Abstract
In human spermatozoa, the arrangement of chromosomes is non-random. Characteristic features are association of centromeres in the interior chromocenter and peripheral location of telomeres. In this paper, we have investigated the highest level of order in DNA packing in sperm ^ absolute and relative intranuclear chromosome positioning. Asymmetrical nuclear shape, existence of a defined spatial marker, and the haploid complement of chromosomes facilitated an experimental approach using in situ hybridization. Our results showed the tendencyfor non-random intranuclear location of individual chromosome territories. Moreover, centromeres demonstrated specific intranuclear position, and were located within a limited area of nuclear volume. Additionally, the relative positions of centromeres were non-random; some were found in close proximity, while other pairs showed significantly greater intercentromere distances. Therefore, a unique and specific adherence mayexist between chromosomes in sperm. The observed chromosome order is discussed in relation to sperm nuclei decondensation, and reactivation during fertilization.
Keywords: centromere, chromosome position, human, sperm
Introduction
There is a growing body of evidence that subnuclear confinement of a gene contributes directly to its expression (reviewed in Cockell & Gasser 1999, Francastel et al. 1999, Dundr & Misteli 2001). It is tempting to speculate that activities of genes on specific chromosomes are influenced dynamically by their chromosomal and nuclear position (Blobel 1985, Heslop-Harrison 1996, Croft et al. 1999). Indeed, spatial arrangement of chromosomes both during prometaphase/metaphase and in interphase is apparently non-random (Cremer & Cremer 2001, Parada & Misteli 2002, Marshall 2002, Gasser 2002). An ordered relative position of chromosomes during prometaphase/metaphase has been observed both in plant (Ashley 1979, Oud et al. 1989) and in animal (Leitch et al. 1994, Nagele et al. 1995) cells. This issue remains controversial since another study executing a similar experimental approach (Allison & Nestor 1999) did not establish order in relative chromosome position in the metaphase ring. Although a significant passive diffusion of chromosomes does occur within the interphase nuclei as a whole, chromosomes are largely constrained within a limited subregion of the nucleus (Marshall et al. 1997). Chromosomes, which are adjacent in mitotic rosettes, are similarly positioned in interphase nuclei (Nagele et al. 1999). For example, in human fibroblasts, the positioning of chromosomes at G1 in daughter cells is closely correlated and is mainly determined by their configuration at mitosis (Sun & Yokota 1999). Recent data indicate non-random radial positioning of chromosomes (Sun et al. 2000, Cremer et al. 2001, Boyle et al. 2001, Tanabe et al. 2002). Chromosomes are distributed according to their size (mass): smaller chromosomes are positioned more centrally than larger ones (Sun et al. 2000). Furthermore, most gene-dense chromosomes localize more internally (Croft et al. 1999, Cremer et al. 2001, Boyle et al. 2001). The crucial question concerns the mechanisms underlying the institution and maintenance of chromosome order during passage through the cell cycle and between generations. These mechanisms are still obscure (Bickmore & Chubb 2003). As a surprising possibility, a permanent physical link between chromosomes has been discussed (Nagele et al. 1995) and, in some cases, experimentally evidenced (Maniotis et al. 1997, Bojanowski et al. 1998, Dozortsev et al. 2000, Saifitdinova et al. 2001).
The spermatozoa are highlydifferentiated cells that result from a multi-step process of spermatogenesis. During the stages following meiosis, histones are graduallyremoved from the chromosomes and replaced byhighlybasic transitional proteins, and then byprotamines (Wouters-Tyrou et al. 1998). Concomitantly, chromatin undergoes structural reorganization; DNA becomes supercondensed and genetically inactive (Balhorn 1982, Ward & Zalensky 1996). Recent studies demonstrated specific and non-random chromosome architecture in sperm nuclei. Characteristic features of sperm architecture are:
Similar to somatic cells, individual chromosomes occupydistinct territories (Haaf & Ward 1995, Zalensky et al. 1995);
Centromeres belonging to non-homologous chromosomes are collected into a compact chromocenter buried within a nuclear volume (Zalensky et al. 1993, 1995, Haaf & Ward 1995, Hoyer-Fender et al. 2000); and
Telomeres are localized at the nuclear periphery where theyinteract in dimers and tetramers (Zalensky et al. 1995, 1997, Meyer-Ficca et al. 1998, Hazzouri et al. 2000).
Similar principles of chromosome spatial organization in sperm retained in other mammals, as indicated by data for bovine (Powell et al. 1990, Zalensky et al. 1997), mouse (Haaf & Ward 1995, Jennings & Powell 1995, Meyer-Fica et al. 1998), pig, horse, and rat (Zalensky et al. 1997).
Furthermore, evidence for a specific spatial arrangement (positioning) of chromosomes in sperm starts to emerge (reviewed in Greaves et al. 2003). In the fibrillar sperm head of monotreme (Watson et al. 1996, Greaves et al. 2003) and marsupial (Greaves et al. 2003) mammals, chromosomes are grouped tandemlyhead-to-tail and in a preciselydefined order. In non-mammal species, limited order in chromosome organization was shown in planarians (Joffe et al. 1998), and no order was found in chicken spermatozoa (Solovei et al. 1988).
Almost 30 years ago, using C-banding Geraedts and Pearson (1975) showed that in human spermatozoa chromosome 1 (CHR 1) and CHR Y are often found in close proximity, whereas positions of CHR 9 and CHR Y are not correlated. More recently, data using fluorescent in situ hybridization (FISH) started to emerge indicating specific chromosome location in human sperm nuclei (Luetjens et al. 1999, Hazzouri et al. 2000, Sbracia et al. 2002). Tilgen and co-authors (2001) studied the arrangement of five chromosome pairs in human spermatozoa and each pair was found to have a non-random spatial distribution. Thus, the existence of a defined chromosome positioning in human sperm nuclei seems possible, although our knowledge is still very limited.
In this study, we analyzed absolute and relative intranuclear position of chromosomes in mature human sperm. We used several approaches based on FISH with whole-chromosome painting and chromosome-specific centromere DNA probes. We present new evidence for the existence of ordered spatial localization of individual chromosomes within nuclei.
Materials and methods
Sperm cells were collected from semen obtained from donors with proven fertility. Cells were washed with PBS, fixed in 0.5% formaldehyde/PBS for 5 min, and permeabilized by treatment with 0.2% Tween-20, PBS for 5 min at room temperature. After washing the detergent, cells were resuspended in 0.01 mg/ml Heparin, 1 mmol/L DTT, PBS. After 30-min incubation at room temperature, cells were loaded on microscopic slide. Treatment with DTT/Heparin induces uniform nuclear swelling while preserving nuclear shape. Such swelling is an essential step to perform FISH analysis in human sperm cells and has been described earlier (Zalensky et al. 1993, 1995).
Fluorescent in situ hybridization
Methods of FISH in application to sperm cells have been described in detail earlier (Zalensky et al. 1993, 1995, 1997). Briefly, microscopic slides with the cells pretreated as described in the previous section were denatured in 70% formamide, 2 × SSC, dehydrated in cold ethanol (70%, 80%, 90%, and 100%) and air-dried. Hybridization conditions and post-hybridization washings depended on the type of DNA probes.
Chromosome painting probes
Biotin-labeled chromosome painting Probes (A.L. Biomedical, Inc.) were hybridized and washed according to the manufacturer's instructions. After post-hybridization washings, the cells were blocked in 3% BSA, 4 × SSC, 0.1% Tween-20 for 15 min at room temperature. The hybridization was detected using Avidin-FITC (Vector) diluted 1: 200 in antibody solution (1% BSA, 4 × SSC, 0.1% Tween-20).
Multicolor FISH with centromere-specific (CEN) probes
For two-color FISH, sperm cells were hybridized overnight at 37°C with a mixture of a biotin (Bio)- and a digoxigenin (Dig)-labeled probe specific to centromeres of two chromosomes (Oncor) in hybridization buffer containing 50% formamide/2 × SSC. For three-color FISH hybridization, the solution was composed of a biotinylated probe for the first centromere, Dig-labeled probe for the second centromere, and a 1: 1 mixture of the biotinylated and Dig-labeled probes for the third centromere. Post-hybridization washings were at 37°C: 15-min wash in 50% formamide/2 × SSC, then two 8-min washes in 2 × SSC. The slides were proceeded to detection/amplification steps after blocking. First, sheep anti-Dig conjugated with FITC was applied; second - rabbit anti-Sheep-FITC, third - goat anti-Rabbit-FITC. Each step was for 20 min at 37°C, all antibodies were from Roche and used at 1: 200 dilution in the antibody solution. This round was followed by detection/amplification of Bio-labeled DNA using Avidin-Texas Red, followed by biotinylated anti-Avidin, and again Avidin-Texas Red. Each step was for 20 min at 37°C, all chemicals and antibodies were from Vector and used at 1: 200 dilution. Between each step, the slides were washed in 4 × SSC, 0.1% Tween-20 three times for 5 min at 37°C.
Alternatively, fast FISH protocol was used for directly-labeled CEN probes (Aquarius): after 1 h hybridization at 37°C slides were washed in 0.25 × SSC at 72°C for 2 min, then for 30 s in 2 × SSC/0.05% Tween-20 at room temperature.
Total nuclear DNA was counterstained with DAPI or propidium iodide, and the slides were mounted using FluoroGuard (BioRad Laboratories).
Microscopy and data analysis
Hybridization results were visualized using a Leitz Ortholux microscope with an oil immersion objective 60×, 1.4NA. In most experiments, we used a triple band-pass filter set that allows simultaneous detection of Texas Red, FITC, and DAPs. Images were collected using a digital color camera (MagnaFire, Optronics Inc.) and processed with Adobe Photoshop 7.0 software. Measurements of distances in digitized images were performed in Sigma Scan Pro 5.0 software for individual sperm nuclei that have an unequivocal pattern of hybridization and a good morphology (absence of clamping, overlap, Âwell-defined outline, etc.).
In each experiment, 200^500 cells were analyzed. Mean values, standard errors, and frequency distributions were determined using the Microsoft Excel or Prism (GraphPad Inc.) software. To test non-random distribution of the individual centromeres or pair-wise intercentromere distances in the two-dimensional space of the sperm head, we used the nonparametric Kolmogorov-Smirnov test. This test estimates the fit of the experimental data to the expected theoretical distribution. In our case, we compared data for each individual centromere position (or intercentromere distance) against the uniformlydistributed data set of the same sample size.
Geometry of the human sperm nucleus and parameters describing chromosome positioning
Nuclei in human sperm cells have an ellipsoid shape with its long axis stretching from the point of the sperm tail attachment to the acrosome. After deposition on a microscope slide, nuclei flatten (Zalensky et al. 1995) and their visible projection could be well approximated by an ellipse (Figure 1), with the posterior part clearly marked by the sperm tail. The sperm tail may be easily observed using visible light or epifluorescence microscopy due to nonspecific staining of a midpiece during FISH/immunolocalization procedures (Figures 2A and 3B). Human sperm nuclei have a line of symmetry that coincides with the long ellipse axis (L-L′ in Figure 1A). Because of this, the cells landing on the slide surface are random relative to rotation around L-L′. Therefore, we anticipated that the position of a given chromosome or its domain along the short axis l-l′ may be determined with limited certainty (Figures 1A, B).
Figure 1.
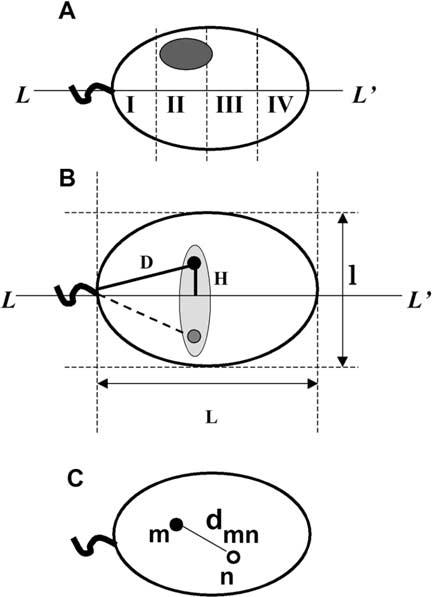
Geometrical parameters of human sperm cell used to determine chromosome positioning. A. Schematic representation of a sperm cell nuclei approximated by an ellipse with the axis of symmetry L-L′. Shaded oval shows an area of hybridization with a chromosome-painting probe. Sperm nucleus is divided into four zones I-IV starting from the basal side that is determined by tail attachment site. B. Parameters, which have been used to describe intranuclear localization of the chromosome-specific centromere hybridization signals. D - distance from the tail attachment point, H - elevation over the long axis. L and l are lengths of the long and short axes. Because of anticipated freedom of rotation along L-L′ we expected random distribution of signals within a schematically shown shaded area. Filled and shaded circles are experimentally observed mirror positions of a centromere. C. Scheme of the sperm nucleus image after multicolor hybridization with two centromere-specific probes. Distances between centers of the FISH signals (dmn) were used to determine relative positioning of centromeres within nuclei.
Figure 2.
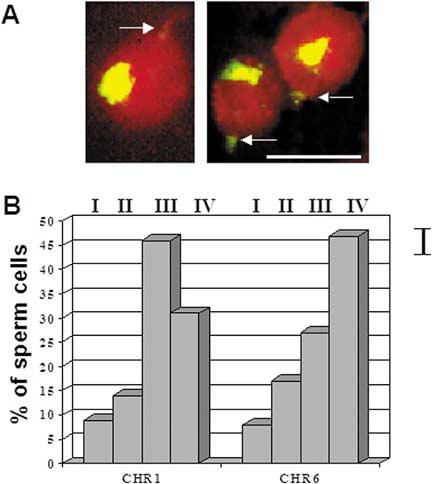
Determination of the intranuclear position of chromosomes in human sperm using FISH with whole chromosome painting probes. A. Representative hybridization pattern: chromosome 6 territory (yellow/green), total nuclear DNA counterstained with propidium iodide (red). Arrows show tail attachment points. Bar corresponds to 5 μm. B. Diagram of the distribution of chromosome FISH signals detected in nuclear sectors [I (basal) - IV (apical)] as defined in Figure 1A. Bar on the right shows confidence interval.
Figure 3.
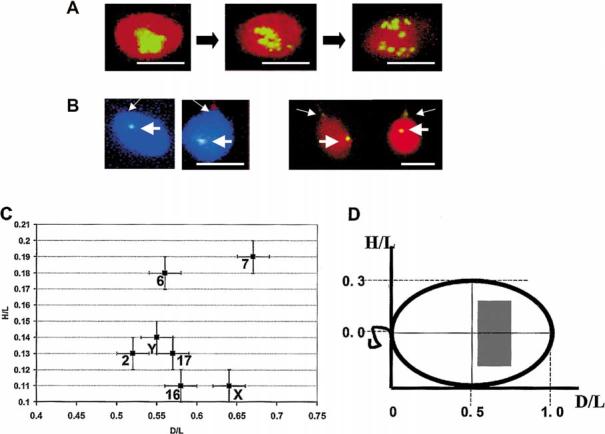
Determination of the intranuclear position of chromosomes using hybridization with centromere-specific probes. A. FISH localization of centromeres of all nonhomologous chromosomes in sperm nuclei using alphoid DNA probe. Typical patterns of FISH demonstrating dispersion of sperm chromocenter induced by cell swelling with Heparin in the presence of 10 mmol/L DTT. Panels from left to right correspond to 0 mg/ml, 0.01 mg/ml, and 0.1 mg/ml Heparin. B. Representative FISH patterns after hybridization with chromosome-specific centromere probes (thick arrows), left panels - CEN Y, total DNA counterstained with DAPI (blue); right panels - CEN 17, total DNA counterstained with PI (red). Thin arrows show tail attachment points. Bars in A and B correspond to 5 μm. C. Localization of individual centromeres within sperm nuclei. Each spot have the mean normalized coordinates experimentally determined (as explained in Methods, Figure 1B) for each chromosome-specific centromere. Bars show standard errors. D. Schematic view of the sperm nucleus showing the area within which all studied centromeres were localized (shaded rectangular). The same system of coordinates as in C is used.
FISH signals obtained using whole chromosome probes are large (Figure 1A), therefore, the intranuclear position of a chromosome maybe determined onlyroughly. We used the following procedure. Nuclear area was subdivided in four zones I-IV (Figure 1A), hits of the FISH signals were assigned to these zones, and percentage of hits into each zone from the total number of FISH signals were calculated for each painting probe.
The intranuclear position of centromeres was determined in a semi-quantitative way. Several geometrical parameters were measured within the sperm nucleus (Figure 1B): lengths of long (L) and small (l) axes, distances from hybridization signal to tail attachment point (D) and to the long axis (H). L and l describe nuclear size, their ratio l/L nuclear shape (roundness). The intranuclear position of hybridization signal was described in coordinates D and H (Figure 1B). To account for slight differences in nuclear swelling for individual cells normalized values D/L and H/L were calculated, averaged, and used to describe the absolute intranuclear position of a selected centromere.
To determine the relative intranuclear position of centromeres, distances dmn between centers of hybridization spots were measured (Figure 1C) and their normalized values dmn/L were used to account for slight variations in the nuclei sizes.
Results
Localization of chromosome territories
FISH using whole-chromosome painting probes showed that the territorial organization of chromosomes known for interphase nuclei is preserved in the human sperm (Haaf & Ward 1995, Zalensky et al. 1995). Because of the large and diffuse hybridization signals produced by the painting probes only a qualitative description of the chromosome position along the anterior-posterior axis L-L′ (Figure 1A) may be obtained. In this work, an intranuclear position of the chromosomes 1, 6, and X was analyzed. Figure 2A shows examples of hybridization patterns. Figure 2B presents the distribution of the chromosome territories along L-L′. In 46% of cells, the CHR 1 was found in the zone III and the CHR 6 is preferentially localized in the zone IV (47% of cells). Both chromosomes showed strong preferential location, in more than 70% of cells these chromosomes were found in the acrosomal half of the sperm nucleus (sectors III + IV). Thus, the data of Figure 2B indicate non-random positioning of chromosomes: CHR 6 has a tendency towards more anterior localization than CHR 1, and both are rarely found in the basal half of the nucleus. In the similar experiments, sex CHR X was found in zone IV in 67% of cells, thus this chromosome demonstrated the extreme anterior position.
Sperm chromocenter and centromere positioning
Centromeres (CEN) are traditionally used as markers describing nuclear positioning of the chromosomes in somatic cells (Nagele et al. 1999, Sun & Yokota 1999, Sun et al. 2000). In the human sperm nucleus, centromeres are collected in a compact chromocenter (Zalensky et al. 1993, 1995, Hoyer-Fender et al. 2000). The chromocenter may be visualized either by CENP-A immunolocalization or by FISH using alpha-satellite probe; both approaches illuminate the centromeres of all chromosomes (Zalensky et al. 1993). Controlled nuclear swelling that may be achieved by treatment of the sperm cells with Heparin/DTT (Zalensky et al. 1993) induces gradual disassembly of the chromocenter into linear arrays, and finally into individual centromeres (Figure 3A).
Specific adherence between the centromeres of nonhomologous chromosomes in human sperm mayresult in their fixed and non-random localization within nuclei. To test if such an order exists, we have used FISH with chromosome-specific centromeric probes to localize individual centromeres in the sperm cells (Figure 3B). We used standard conditions of nuclei swelling in 0.01 mg/ml Heparin, 10 mmol/L DTT for 30 min at room temperature. These conditions result in a slight dispersion of the compact chromocenter shown in middle panel of Figure 3A. For each of the sperm nuclei the normalized distances D/L and H/L (see scheme Figure 1B) were used as indicators of the centromere (chromosome) position (Table 1). Standard errors of the mean for both parameters are rather small, pointing to tight distribution. The t-test showed that differences between population means were statistically significant. For example, for sets of D/L values measured for centromere of chromosome 2 (CEN 2) and CEN 17 t = 2.62 and p = 0.009. To demonstrate that the position of a given centromere is non-random we performed a Kolmogorov-Smirnov (K-S) test using a uniform distribution as a theoretical expectation (see Materials and methods for details). The K-S test statistic was as follows: the cumulative distribution D varies from 0.24 to 0.62 for different data sets with corresponding p values 40.01. Therefore, centromeres distributions in minimally swollen sperm nuclei (Table 1) are statistically different from uniform, indicating that each centromere occupies a defined position.
Table 1.
Centromere positioning within human sperm nuclei. Mean normalized coordinates of centromere FISH signals and standard errors (SE) are shown. Mean dimensions of sperm nuclei studied were L=46.6±1.0, l=32.5±0.6, roundness l/L=0.7, see Figure 1 for the definition of parameters.
| CHR | 17 | 2 | 16 | X | Y | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Mean D/L | 0.57 | 0.52 | 0.58 | 0.64 | 0.55 | 0.56 | 0.67 |
| SE | 0.01 | 0.02 | 0.02 | 0.03 | 0.03 | 0.02 | 0.04 |
| Mean H/L | 0.13 | 0.13 | 0.11 | 0.11 | 0.14 | 0.18 | 0.19 |
| SE | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 |
Figure 3C provides another view of the same data. Here, a point with mean coordinates (H/L; D/L) shows the position of a given centromere. Note that a mirror position for each centromere is equallyprobable since we cannot distinguish between the two in the experiment. Surprisingly, we do not observe spreading in H/L coordinate of centromeres which has been expected because of assumed rotational symmetry along L-L′ (Figure 1). Therefore, there are limitations to how sperm cells maydeposit onto the surface of the microscopic slide. Apparently, only two positions are possible, similar to those realized during coin tossing. This indicates the existence of the dorsoventral polarityof human sperm cells.
Data presented in Figure 3C demonstrate that CEN 7 and CEN 6 are positioned most peripherally among seven centromeres studied; CEN X and CEN 16 are located most closelyto the long nuclear axis; CEN 7 and CEN X have most anterior positions.
In Figure 3D, the area of centromere localization is shown in the ‘unit’ sperm nuclei with length D/L = 1 and width H/L = 0.7 (experimentally determined maximal value). It is noteworthythat, in slightlydecondensed sperm cells, all centromeres are grouped within a limited area of nuclei shown bythe shaded rectangle. This area is centrallylocated and shifted towards the anterior. Such compact localization corresponds to the condensed chromocenter that was established in earlier studies bylabeling of all human centromeres (Zalensky et al. 1993, 1995).
Non-random relative localization of individual centromeres
The data above indicate that in human sperm nuclei, localization of the chromosomes, as judged by localization of their centromeres, is non-random. Therefore, it can be expected that the relative positioning of chromosomes might be non-random as well. To investigate the question we performed either two- or three-color FISH using combinations of chromosome-specific CEN probes (Figure 4A, B). Normalized intercentromere (CEN-CEN) distances dmn/L were measured in digital images as described in Methods. Table 2 presents the mean values calculated for each pair. It is seen that CEN 16 and Y, 16 and 15, 17 and 7 on average are located much closer to each other than CEN 16 and 17, or 16 and 18. Figure 4C gives a different representation of data. It is clear, for example, that CEN 17 is located much closer to CEN 7 than to CEN 15. Figure 4D provides an example of distance frequency distribution for two chromosome pairs 16/Y and 16/6. The frequency distributions are almost symmetrical, with the values of the mean distance equal to the median distance, and the difference observed between two chromosomal pairs is statistically significant. Thus, chromosome 16 is positioned closer to chromosome Y than to chromosome 6 in the majority of the sperm cells. Differences between the observed intracentromere distances with those expected by chance were estimated using a nonparametric statistical test as described in Methods. The cumulative distribution D in the K-S test varied from 0.11 to 0.18 for different data sets with a corresponding p value 40.03. In summary, the data of Table 2 and Figure 4 demonstrate preferences for specific relative positions of chromosomes in sperm nuclei.
Figure 4.
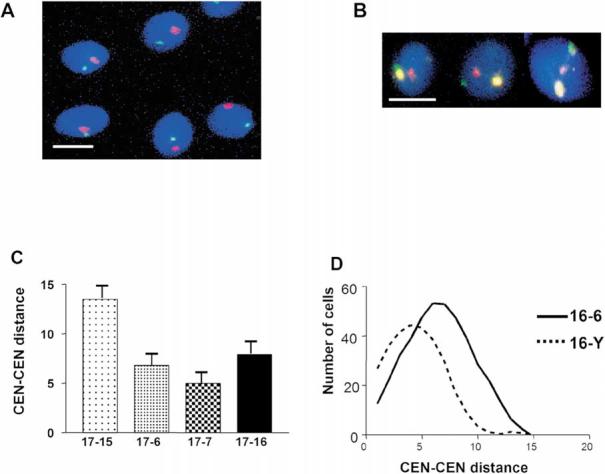
Determination of relative position of chromosomes using FISH with chromosome-specific centromere probes. Typical patterns of centromere localization by multicolor FISH which were used to measure intercentromere distances dmn, total DNA was counterstained with DAPI (blue). A. CEN16 (green), CEN18 (red). B. CEN16 (green), CEN17 (yellow) CEN X (red). Bars in A and B correspond to 5 μm. C. Chart of intercentromere distances between chromosome 17 and four selected chromosomes. Bars indicate standard errors. D. Frequency distribution plot for intercentromere distances for the chromosome pairs 16-6 and 16-Y.
Table 2.
Mean normalized distances between centromeres of selected chromosomes in human sperm nuclei (note that relative units of distance are different from those in Table 1).
| 16–6 | 16–7 | 16–15 | 16–17 | 16–18 | 16–X | 16–Y | 17–6 | 17–7 | 17–15 | 17–X | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean dmn/L | 6.4 | 7.1 | 5.4 | 7.9 | 6.5 | 5.8 | 4.1 | 6.8 | 5.0 | 5.7 | 6.8 |
| SE | 0.2 | 0.3 | 0.4 | 0.5 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.4 |
Discussion
Here we present three lines of evidence for a preferred, non-random spatial localization of the chromosomes within the nucleus of human spermatozoa. The partially asymmetrical nuclear shape, an existence of a defined spatial marker (tail attachment point), and haploid complement of chromosomes facilitated the experimental approach.
In the initial experiments, indications of an ordered chromosome positioning were obtained using FISH with whole chromosome painting probes (Figure 2). The apical half of sperm nuclei was the preferred area of localization for three chromosomes studied (CHR 1, CHR 6, and CHR X). Among these three chromosomes, CHR 6 had a tendencyto occupythe anterior position. Earlier, preferential localization of CHR X in the sub-acrosomal part of sperm was shown using a similar approach (Luetjens et al. 1999, Hazzouri et al. 2000). CHR 18 was found in the basal half in 60% of sperm cells (Luetjens et al. 1999), while CHR 13 did not demonstrate anyrestricted positioning (Hazzouri et al. 2000). It is noteworthythat sex chromosome X was found in a position close to the place of the first contact between sperm and egg, not onlyin humans but also in evolutionary distant marsupial and monotreme mammals (Greaves et al. 2003).
Because of the large size and diffuse shape of hybridization signals produced by the painting probes, it is di/cult to describe and evaluate chromosome position in quantitative terms. Therefore, in most of the experiments we used FISH with chromosome-specific centromeric probes. These DNA probes produce bright, spatially-confined hybridization signals (Figures 3A and 4A, B) and have been commonlyused to studychromosome positioning in cells of different types (Nagele et al. 1995, Sun & Yokota 1999, Gurevitch et al. 2001). Data presented in Table 1 and Figure 3C demonstrate that a selected position in the human sperm nucleus maybe attributed to each centromere. Analysis of the absolute intranuclear position of seven centromeres in sperm cells shows that they are located within a restricted space (Figure 3D). This is in good agreement with the result by Gurevitch and co-authors (2001). These authors showed that in human sperm, centromeres of five acrocentric chromosomes were located close to each other in an area occupying 8% of the nuclear mass. Such a compact gathering of centromeres presents a chromocenter (Zalensky et al. 1993) which is located centrallyand shifted towards the apical side of the sperm nuclei (Figure 3D). It also suggests that the association of nonhomologous centromeres into the sperm chromocenter is non-random. We have previouslyshown (Zalensky et al. 1993) that the human sperm chromocenter is composed of several linear arrays of centromeres that maybe visualized in mildlydecondensed sperm nuclei. Data in the present studyindicate that a specific linear order of centromeres may exist within these arrays.
Final evidence for the ordered positioning of chromosomes in human sperm came from the investigation of relative centromere localization. The data of Table 2 and Figure 4 demonstrate that distances between selected pairs of centromeres are non-random. The shortest mean distance among 11 centromere pairs studied was observed for the pair 16^Y. In a similar study(Tilgen et al. 2001), distances between another set of chromosome pairs (Y-1, Y-9, Y-16, X-1, X-9) were evaluated and each pair was found to have a nonrandom spatial distribution. The authors found that the Y-16 pair exhibits the smallest mean distance, which coincides with our finding for the same pair. Proximitybetween some nonhomologous chromosomes maydepend on fusion between heterochromatic regions. The data of Tilgen and co-authors (2001) indicate that the relative proximity(positioning) of chromosomes cannot be explained byassociations between heterochromatin blocks. Unique and specific adherence between centromeres in sperm may result in the observed non-random absolute localization of chromosomes (Table 1, Figure 3C).
The factors governing the ordered packaging of chromosomes in sperm and the mechanisms underlying the establishment and maintenance of such order are obscure. The possibilityof the universal spatial ordering of chromosomes that is transmitted through germline cells has been suggested in publications by Nagele and co-authors (1995, 1998). In view of this hypothesis, it would be appealing to compare the relative positioning of chromosomes in sperm and somatic cells.
Non-random positioning of chromosomes in the human sperm nucleus is yet another additional element of the well-defined genome architecture observed earlier (Zalensky et al. 1993, 1995, 1997, Haaf & Ward 1995, Hazzouri et al. 2000, Gurevitch et al. 2001, Tilgen et al. 2001). In the course of fertilization, the sperm chromosomes are withdrawn from the nucleus and remodeled. This latter process involves changes in higher-order chromatin structure, removal of the protamines, the accumulation of histones, and restoration of transcriptional competence (reviewed by McLay & Clarke 2003). It is suggested that these dynamic processes are dependent both on the environment in oocyte cytoplasm and on the precise topology of sperm chromosomes (Haaf 2001). Delayed decondensation of the sub-acrosomal nuclear region was observed following intracytoplasmic sperm injection (Terada et al. 2000), the procedure routinely used in assisted reproduction. Thus chromosomes, which are preferentiallylocalized at this apical region of the nucleus, would be remodeled and activated abnormally. In this context, further studies of sperm nuclear organization including chromosome positioning are not of theoretical importance only. Further experiments examining chromosome positioning in sperm and during fertilization are underwayin this laboratory.
Acknowledgement
The National Institute of Health grant R01 HD-42748 to A.O.Z has supported this work.
References
- Allison DC, Nestor AL. Evidence for a relatively random array of human chromosomes on the mitotic ring. J Cell Biol. 1999;145:1–14. doi: 10.1083/jcb.145.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T. Specific end-to-end attachment of chromosomes in Ornithogalum virens. J Cell Sci. 1979;38:357–367. doi: 10.1242/jcs.38.1.357. [DOI] [PubMed] [Google Scholar]
- Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, Chubb JR. Chromosome position: now, where was I? Curr Biol. 2003;13:357–359. doi: 10.1016/s0960-9822(03)00276-8. [DOI] [PubMed] [Google Scholar]
- Bojanowski K, Maniotis AJ, Plisov S, Larsen AK, Ingber DE. DNA topoisomerase II can drive changes in higher order chromosome architecture without enzymatically modifying DNA. J Cell Biochem. 1998;69:127–142. doi: 10.1002/(sici)1097-4644(19980501)69:2<127::aid-jcb4>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerinmutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Gasser SM. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cremer M, von Hase J, Volm T, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozortsev D, Coleman A, Nagy P, et al. Nucleoli in a pronuclei-stage mouse embryo are represented by major satellite DNA of interconnecting chromosomes. Fertil Steril. 2000;73:366–371. doi: 10.1016/s0015-0282(99)00491-4. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C, Walters MC, Groudine M, Martin DI. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centromeric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- Geraedts JP, Pearson PL. Spatial distribution of chromosomes 1 and Y in human spermatozoa. J. Reprod Fertil. 1975;45:515–417. doi: 10.1530/jrf.0.0450515. [DOI] [PubMed] [Google Scholar]
- Greaves IK, Rens W, Ferguson-Smith MA, Griffin D, Marshall-Graves JA. Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res. 2003;11:503–512. doi: 10.1023/a:1024982929452. [DOI] [PubMed] [Google Scholar]
- Gurevitch M, Amiel A, Ben-Zion M, Fejgin M, Bartoov B. Acrocentric centromere organization within the chromocenter of the human sperm nucleus. Mol Reprod Dev. 2001;60:507–516. doi: 10.1002/mrd.1116. [DOI] [PubMed] [Google Scholar]
- Haaf T. The battle of the sexes after fertilization: behaviour of paternal and maternal chromosomes in the early mammalian embryo. Chromosome Res. 2001;9:263–271. doi: 10.1023/a:1016686312142. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ward DC. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res. 1995;219:604–611. doi: 10.1006/excr.1995.1270. [DOI] [PubMed] [Google Scholar]
- Hazzouri M, Rousseaux S, Mongelard F, et al. Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev. 2000;55:307–315. doi: 10.1002/(SICI)1098-2795(200003)55:3<307::AID-MRD9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS. Comparative analysis of plant genome architecture. Symp Soc Exp Biol. 1996;50:17–23. [PubMed] [Google Scholar]
- Hoyer-Fender S, Singh PB, Motzkus D. The murine heterochromatin protein M31 is associated with the chromocenter in round spermatids and is a component of mature spermatozoa. Exp Cell Res. 2000;254:72–79. doi: 10.1006/excr.1999.4729. [DOI] [PubMed] [Google Scholar]
- Jennings C, Powell D. Genome organization in the murine sperm nucleus. Zygote. 1995;3:123–131. doi: 10.1017/s0967199400002495. [DOI] [PubMed] [Google Scholar]
- Joffe BI, Solovei IV, Macgregor HC. Ordered arrangement and rearrangement of chromosomes during spermatogenesis in two species of planarians (Plathelminthes) Chromosoma. 1998;107:173–183. doi: 10.1007/s004120050294. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Brown JK, Mosgoller W, Schwarzacher T, Heslop-Harrison JS. The spatial localization of homologous chromosomes in human fibroblasts at mitosis. Hum Genet. 1994;93:275–280. doi: 10.1007/BF00212022. [DOI] [PubMed] [Google Scholar]
- Luetjens CM, Payne C, Schatten G. Non-random chromosome positioning in human sperm and sex chromosome anomalies following intracytoplasmic sperm injection. Lancet. 1999;353:1240. doi: 10.1016/S0140-6736(99)80059-2. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Bojanowski K, Ingber DE. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J Cell Biochem. 1997;65:114–130. [PubMed] [Google Scholar]
- Marshall WF. Order and disorder in the nucleus. Curr Biol. 2002;12:185–192. doi: 10.1016/s0960-9822(02)00724-8. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca M, Muller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111:1363–1370. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- Nagele R, Freeman T, McMorrow L, Lee HY. Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science. 1995;270:1831–1835. doi: 10.1126/science.270.5243.1831. [DOI] [PubMed] [Google Scholar]
- Nagele RG, Freeman T, Fazekas J, Lee KM, Thomson Z, Lee HY. Chromosome spatial order in human cells: evidence for early origin and faithful propagation. Chromosoma. 1998;107:330–338. doi: 10.1007/s004120050315. [DOI] [PubMed] [Google Scholar]
- Nagele RG, Freeman T, McMorrow L, Thomson Z, Kitson-Wind K, Lee H. Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J Cell Sci. 1999;112:525–535. doi: 10.1242/jcs.112.4.525. [DOI] [PubMed] [Google Scholar]
- Oud JL, Mans A, Brakenhoff GJ, van Der Voort HT, van Spronsen EA, Nanninga N. Three-dimensional chromosome arrangement of Crepis capillaris in mitotic prophase and anaphase as studied by confocal scanning laser microscopy. J Cell Sci. 1989;92:329–339. doi: 10.1242/jcs.92.3.329. [DOI] [PubMed] [Google Scholar]
- Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12:425–432. doi: 10.1016/s0962-8924(02)02351-6. [DOI] [PubMed] [Google Scholar]
- Powell D, Cran DG, Jennings C, Jones R. Spatial organization of repetitive DNA sequences in the bovine sperm nucleus. J Cell Sci. 1990;97:185–191. doi: 10.1242/jcs.97.1.185. [DOI] [PubMed] [Google Scholar]
- Saifitdinova AF, Derjusheva SE, Malykh AG, Zhurov VG, Andreeva TF, Gaginskaya ER. Centromeric tandem repeat from the chaffinch genome: isolation and molecular characterization. Genome. 2001;44:96–103. doi: 10.1139/gen-44-1-96. [DOI] [PubMed] [Google Scholar]
- Sbracia M, Baldi M, Cao D, et al. Preferential location of sex chromosomes, their aneuploidy in human sperm, and their role in determining sex chromosome aneuploidy in embryos after ICSI. Hum Reprod. 2002;17:320–324. doi: 10.1093/humrep/17.2.320. [DOI] [PubMed] [Google Scholar]
- Solovei IV, Joffe BI, Hori T, Thomson P, Mizuno S, Macgregor HC. Unordered arrangement of chromosomes in the nuclei of chicken spermatozoa. Chromosoma. 1998;107:184–188. doi: 10.1007/s004120050295. [DOI] [PubMed] [Google Scholar]
- Sun HB, Yokota H. Correlated positioning of homologous chromosomes in daughter fibroblast cells. Chromosome Res. 1999;7:603–610. doi: 10.1023/a:1009279918034. [DOI] [PubMed] [Google Scholar]
- Sun HB, Shen J, Yokota H. Size-dependent positioning of human chromosomes in interphase nuclei. Biophys J. 2000;79:184–190. doi: 10.1016/S0006-3495(00)76282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat Res. 2002;504:37–45. doi: 10.1016/s0027-5107(02)00077-5. [DOI] [PubMed] [Google Scholar]
- Terada Y, Luetjens CM, Sutovsky P, Schatten G. Atypical decondensation of the sperm nucleus, delayed replication of the male genome, and sex chromosome positioning following intracytoplasmic human sperm injection (ICSI) into golden hamster eggs: does ICSI itself introduce chromosomal anomalies? Fertil Steril. 2000;74:454–460. doi: 10.1016/s0015-0282(00)00671-3. [DOI] [PubMed] [Google Scholar]
- Tilgen N, Guttenbach M, Schmid M. Heterochromatin is not an adequate explanation for close proximity of interphase chromosomes 1-Y, 9-Y, and 16-Y in human spermatozoa. Exp Cell Res. 2001;265:283–287. doi: 10.1006/excr.2001.5193. [DOI] [PubMed] [Google Scholar]
- Ward WS, Zalensky AO. The unique, complex organization of the transcriptionally silent sperm chromatin. Crit Rev Eukaryot Gene Expr. 1996;6:139–147. doi: 10.1615/critreveukargeneexpr.v6.i2-3.30. [DOI] [PubMed] [Google Scholar]
- Watson JM, Meyne J, Graves JA. Ordered tandem arrangement of chromosomes in the sperm heads of monotreme mammals. Proc Natl Acad Sci USA. 1996;93:10200–10205. doi: 10.1073/pnas.93.19.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters-Tyrou D, Martinage A, Chevaillier P, Sautiere P. Nuclear basic proteins in spermiogenesis. Biochimie. 1998;80:117–128. doi: 10.1016/s0300-9084(98)80018-7. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Breneman JW, Zalenskaya IA, Brinkley BR, Bradbury EM. Organization of centromeres in the decondensed nuclei of mature human sperm. Chromosoma. 1993;102:509–518. doi: 10.1007/BF00368344. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Allen MJ, Kobayashi A, Zalenskaya IA, Bradbury EM. Well-defined genome architecture in the human sperm nucleus. Chromosoma. 1995;103:577–590. doi: 10.1007/BF00357684. [DOI] [PubMed] [Google Scholar]
- Zalensky AO, Tomilin NV, Zalenskaya IA, Teplitz RL, Bradbury EM. Telomere-telomere interactions and candidate telomere binding protein(s) in mammalian sperm cells. Exp Cell Res. 1997;232:29–41. doi: 10.1006/excr.1997.3482. [DOI] [PubMed] [Google Scholar]


