, EGF up-regulates CD44-dependent cell migration [18], although the mechanism that regulates the migration remains to be fully elucidated.
In the present study, we examined the involvement of CD44 cleavage in EGF-induced cell migration. We demonstrate that EGF induces CD44 cleavage in tumour cells and enhances cell migration. By using an RNAi (RNA interference) technique, we show direct evidence that EGF-induced CD44 cleavage is regulated by Rac GTPase and is mainly mediated by ADAM10.
EXPERIMENTAL
Reagents
Human recombinant EGF was purchased from Boehringer Mannheim. Tyrphostin A25 and AG1478 were purchased from Sigma. A pAb (polyclonal antibody) against the cytoplasmic domain of CD44, anti-CD44cyo pAb [19] was provided by Dr Hideyuki Saya (Kumamoto University, Kumamoto, Japan). Anti-CD44 mAb (monoclonal antibody) IM7 was purified from rat B-cell hybridoma obtained from the A.T.C.C. (Manassas, VA, U.S.A.) Anti-CD44 mAb BRIC235 was purchased from the International Blood Group Reference Laboratory (Bristol, U.K.). Anti-β-tubulin mAb and anti-ADAM10 pAb were purchased from Oncogene Research Products. Anti-Rac1 mAb was purchased from Upstate Biotechnology. Anti-ERK (extracellular-signal-regulated kinase) pAb and anti-phosphorylated ERK pAb were obtained from Cell Signaling Technology. A MEK [MAPK (mitogen-activated protein kinase)/ERK kinase] inhibitor PD98059 was purchased from Calbiochem.
Cell culture and transfection
The human glioblastoma cell line U251MG was grown in DMEM (Dulbecco's modified Eagle's medium)/F-12 medium (Invitrogen) supplemented with 10% foetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin at 37 °C in an atmosphere containing 5% CO2. Introduction of the plasmid pEF-BOS-myc-N17Rac1, kindly provided by Dr Yoshimi Takai (Osaka University, Suita, Japan), was performed as follows: the cells were seeded in 24-well plates at 2.5×104 cells/well and were transfected with plasmid using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. siRNA (small interfering RNA) was introduced into the cells using Lipofectamine™ 2000. siRNAs used were as follows: Rac1, 5′-GAGGAAGAGAAAAUGCCUGdTdT-3′ and 5′-CAGGCAUUUUCUCUUCCUCdTdT-3′; and scrambled irrelevant siRNA, 5′-GCGCGCUUUGUAGGAUUCGdTdT-3′ and 5′-CGAAUCCUACAAAGCGCGCdTdT-3′ (Dharmacon Research). Sequences of the siRNA used for ADAM10 knockdown have been described previously [6]. The silencing efficiency of Rac1 and ADAM10 was determined by Western blotting using their specific antibodies.
Western blot analysis
Cells were seeded in a 24-well plate at 5×104 cells per well and were cultured overnight. After serum starvation for 4 h and pre-incubation with 10 μM MG132 (benzyloxycarbonyl-leucinyl-leucinyl-leucinal) (Peptide Institute) for 30 min, the cells were incubated in the presence of EGF or other agents, and then directly lysed with SDS sample buffer (2% SDS, 10% glycerol, 0.1 M dithiothreitol, 120 mM Tris/HCl, pH 6.8, 0.02% Bromophenol Blue). Samples extracted from equal numbers of cells were subjected to SDS/PAGE and transferred to a PVDF filter. For detection of CD44 cleavage, the filter was probed with anti-CD44cyto pAb and HRP (horseradish peroxidase)-conjugated anti-rabbit IgG and developed with ECL® (enhanced chemiluminescence) Western blotting detection reagents (Amersham Biosciences). For detection of β-tubulin, the filter was probed with anti-β-tubulin mAb and HRP-conjugated anti-mouse IgG. For the inhibition study, tyrphostin A25 or AG1478 was added 1 h before incubation with EGF. The ‘cleavage index’ shows the CD44 cleavage product band intensity and was expressed as a percentage of untreated cells. Values were normalized to β-tubulin band intensity. Quantitative analysis was performed by densitometry using NIH (National Institutes of Health) Image software.
For detection of phosphorylated ERK MAPK, the filter was probed with anti-phosphorylated ERK pAb and HRP-conjugated anti-rabbit IgG. For detection of total ERK, the filter was probed with anti-ERK pAb and HRP-conjugated anti-rabbit IgG. In experiments with PD98059, the cells were pre-incubated with PD98059 for 30 min before the incubation with EGF.
Immunofluorescence microscopy
Cells incubated with or without 10 ng/ml EGF were fixed with 4% (w/v) paraformaldehyde, permeabilized with 0.2% (v/v) Triton X-100 and blocked with 1% (w/v) BSA. Thereafter, the cells were incubated with anti-CD44cyto pAb or normal rabbit IgG followed by incubation with Alexa Fluor® 594-conjugated goat anti-rabbit IgG and Alexa Fluor® 488-conjugated phalloidin (both Molecular Probes). Fluorescence images were acquired using a TCS SL confocal laser-scanning microscope (Leica Microsystems).
FRET (fluorescence resonance energy transfer) assay
In situ detection of Rac1 activation was carried out using FRET probes as described previously [6]. Briefly, cells cultured on glass coverslips were transfected with the plasmid pEYFP-Rac1 [6]. After incubation for 24 h followed by serum starvation for 2 h, the cells were microinjected into the cytoplasm with BODIPY® TR–GTP (Molecular Probes). After 10 min, the cells were incubated with 10 ng/ml EGF, and were observed using an IX70 inverted microscope (Olympus) equipped with a CSU10 confocal laser-scanner unit (Yokogawa Electronic). The cells were excited with a 488-nm Ar+ laser (Uniphase), and images were obtained using a MicroMAX 512BFT cooled charge-coupled device camera (Princeton Scientific Instruments) through 515–550-nm and 615–645-nm filters for YFP (yellow fluorescent protein) and FRET channels respectively. Calculation of FRET signals using the ‘corrected FRET’ method [20] and conversion into multicolour images were performed using MetaMorph software (Universal Imaging Co., Downingtown, PA, U.S.A.). FRET intensity was displayed using a colour spectrum as indicated in the bar of Figure 2.
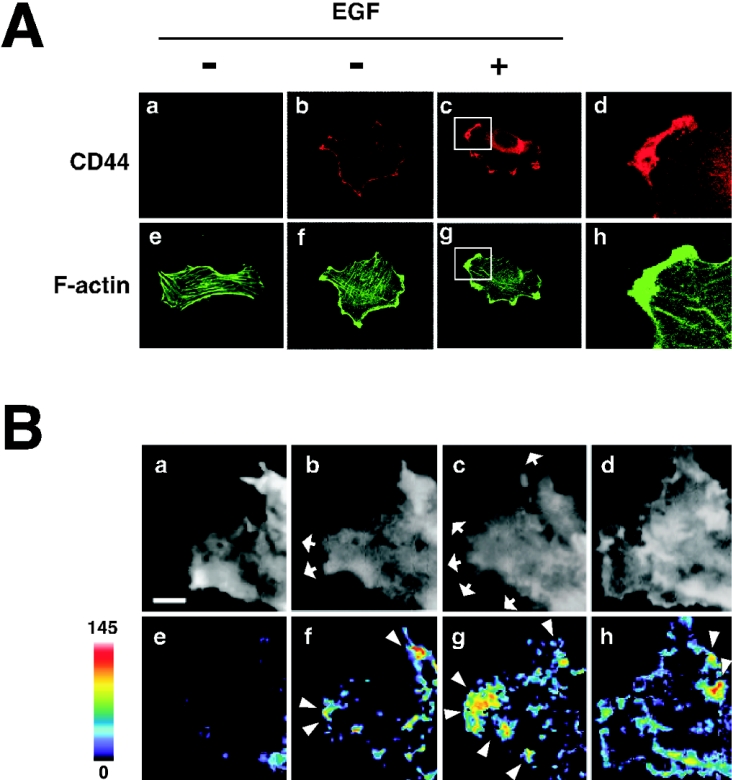
Figure 2. EGF induces redistribution of CD44 to the lamellipodial membrane protrusion area and peripheral activation of Rac1.
(A) Immunocytochemical staining of CD44. U251MG cells incubated with (panels c and g) or without (panels a, b, e and f) 10 ng/ml EGF were stained with anti-CD44cyto pAb (panels b and c) or a control antibody (panel a). The cells were also stained with Alexa Fluor® 488-conjugated phalloidin (panels e, f and g). Panels d and h are enlargements of the regions indicated in panels c and g respectively. (B) In situ detection of Rac1 activation. U251MG cells were transfected with YFP–Rac1 followed by microinjection with BODIPY® TR–GTP. The cells were treated with 10 ng/ml EGF, and the images after 0 (panels a and e; before EGF treatment), 5 (panels b and f), 10 (panels c and g) and 30 min (panels d and h) were acquired through the YFP (panels a–d) and the FRET (panels e–h) channels under confocal microscopy. FRET signal intensity from 0 to 145 is displayed using pseudocolour as indicated by the scale bar. Arrowheads indicate the areas where intense FRET signal was detected, and arrows show the areas of outgrowth. Scale bar, 2 μm.
Pull-down assay
Pull-down assay for detection of the active form of Rac1 was performed as described previously [6].
Migration assay
Cell migration was assayed in Transwell chambers (Corning) with 6.5-mm-diameter polycarbonate filters of 8-μm pore size. The filters were coated with 100 μg/ml hyaluronan (human umbilical cord) (Sigma) or 4 μg/ml human plasma fibronectin (Cappel) at 4 °C overnight. The lower compartments of the chambers were filled with 0.6 ml of DMEM/F-12 medium containing 0.1% (w/v) BSA, and the filters were placed into the chamber. Cells treated with or without siRNA were pre-incubated in the presence or absence of 10 μg/ml BRIC235 mAb for 10 min. A 100 μl aliquot of the cell suspension (2×104 cells) was added to the upper compartments and incubated at 37 °C for 24 h in an atmosphere containing 5% CO2. After the cells on the upper surface of the filters were removed with cotton swabs, the cells on the lower surface were stained with haematoxylin and eosin, and were counted under a light microscope in five defined high-power fields (×200 magnification).
RESULTS
EGF induces CD44 cleavage in tumour cells
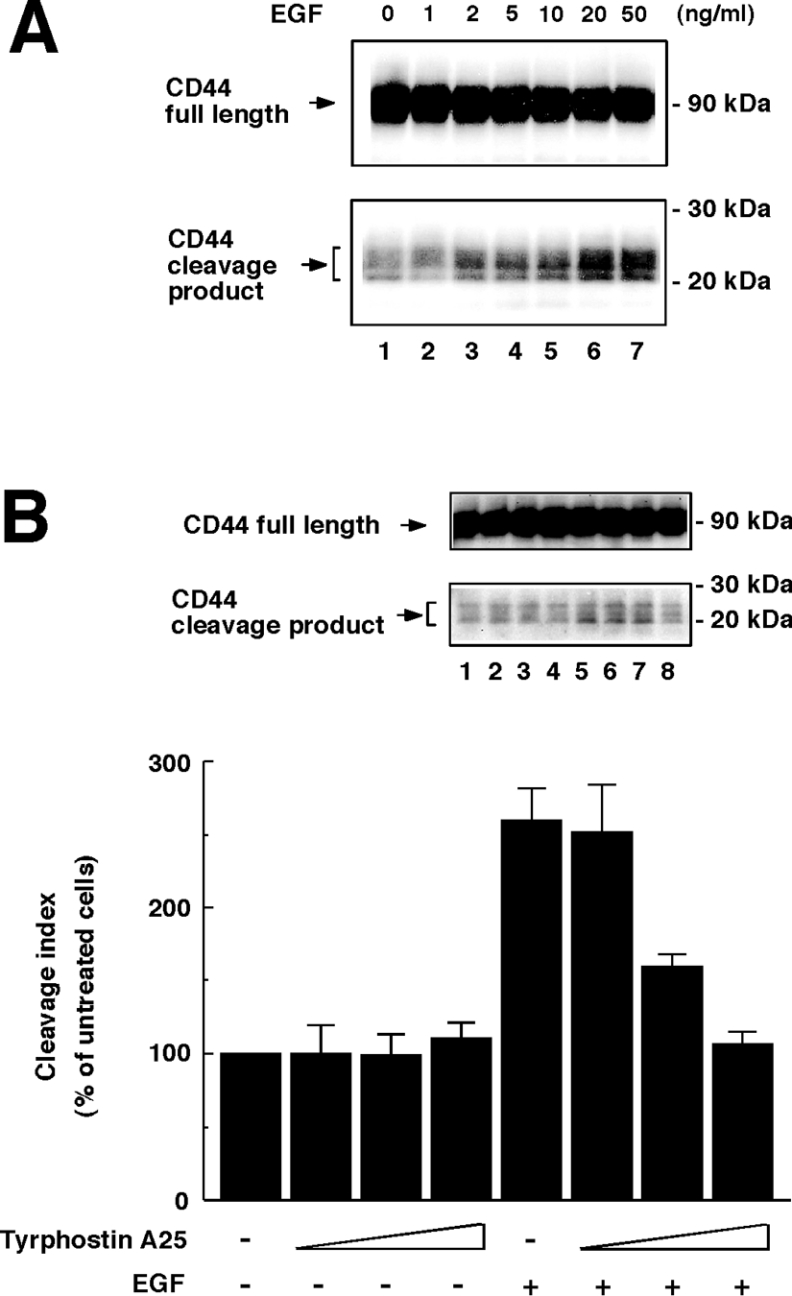
We reported previously that engagement of CD44 up-regulates CD44 cleavage from tumour cells [6]. In the present study, we determined whether EGF induces CD44 cleavage as a physiological inducer. For our studies, we selected U251MG, a highly invasive human glioma cell line, because this cell line was shown previously to have high CD44-cleavage activity [6,19,21] and exhibited CD44-dependent cell migration in response to EGF [18]. U251MG cells expressed EGFR abundantly (results not shown) in agreement with previous reports [15,18]. When U251MG cells were incubated in the presence of EGF, CD44-cleavage products of approx. 20–25 kDa were clearly detected in the cell lysates by Western blotting (Figure 1A). The induction of CD44 cleavage by EGF was dose-dependent and reached a plateau at 20 ng/ml. Similar results were obtained in MIA PaCa-2 human pancreatic carcinoma cells (results not shown). The EGF-induced CD44 cleavage was significantly inhibited by tyrphostin A25, an agent that inhibits EGFR kinase activity (Figure 1B), indicating that kinase activity of EGFR was critical for the CD44 cleavage.
Figure 1. EGF induces ectodomain cleavage of CD44.
(A) EGF induces CD44 cleavage in U251MG cells in a dose-dependent manner. U251MG cells were incubated in the presence of EGF at the indicated concentrations for 2 h. To detect the CD44-cleavage product, the cells were lysed and subjected to Western blot analysis using anti-CD44cyto pAb. (B) Effects of tyrphostin A25 on EGF-induced CD44 cleavage. U251MG cells were pre-incubated in the absence (lanes 1 and 5) or presence (lanes 2 and 6, 1 μg/ml; lanes 3 and 7, 5 μg/ml; lanes 4 and 8, 20 μg/ml) of tyrphostin A25 for 1 h and were subsequently treated with 10 ng/ml EGF (lanes 5–8) or left untreated (lanes 1–4) for 2 h. The cleavage index represents the results of quantitative analysis of the CD44-cleavage product bands. Results are means±S.D. for three independent experiments.
EGF-induced CD44 cleavage is mediated by Rac GTPase
The U251MG cells treated with EGF showed intense membrane ruffling and preferential localization of CD44 in the lamellipodia (Figure 2A). The staining of CD44 was specific because no obvious signal was detected when a control normal IgG was used instead (Figure 2A, panel a). It has been well documented that EGF can induce activation of Rac GTPase and formation of lamellipodia [22]. Thus we investigated the involvement of Rac in EGF-induced CD44 cleavage. We reported previously a method based on FRET that enables us to observe spatial and temporal regulation of Rac1 activity [6]. Using this method, we observed activation of Rac1 induced by EGF stimulation. When U251MG cells harbouring YFP–Rac1 and BODIPY® TR–GTP were imaged for FRET (615–645 nm), few FRET signals were detected before stimulation (Figure 2B, panel e). When the cells were stimulated with EGF, prominent membrane protrusion was initiated (Figure 2B, panels b–d) and a distinct FRET signal appeared at regions, mainly of the leading edge, of the cells within 5 min (Figure 2B, panels f–h). It is of note that the area with intense FRET signal (Figure 2B, arrowheads) exhibits lamellipodial outgrowth (Figure 2B, arrows), where CD44 and F-actin were accumulated and co-localized (Figure 2A, panels d and h). These results indicate that EGF promotes Rac1 activation and membrane protrusion at the peripheral leading edges, and suggest that these may lead to CD44 cleavage at the ruffling area.
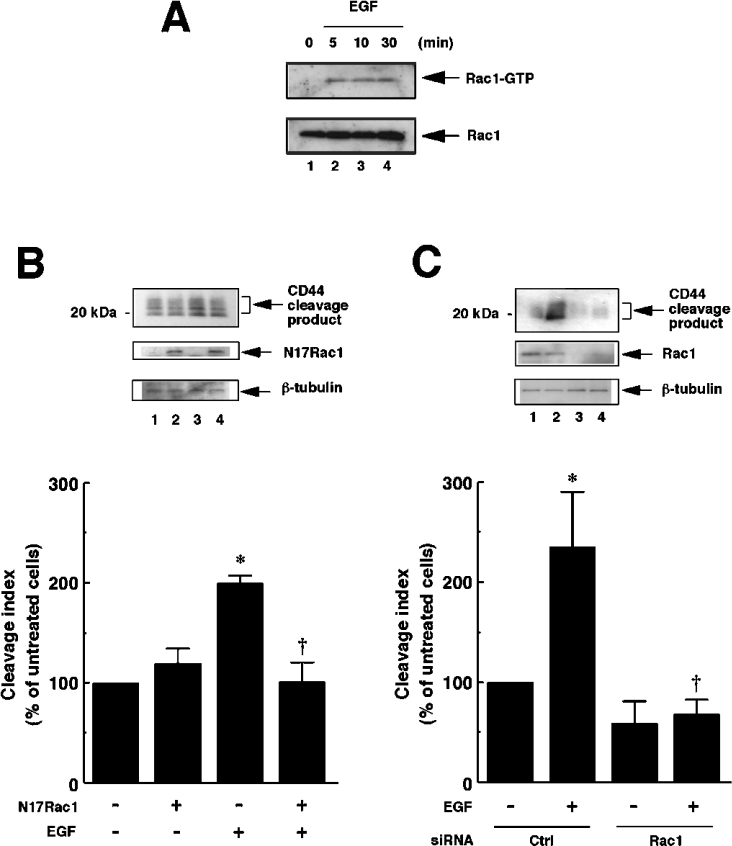
We next detected activation of Rac1 by using the pull-down assay for GTP-bound Rac1 (active Rac1), using a recombinant PBD (p21-binding domain) of PAK1 (p21-activated kinase 1) fused with GST (glutathione S-transferase). As shown in Figure 3(A), the amount of Rac1–GTP was markedly increased within 5 min of incubation with EGF and persisted for at least 30 min. This time course corresponds to the results shown in Figure 2(B), confirming that Rac1 activation is promoted in response to EGF in U251MG cells.
Figure 3. EGF-induced CD44 cleavage is mediated by Rac GTPase.
(A) EGF induces Rac1 activation in U251MG cells. U251MG cells were incubated with 10 ng/ml EGF for the indicated time periods. The cells were then lysed, and the extracts were incubated with glutathione–agarose-bound GST–PBD. The bound Rac1 protein (Rac1–GTP, upper panel) was detected by the pull-down assay. Lower panel, total cellular Rac1 protein. (B) Effect of dominant-negative form of Rac1 (N17Rac1) on CD44 cleavage enhanced by EGF. U251MG cells were transfected with the expression plasmid for N17Rac1 (pEF-BOS-myc-N17Rac1; lanes 2 and 4) or an empty vector (pEF-BOS; lanes 1 and 3). After incubation with (lanes 3 and 4) or without (lanes 1 and 2) 10 ng/ml EGF for 2 h, the cells were lysed and subjected to Western blot analysis using anti-CD44cyto pAb (top panel) or anti-β-tubulin mAb (bottom panel). Expression of N17Rac1 was detected by Western blotting using anti-Rac1 mAb (middle panel). The histogram shows the results of quantitative analysis of the CD44 cleavage product bands as means±S.D. for three independent experiments. (C) Effect of knockdown of endogenous Rac1 on CD44 cleavage enhanced by EGF. U251MG cells were transfected with siRNA for Rac1 (lanes 3 and 4) or control scrambled siRNA (Ctrl; lanes 1 and 2). After incubation with (lanes 2 and 4) or without (lanes 1 and 3) 10 ng/ml EGF for 2 h, the cells were lysed and subjected to Western blot analysis using anti-CD44cyto pAb (top panel) or anti-β-tubulin mAb (bottom panel). Knockdown of Rac1 was monitored by Western blotting using anti-Rac1 mAb (middle panel). The histogram shows the results of quantitative analysis of the CD44 cleavage product bands as means±S.D. for three independent experiments. *, P<0.01 compared with untreated cells; †, P<0.01 compared with EGF-treated and mock-transfected cells (ANOVA with Dunnett's post hoc test).
To determine whether Rac1 activation is required for EGF-induced CD44 cleavage, we introduced a dominant-negative form of Rac1, N17Rac1, into U251MG cells. As shown in Figure 3(B), the CD44 cleavage enhanced upon EGF stimulation was abrogated in the cells transfected with N17Rac1. We next attempted to obtain direct evidence that Rac1 is required for EGF-induced CD44 cleavage. To this end, we used an RNAi technique and achieved efficient suppression of the Rac1 expression by introduction of Rac1-specific siRNA (Figure 3C, middle panel). In the Rac1-knockdown cells, CD44 cleavage was abrogated even after the treatment with EGF (Figure 3C, top panel). These results confirmed that Rac1 is required for the EGF-induced CD44 cleavage as a key regulatory molecule.
Relationship between EGF- and CD44-induced cleavage of CD44
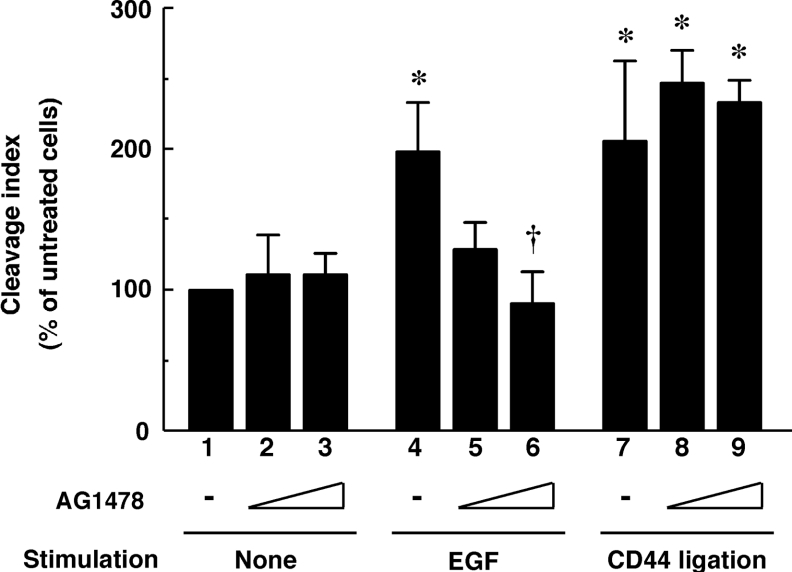
We reported previously that Rac1 was also involved in CD44 cleavage induced by CD44 ligation [6]. Thus the relationship between EGF- and CD44-induced ectodomain cleavage of CD44 is an interesting aspect in terms of intracellular signalling. To distinguish these two signalling pathways, we tested the effects of AG1478, a selective inhibitor of EGFR phospholylation. As shown in Figure 4, AG1478 significantly inhibited the EGF-induced CD44 cleavage at the concentration of 1 μM, whereas it did not apparently inhibit the cleavage induced by CD44 ligation. These results suggest that EGF regulates CD44 cleavage through a new mechanism distinct from that of CD44 ligation signals.
Figure 4. Difference between EGF- and CD44-induced CD44 cleavage.
U251MG cells were pre-incubated in the absence (lanes 1, 4 and 7) or presence (lanes 2, 5 and 8, 0.1 μM; lanes 3, 6 and 9, 1 μM) of AG1478 for 1 h and subsequently treated with 10 ng/ml EGF (lanes 4–6) or 1 μg/ml IM7 mAb (lanes 7–9), or left untreated (lanes 1–3) for 1 h. The cleavage index was calculated by densitometric analysis as described under the Experimental section. Results are means±S.D. for three independent experiments. *, P<0.01 compared with bar 1; †, P<0.01 compared with bar 4 (ANOVA with Dunnett's post hoc test).
EGF-induced CD44 cleavage is dependent on MAPK activation
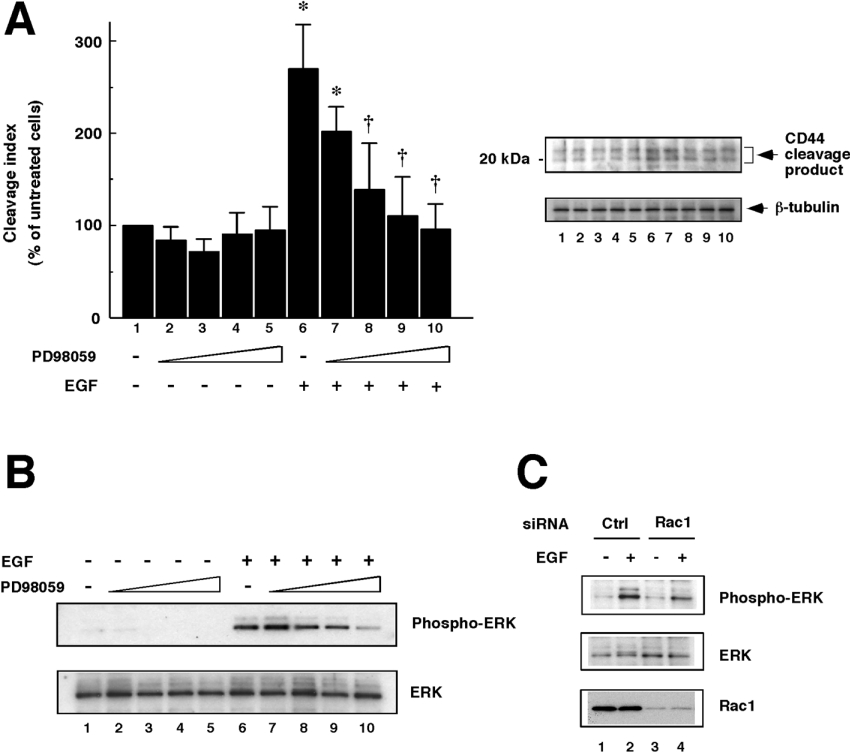
Several reports have indicated that ectodomain cleavage of membrane proteins such as L-selectin and HB-EGF (heparin-binding EGF-like growth factor) was abrogated by inhibitors of ERK/MAPK signalling [23,24], suggesting that MAPK may act as a mediator in the regulation of the cleavage of these molecules. These reports prompted us to examine whether EGF-induced CD44 cleavage is dependent on such MAPK activities. As shown in Figure 5(A), the MEK inhibitor PD98059 abrogated the CD44 cleavage induction by EGF in a dose-dependent manner. Under these conditions, ERKs were prominently activated in response to EGF, and the activation was inhibited by PD98059 dose-dependently (Figure 5B). In contrast, CD44 cleavage induced by its ligation was not affected by PD98059 (results not shown). These results demonstrate a novel aspect of EGF-induced CD44 cleavage, suggesting that the cleavage is regulated in an ERK/MAPK pathway-dependent manner.
Figure 5. Involvement of ERK/MAPKs in EGF-induced CD44 cleavage.
(A) Effect of the MEK inhibitor PD98059 on EGF-induced CD44 cleavage. U251MG cells were pre-treated with 0 (lanes 1 and 6), 0.5 (lanes 2 and 7), 5 (lanes 3 and 8), 50 (lanes 4 and 9) or 100 (lanes 5 and 10) μM PD98059 for 30 min. The cells were treated with (lanes 6–10) or without (lanes 1–5) 10 ng/ml EGF for 1 h, and then lysed. CD44 cleavage was detected by Western blot analysis (upper panel). β-Tubulin was detected as an internal control (lower panel). The histogram shows the results of quantitative analysis of the CD44 cleavage product bands as means±S.D. for three independent experiments. *, P<0.01 compared with bar 1; †, P<0.01 compared with bar 6 (ANOVA with Dunnett's post hoc test). (B) Detection of activation of ERK/MAPKs. The cell lysates prepared as described above were subjected to Western blot analysis using anti-phosphorylated ERK pAb (upper panel) or anti-ERK pAb (lower panel). (C) Dependence of EGF-induced ERK/MAPK activation on Rac1. U251MG cells transfected with siRNA for Rac1 (lanes 3 and 4) or control scrambled siRNA (Ctrl; lanes 1 and 2) were incubated with (lanes 2 and 4) or without (lanes 1 and 3) 10 ng/ml EGF for 1 h. Thereafter, the cells were lysed and subjected to Western blot analysis using anti-phosphorylated ERK pAb (top panel) or anti-ERK pAb (middle panel). Bottom panel, detection of Rac1.
While hitherto Rac and ERK appear to be involved in EGF-induced CD44 cleavage, it remains to be elucidated how these molecules are regulated. To address this issue, ERK activation was determined in Rac1-knockdown cells. As shown in Figure 5(C), ERK activation was still induced upon EGF stimulation in Rac1-knockdown cells, and the induction level seemed slightly lower than in the control cells. These results suggest that EGF-induced CD44 cleavage might be regulated by the Rac→ERK pathway downstream of EGF transactivation signals.
ADAM10 is involved in EGF-induced CD44 cleavage
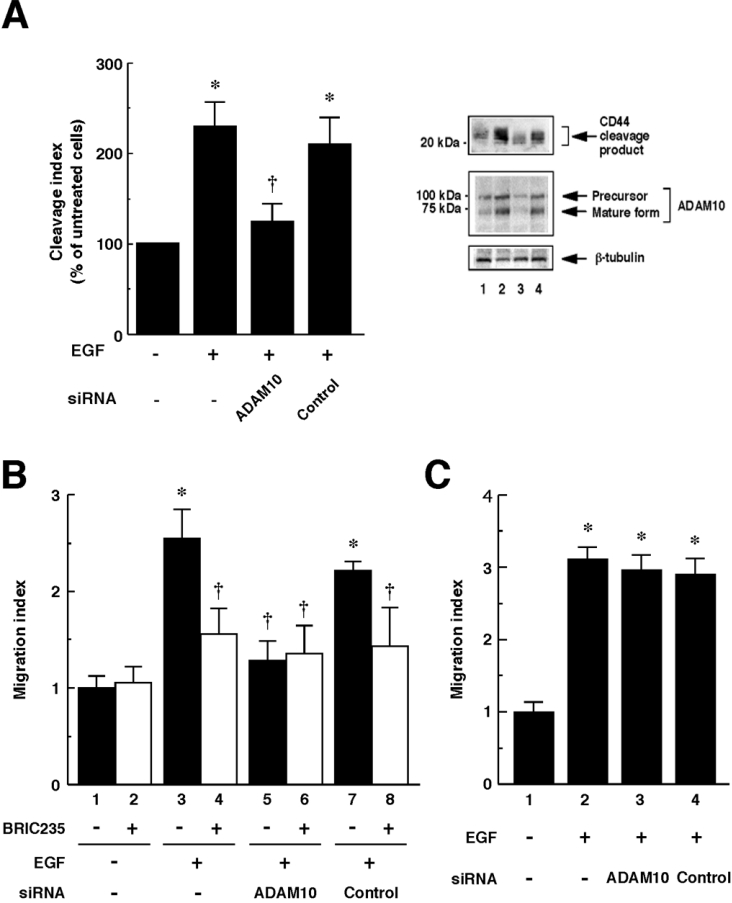
We previously reported that ADAM10/Kuzbanian is involved in CD44 cleavage induced by CD44 ligation [6]. In the present study, we determined whether the same protease is also involved in EGF-induced CD44 cleavage. As shown in Figure 6(A), siRNA to ADAM10 suppressed EGF-induced CD44 cleavage. Efficient knockdown of ADAM10 was monitored by Western blot analysis (Figure 6A, middle panel). To determine whether EGF promotes CD44-dependent cell migration through ADAM10-mediated CD44 cleavage, we performed Boyden chamber-type migration assays. As shown in Figure 6(B), EGF enhanced cell migration on hyaluronan. The enhanced migration was inhibited by pre-treatment with an anti-CD44 blocking mAb BRIC235, indicating that the migration enhanced by EGF was dependent on the CD44–hyaluronan interactions. The enhanced migration on hyaluronan was inhibited by introduction of siRNA to ADAM10 (Figure 6B), whereas the migration on an alternative extracellular matrix substrate, fibronectin, was not significantly affected by ADAM10 siRNA (Figure 6C), indicating that ADAM10 is involved in CD44-mediated cell migration enhanced by EGF. Taken together, these results suggest that EGF-induced CD44 cleavage mediated by ADAM10 is critical for the up-regulation of cell migration through regulating CD44-hyaluronan interactions.
Figure 6. EGF-induced CD44 cleavage and CD44-dependent cell migration are mediated by ADAM10.
(A) Effect of ADAM10 siRNA on EGF-induced CD44 cleavage. U251MG cells were transfected with ADAM10 siRNA (lane 3) or control siRNA (lane 4), and left untreated (lane 1) or treated with 10 ng/ml EGF for 2 h (lanes 2–4). CD44 cleavage product was detected by Western blot analysis (top panel). Blocking of endogenous ADAM10 expression by siRNA was monitored by Western blot analysis using anti-ADAM10 pAb (middle panel). β-Tubulin was detected as an internal control (bottom panel). The histogram shows the results of quantitative analysis of the CD44 cleavage product bands as means±S.D. for three independent experiments. (B) Effects of EGF on the cell migration on hyaluronan. The U251MG cells transfected with ADAM10 siRNA (bars 5 and 6) or control siRNA (bars 7 and 8) or the cells left untransfected (bars 1–4) were pre-incubated in the presence (bars 2, 4, 6 and 8) or absence (bars 1, 3, 5 and 7) of 10 μg/ml BRIC235 mAb for 10 min, and then incubated at 37 °C for 24 h in the absence (bars 1 and 2) or presence (bars 3–8) of 10 ng/ml EGF in the hyaluronan-coated Transwell chambers. Results are mean ratios of the control migration, expressed as means±S.D. for triplicate determinations. (C) Effects of EGF on the cell migration on fibronectin. The U251MG cells transfected with ADAM10 siRNA (bar 3) or control siRNA (bar 4) or the cells left untransfected (bars 1–2) were incubated for 24 h in the absence (bar 1) or presence (bars 2–4) of 10 ng/ml EGF in the fibronectin-coated Transwell chambers. Results are mean ratios to the control migration, expressed as means±S.D. for triplicate determinations. *, P<0.01 compared with untreated cells; †, P<0.01 compared with only EGF-treated cells (ANOVA with Dunnett's post hoc test).
DISCUSSION
CD44 cleavage has been reported to have some links with tumour cell migration in vitro [5,6,19,25,26] and is detected in human tumour tissues such as gliomas [27], suggesting that CD44 cleavage may play an important role in the pathogenesis of tumours. However, physiological factors that induce CD44 cleavage in vivo remain mostly uncharacterized, although we recently identified hyaluronan oligosaccharides as such factors [5,6]. The present study shows that EGF can also act as a physiological inducer of CD44 cleavage (Figure 1). Rac1 is activated by EGF, and is indispensable for EGF-induced CD44 cleavage (Figures 2 and 3). We also found that other agonists of Rho family GTPases, platelet-derived growth factor and bradykinin, could induce CD44 cleavage in U251MG glioma cells, and lysophosphatidic acid suppressed it (results not shown), suggesting that various soluble factors are involved in tumour cell invasion by regulating CD44 cleavage through GTPases.
In the present study, we have demonstrated that the MEK inhibitor PD98059 suppressed EGF-induced ERK activation and CD44 cleavage (Figure 5). While ERK/MAPK pathways are considered to mediate gene expression and proliferation, the cleavage occurred within 10 min (results not shown). These results are similar to the cases of L-selectin and HB-EGF cleavage [23,24]. These suggest that ERK appears to be involved in CD44 cleavage by activation of the cleavage enzyme through phosphorylation, but not by induction of gene expression for de novo protein synthesis. It has been reported that ERK phosphorylates the cytoplasmic region of ADAM17/tumour necrosis factor α-converting enzyme and the phosphorylation may regulate the ectodomain cleavage of TrkA neurotrophin receptor [28]. The minimum consensus sequence for phosphorylation by ERK is (Thr/Ser)-Pro [29,30], and human ADAM10 contains a Thr703-Pro sequence in its cytoplasmic region. Considering these facts, it is feasible that a phosphorylation event in ADAM10 may play a role in the regulation of CD44 cleavage upon EGF stimulation.
Knockdown of the ADAM10 expression by RNAi revealed that the enzyme responsible in EGF-induced CD44 cleavage is ADAM10 (Figure 6A). Knockdown of ADAM10 also abrogated the migratory properties on hyaluronan enhanced by EGF (Figure 6B). These results suggest that ADAM10-mediated CD44 cleavage might enhance tumour cell migration in response to EGF. Although ADAM10 might be activated by EGF (Figure 6A), no detectable changes were observed in the overall expression level of ADAM10 after 24 h of EGF treatment (results not shown). Several membrane proteins such as c-Met, syndecan-1 and syndecan-4 were reported to be cleaved upon EGF stimulation, and the cleavage was inhibited by TIMP-3 (tissue inhibitor of metalloproteinases-3) and not by TIMP-1 or TIMP-2 [31–33]. Considering the fact that in vitro ADAM10 activity is inhibited by TIMP-1 and TIMP-3, but not by TIMP-2 [34], the cleavage enzyme(s) for c-Met, syndecan-1 and syndecan-4 seems distinct from that for CD44. We reported previously that the activation of EGFR by binding of EGF could be detected at the single-molecule level by using fluorescent dye-labelled probes [35–37]. The technology we developed allowed us to visualize real-time location and movements of each molecule during biological reactions. Therefore application of this technology might pave the way to analyse the mechanisms of the EGF-induced ectodomain cleavage of membrane proteins including CD44.
Acknowledgments
This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Dr Y. Takai and Dr H. Saya for their gifts of reagents.
References
- 1.Clark R. A., Alon R., Springer T. A. CD44 and hyaluronan-dependent rolling interactions of lymphocytes on tonsillar stroma. J. Cell Biol. 1996;134:1075–1087. doi: 10.1083/jcb.134.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murai T., Sougawa N., Kawashima H., Yamaguchi K., Miyasaka M. CD44–chondroitin sulfate interactions mediate leukocyte rolling under physiological flow conditions. Immunol. Lett. 2004;93:163–170. doi: 10.1016/j.imlet.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Aruffo A., Stamenkovic I., Melnik M., Underhill C. B., Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 4.Günthert A. R., Sträter J., von Reyher U., Hanne C., Joos S., Koretz K., Moldenhauer G., Krammer P. H., Moller P. Early detachment of colon carcinoma cells during CD95(APO-1/Fas)-mediated apoptosis. I. De-adhesion from hyaluronate by shedding of CD44. J. Cell Biol. 1996;134:1089–1096. doi: 10.1083/jcb.134.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugahara K. N., Murai T., Nishinakamura H., Kawashima H., Saya H., Miyasaka M. Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J. Biol. Chem. 2003;278:32259–32265. doi: 10.1074/jbc.M300347200. [DOI] [PubMed] [Google Scholar]
- 6.Murai T., Miyazaki Y., Nishinakamura H., Sugahara K. N., Miyauchi T., Sako Y., Yanagida T., Miyasaka M. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J. Biol. Chem. 2004;279:4541–4550. doi: 10.1074/jbc.M307356200. [DOI] [PubMed] [Google Scholar]
- 7.Wells A., Kassis J., Solava J., Turner T., Lauffenburger D. A. Growth factor-induced cell motility in tumor invasion. Acta Oncol. 2002;41:124–130. doi: 10.1080/028418602753669481. [DOI] [PubMed] [Google Scholar]
- 8.Libermann T. A., Razon N., Bartal A. D., Yarden Y., Schlessinger J., Soreq H. Expression of epidermal growth factor receptors in human brain tumors. Cancer Res. 1984;44:753–760. [PubMed] [Google Scholar]
- 9.Libermann T. A., Nusbaum H. R., Razon N., Kris R., Lax I., Soreq H., Whittle N., Waterfield M. D., Ullrich A., Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature (London) 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 10.Libermann T. A., Nusbaum H. R., Razon N., Kris R., Lax I., Soreq H., Whittle N., Waterfield M. D., Ullrich A., Schlessinger J. Amplification and overexpression of the EGF receptor gene in primary human glioblastomas. J. Cell Sci. Suppl. 1985;3:161–172. doi: 10.1242/jcs.1985.supplement_3.16. [DOI] [PubMed] [Google Scholar]
- 11.Schlegel J., Stumm G., Brändle K., Merdes A., Mechtersheimer G., Hynes N. E., Kiessling M. Amplification and differential expression of members of the erbB-gene family in human glioblastoma. J. Neuro–Oncol. 1994;22:201–207. doi: 10.1007/BF01052920. [DOI] [PubMed] [Google Scholar]
- 12.Schlegel J., Merdes A., Stumm G., Albert F. K., Forsting M., Hynes N., Kiessling M. Amplification of the epidermal-growth-factor-receptor gene correlates with different growth behaviour in human glioblastoma. Int. J. Cancer. 1994;56:72–77. doi: 10.1002/ijc.2910560114. [DOI] [PubMed] [Google Scholar]
- 13.Shinojima N., Tada K., Shiraishi S., Kamiryo T., Kochi M., Nakamura H., Makino K., Saya H., Hirano H., Kuratsu J., et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- 14.Lund-Johansen M., Bjerkvig R., Humphrey P. A., Bigner S. H., Bigner D. D., Laerum O. D. Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res. 1990;50:6039–6044. [PubMed] [Google Scholar]
- 15.Pedersen P. H., Ness G. O., Engebraaten O., Bjerkvig R., Lillehaug J. R., Laerum O. D. Heterogeneous response to the growth factors [EGF, PDGF (bb), TGF-α, bFGF, IL-2] on glioma spheroid growth, migration and invasion. Int. J. Cancer. 1994;56:255–261. doi: 10.1002/ijc.2910560219. [DOI] [PubMed] [Google Scholar]
- 16.Berens M. E., Rief M. D., Shapiro J. R., Haskett D., Giese A., Joy A., Coons S. W. Proliferation and motility responses of primary and recurrent gliomas related to changes in epidermal growth factor receptor expression. J. Neuro–Oncol. 1996;27:11–22. doi: 10.1007/BF00146079. [DOI] [PubMed] [Google Scholar]
- 17.Tysnes B. B., Haugland H. K., Bjerkvig R. Epidermal growth factor and laminin receptors contribute to migratory and invasive properties of gliomas. Invasion Metastasis. 1997;17:270–280. [PubMed] [Google Scholar]
- 18.Monaghan M., Mulligan K. A., Gillespie H., Trimble A., Winter P., Johnston P. G., McCormick D. Epidermal growth factor up-regulates CD44-dependent astrocytoma invasion in vitro. J. Pathol. 2000;192:519–525. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH784>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto I., Kawano Y., Tsuiki H., Sasaki J., Nakao M., Matsumoto M., Suga M., Ando M., Nakajima M., Saya H. CD44 cleavage induced by a membrane-associated metalloprotease plays a critical role in tumor cell migration. Oncogene. 1999;18:1435–1446. doi: 10.1038/sj.onc.1202447. [DOI] [PubMed] [Google Scholar]
- 20.Gordon G. W., Berry G., Liang X. H., Levine B., Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto I., Kawano Y., Matsumoto M., Suga M., Kaibuchi K., Ando M., Saya H. Regulated CD44 cleavage under the control of protein kinase C, calcium influx, and the Rho family of small G proteins. J. Biol. Chem. 1999;274:25525–25534. doi: 10.1074/jbc.274.36.25525. [DOI] [PubMed] [Google Scholar]
- 22.Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 23.Fan H., Derynck R. Ectodomain shedding of TGF-α and other transmembrane proteins is induced by receptor tyrosine kinase activation and MAP kinase signaling cascades. EMBO J. 1999;18:6962–6972. doi: 10.1093/emboj/18.24.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gechtman Z., Alonso J. L., Raab G., Ingber D. E., Klagsbrun M. The shedding of membrane-anchored heparin-binding epidermal-like growth factor is regulated by the Raf/mitogen-activated protein kinase cascade and by cell adhesion and spreading. J. Biol. Chem. 1999;274:28828–28835. doi: 10.1074/jbc.274.40.28828. [DOI] [PubMed] [Google Scholar]
- 25.Kawano Y., Okamoto I., Murakami D., Itoh H., Yoshida M., Ueda S., Saya H. Ras oncoprotein induces CD44 cleavage through phosphoinositide 3-OH kinase and the rho family of small G proteins. J. Biol. Chem. 2000;275:29628–29635. doi: 10.1074/jbc.M002440200. [DOI] [PubMed] [Google Scholar]
- 26.Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001;153:893–904. doi: 10.1083/jcb.153.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto I., Tsuiki H., Kenyon L. C., Godwin A. K., Emlet D. R., Holgado-Madruga M., Lanham I. S., Joynes C. J., Vo K. T., Guha A., et al. Proteolytic cleavage of the CD44 adhesion molecule in multiple human tumors. Am. J. Pathol. 2002;160:441–447. doi: 10.1016/S0002-9440(10)64863-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díaz-Rodríguez E., Montero J. C., Esparís-Ogando A., Yuste L., Pandiella A. Extracellular signal-regulated kinase phosphorylates tumor necrosis factor α-converting enzyme at threonine 735: a potential role in regulated shedding. Mol. Biol. Cell. 2002;13:2031–2044. doi: 10.1091/mbc.01-11-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark-Lewis I., Sanghera J. S., Pelech S. L. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J. Biol. Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 30.Songyang Z., Lu K. P., Kwon Y. T., Tsai L. H., Filhol O., Cochet C., Brickey D. A., Soderling T. R., Bartleson C., Graves D. J., et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nath D., Williamson N. J., Jarvis R., Murphy G. Shedding of c-Met is regulated by crosstalk between a G-protein coupled receptor and the EGF receptor and is mediated by a TIMP-3 sensitive metalloproteinase. J. Cell Sci. 2001;114:1213–1220. doi: 10.1242/jcs.114.6.1213. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian S. V., Fitzgerald M. L., Bernfield M. Regulated shedding of syndecan-1 and -4 ectodomains by thrombin and growth factor receptor activation. J. Biol. Chem. 1997;272:14713–14720. doi: 10.1074/jbc.272.23.14713. [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald M. L., Wang Z., Park P. W., Murphy G., Bernfield M. Shedding of syndecan-1 and -4 ectodomains is regulated by multiple signaling pathways and mediated by a TIMP-3-sensitive metalloproteinase. J. Cell Biol. 2000;148:811–824. doi: 10.1083/jcb.148.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., Murphy G. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473:275–279. doi: 10.1016/s0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- 35.Sako Y., Minoguchi S., Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 36.Sako Y., Hibino K., Miyauchi T., Miyamoto Y., Ueda M., Yanagida T. Single-molecule imaging of signaling molecules in living cells. Single Mol. 2000;2:159–163. [Google Scholar]
- 37.Ichinose J., Murata M., Yanagida T., Sako Y. EGF signalling amplification induced by dynamic clustering of EGFR. Biochem. Biophys. Res. Commun. 2004;324:1143–1149. doi: 10.1016/j.bbrc.2004.09.173. [DOI] [PubMed] [Google Scholar]