Abstract
Hypoxia has recently been shown to activate the endoplasmic reticulum kinase PERK, leading to phosphorylation of eIF2α and inhibition of mRNA translation initiation. Using a quantitative assay, we show that this inhibition exhibits a biphasic response mediated through two distinct pathways. The first occurs rapidly, reaching a maximum at 1–2 h and is due to phosphorylation of eIF2α. Continued hypoxic exposure activates a second, eIF2α-independent pathway that maintains repression of translation. This phase is characterized by disruption of eIF4F and sequestration of eIF4E by its inhibitor 4E-BP1 and transporter 4E-T. Quantitative RT–PCR analysis of polysomal RNA indicates that the translation efficiency of individual genes varies widely during hypoxia. Furthermore, the translation efficiency of individual genes is dynamic, changing dramatically during hypoxic exposure due to the initial phosphorylation and subsequent dephosphorylation of eIF2α. Together, our data indicate that acute and prolonged hypoxia regulates mRNA translation through distinct mechanisms, each with important contributions to hypoxic gene expression.
Keywords: eIF2α, eIF4F, hypoxia, mRNA translation
Introduction
The presence of hypoxic and anoxic areas in human tumors is well documented, and is prognostic for poor outcome (reviewed in Harris, 2002; Wouters et al, 2002). The clinical importance of tumor hypoxia results from its ability to protect cells against both radiation and chemotherapy and from the fact that it can provide a selection pressure for apoptotically resistant cells (Graeber et al, 1996). Furthermore, the cellular response to hypoxia causes important changes in gene expression that affect cell behavior and influence patient prognosis. There has been particular focus on changes mediated through the family of hypoxia-inducible transcription factors (HIFs). HIF-1 and HIF-2 promote transcription of more than 60 putative downstream genes (for a review see Semenza, 2003) that affect hypoxia tolerance, energy homeostasis, angiogenesis and tumor growth. Although the transcriptional response to hypoxia is clearly very important (Ryan et al, 1998; Tang et al, 2004; Leek et al, 2005), tumor cells also experience short, transient exposures to hypoxia and/or anoxia that occur over time frames too fast for an effective transcriptional response. Transient changes in oxygenation occur owing to the abnormal vasculature found in most tumors, characterized by immature, leaky and improperly formed vessels. Perfusion of these vessels can change dynamically in time, leading to rapid but transient episodes of severe hypoxia in the tumor cells dependent upon them (Bennewith and Durand, 2004; Cardenas-Navia et al, 2004). Consequently, post-transcriptional responses are presumably important for adaptation to cycling oxygenation in tumors.
Control of mRNA translation during hypoxia is emerging as an important cellular response to hypoxia (Koumenis et al, 2002; Koritzinsky et al, 2005; Wouters et al, 2005). As protein synthesis is energy costly, inhibition of mRNA translation may represent an active response to prevent loss of energy homeostasis during hypoxia. Indeed, it has been shown that overall mRNA translation is severely but reversibly inhibited during hypoxia (Koumenis et al, 2002; Erler et al, 2004; Bi et al, 2005) with kinetics that precede ATP depletion (Lefebvre et al, 1993). Furthermore, regulation of mRNA translation can have a significant and rapid impact on individual gene expression. This is because the sensitivity of individual genes to changes in overall translation varies widely and in a manner that reflects the molecular mechanisms responsible for controlling translation (Johannes et al, 1999; Harding et al, 2000). Regulation of gene expression through control of mRNA translation is important during various pathologies including cancer (Holland et al, 2004). The mechanisms responsible for inhibiting translation during hypoxia are not yet fully understood.
We have previously investigated the involvement of the endoplasmic reticulum (ER) kinase PERK in the hypoxia-induced downregulation of protein synthesis (Koumenis et al, 2002). PERK is activated as part of the evolutionarily conserved unfolded protein response (UPR) (reviewed in Schroder and Kaufman, 2005). It phosphorylates eIF2α, a subunit of eIF2, which in its GTP-bound form recruits the aminoacylated tRNA to the 40S ribosomal subunit. The exchange of GDP for GTP is mediated by the guanine nucleotide exchange factor eIF2B. Ser51-phosphorylated eIF2α inhibits eIF2B, resulting in inhibition of translation initiation. eIF2α phosphorylation results in a set of molecular events collectively termed the integrated stress response. These include the inhibition of global mRNA translation in conjunction with induced expression of the transcription factor ATF4 and its downstream target genes (Harding et al, 2003). We showed that hypoxia rapidly activated PERK, which led to reversible phosphorylation of eIF2α (Koumenis et al, 2002). Hypoxia-induced inhibition of protein synthesis was severely attenuated in cells without functional PERK. After prolonged periods of hypoxia, PERK-deficient cells did show partial inhibition, suggesting that protein synthesis is regulated through additional mechanisms.
Another candidate mechanism for inhibiting translation during hypoxia is disruption of the cap-binding protein complex eIF4F, which consists of eIF4E, eIF4A and eIF4G (for recent reviews see Gebauer and Hentze, 2004; Holcik and Sonenberg, 2005). eIF4E participates in a protein bridge between the mRNA and the ribosome by its simultaneous interaction with the mRNA 5′ cap structure and the large scaffolding protein eIF4G, which in turn interacts with eIF3 that is bound to the 40S ribosomal subunit. eIF4E is regulated through a set of binding proteins (4E-BPs) that bind reversibly to eIF4E in their hypophosphorylated form, and this obstructs the interaction between eIF4E and eIF4G. The 4E-BP1 protein becomes hyperphosphorylated in response to a number of stimuli, such as insulin, hormones, growth factors, mitogens and cytokines, as a result of activation of the PI3-kinase/Akt/FRAPmTOR pathway (Hay and Sonenberg, 2004).
It remains unclear to what degree the lack of eIF4F assembly contributes to inhibition of translation during tumor hypoxia. Several studies have investigated the combined consequences of ischemia/reperfusion on eIF4F-related proteins in rat brains (reviewed in DeGracia et al, 2002). Proteolysis of eIF4G was reported during ischemia and reperfusion in vivo (Neumar et al, 1998; Martin de la Vega et al, 2001), but not in neuronal cells cultured in vitro (NGF differentiated PC12 cells) (Martin et al, 2000). The reports addressing the expression and phosphorylation status of eIF4E during ischemia are conflicting, but 4E-BP1 dephosphorylation has been demonstrated both in vivo and in vitro (Martin et al, 2000; Martin de la Vega et al, 2001). The acuteness and complexity of ischemia/reperfusion stress and the high sensitivity of neurons to deprivation and reconstitution of both oxygen and nutrients are distinct properties of this model system and thus difficult to extrapolate to tumor hypoxia. In rat hepatocytes, 4E-BP1 becomes dephosphorylated and associates with eIF4E rapidly (15–60 min) upon mild hypoxia, but this could not explain the observed downregulation of protein synthesis (Tinton and Buc-Calderon, 1999). More recently, it was reported that hypoxia could influence 4E-BP1 phosphorylation by affecting the activity of mTOR (Arsham et al, 2003). Serum-starved and hypoxic human embryonic kidney cells failed to activate mTOR, phosphorylate 4E-BP1 and dissociate 4E-BP1 from eIF4E in response to insulin treatment. Nonetheless, it remains unknown whether hypoxia alone is sufficient to disrupt the eIF4F complex and to what extent this influences overall translation during hypoxia. Here we show that hypoxia induces a biphasic inhibition of mRNA translation characterized by transient phosphorylation of eIF2α and subsequent dissociation of eIF4F. These two mechanisms operate independently of each other and both have important consequences for gene expression during hypoxia.
Results
Kinetics of translation inhibition
To determine the effects of hypoxia on mRNA translation initiation in HeLa cells, we examined the association of ribosomes with mRNA at various time points. In this assay, the number of ribosomes found within the ‘polysomal' fraction of mRNA (mRNA containing two or more ribosomes) is a reflection of de novo protein synthesis. This technique is advantageous to other methods such as 35S incorporation, which requires prior amino-acid starvation, a procedure that can itself influence translation initiation (Kimball and Jefferson, 2000). Figure 1A shows that at all time points examined, hypoxia causes a large decrease in polysomal mRNA and a corresponding increase in free ribosomes and ribosomal subunits. The reduction in translation is not influenced by cell death, as cell viability remains above 90% following 16 h of hypoxia (data not shown). Furthermore, the inhibition of translation is completely reversible upon reoxygenation (data not shown).
Figure 1.
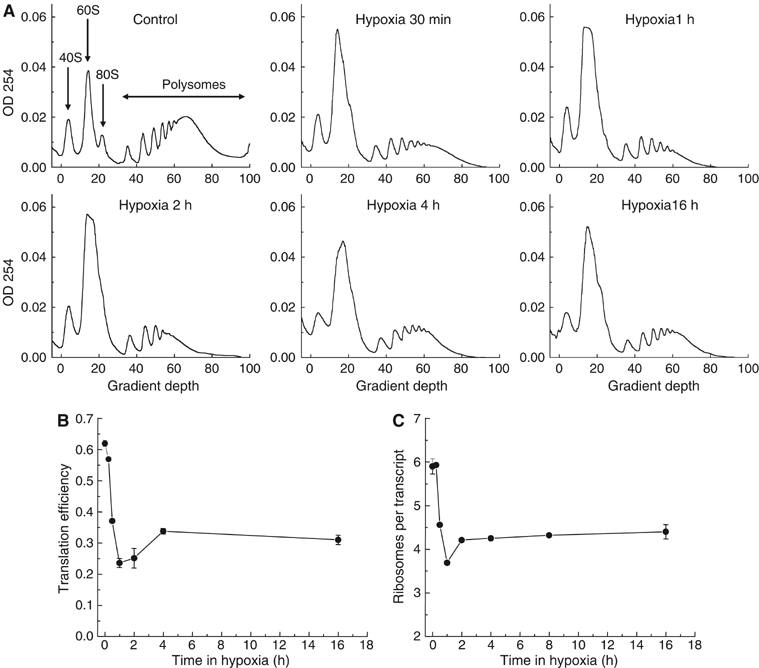
Hypoxia inhibits mRNA translation. HeLa cells were exposed to 0.0% O2 for 0–16 h and cell lysates were separated on a sucrose gradient. (A) The optical density (OD) at 254 nm is shown as a function of gradient depth for each time point. Actively translated mRNA is associated with high-molecular-weight polysomes deep in the gradient. (B) Translation efficiency in HeLa cells as a function of time in 0.0% O2. As a measure of overall translation efficiency, the relative amount of rRNA participating in polysomes was estimated. This fraction is proportional to the integrated area under the curve containing polysomes, as marked in (A). (C) The average number of ribosomes per mRNA in the polysomes as a function of time in 0.0% O2. This was calculated by differential integration of the profiles in (A).
To assess quantitatively overall mRNA translation from the polysome profiles, we calculated the percentage of rRNA participating in polysomes and defined this as the overall translation efficiency. This value is reduced from 62 to 24% after 1 h of hypoxia, and then recovers somewhat stabilizing at ∼30% (Figure 1B). The drop in translation reproducibly exhibited this biphasic response with maximum inhibition after 1–2 h, followed by a small recovery. The magnitude of inhibition is comparable to that observed following complete disruption of the cellular redox environment with 1 mM dithiothreitol (DTT) (17%) (data not shown).
Analysis of the polysome profiles in Figure 1A shows that hypoxia also causes a change in the distribution of the polysomal mRNA, with proportionally less signal in the higher molecular weight fractions. This indicates that the average number of ribosomes per mRNA transcript is also decreased during hypoxia, reflecting a reduction in translation initiation efficiency even for those transcripts that remain translated. From the polysome profiles, we calculated the average number of ribosomes per translated transcript (i.e. mRNAs containing two or more ribosomes) at different time points during hypoxia (Figure 1C). The kinetics of this parameter follow in large part that of the overall translation.
eIF2α regulates translation during acute hypoxia
The eIF2α kinase PERK is at least partly responsible for protein synthesis inhibition during acute hypoxia, as measured by radioactive labeling of newly synthesized proteins (Koumenis et al, 2002). Thus, we hypothesized that the rapid inhibition and subsequent partial recovery in translation is due to changes in eIF2α phosphorylation. Indeed, we found that the phosphorylation of eIF2α is greatest after 1–2 h and then decreases by 8 h of hypoxia in several cell lines (Figure 2A). ATF4 protein levels also increase and then decrease during hypoxia in a manner that mirrors eIF2α phosphorylation. The dynamics of eIF2α phosphorylation and ATF4 protein induction thus correlate with the initial inhibition of translation and its subsequent recovery.
Figure 2.
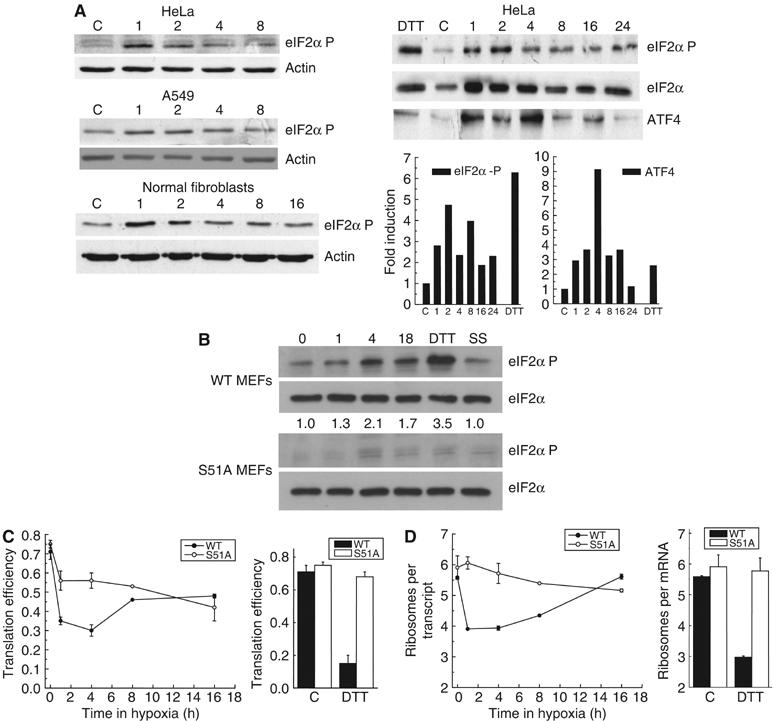
Inhibition of translation during acute hypoxia is dependent on eIF2α. HeLa cells, A549 cells, human normal fibroblasts (NF) and WT or S51A MEFs were exposed to 0.0% O2 for 0–16 h, 1 mM DTT or serum starvation (SS) for 1 h. Cell lysates were separated by SDS–PAGE. Immunoblots for (A) HeLa, A549 and NF or (B) MEFs were performed using antibodies against total or phosphorylated eIF2α, ATF4 and β-actin. In (A), optical densitometry for phosphorylated eIF2α or ATF4 normalized by total eIF2α is also shown. Total eIF2α expression has previously been shown to be constant during hypoxia (Koumenis et al, 2002). (C) Cell lysates were separated on a sucrose gradient, and OD at 254 nm was recorded. Translation efficiency as a function of time in 0.0% O2 in WT and S51A MEFs was estimated as in Figure 1. (D) Average number of ribosomes per mRNA in the polysomes in WT and S51A MEFs as a function of time in 0.0% O2 was calculated as in Figure 1.
To assess the requirement of eIF2α phosphorylation for translation inhibition during hypoxia we examined the response of mouse embryo fibroblasts (MEFs) derived from eIF2α knock-in mice containing an S51A mutation (Scheuner et al, 2001). As expected, these cells were defective in phosphorylation of eIF2α during hypoxia (Figure 2B). The translation efficiency in wild-type (WT) MEFs is similar to that in HeLa cells, with a rapid drop during acute hypoxia followed by a partial recovery (Figure 2C). In contrast, S51A MEFs display a substantial defect in their ability to inhibit translation during the initial phase. Nonetheless, after 16 h of hypoxia, both cell lines show a similar loss in translation efficiency. These data indicate that eIF2α phosphorylation is indeed necessary for inhibition of translation during acute hypoxia, but not at later times.
When the polysome profiles are analyzed in terms of the average number of ribosomes per translated transcript, S51A MEFs exhibit an even stronger defect in their response during acute hypoxia. Despite a small but detectable drop in translation efficiency during the first 4 h of hypoxia (Figure 2C), S51A MEFs show no decrease in the average number of ribosomes per translated transcript (Figure 2D). The same result was found in cells treated with DTT, a known activator of PERK that causes eIF2α phosphorylation. In contrast, WT MEFs show a strong reduction in average ribosomes per transcript during both acute hypoxia and DTT treatment. Interestingly, after 8 h of hypoxia, the average number of ribosomes per translated transcript increases again toward normal levels in WT cells and is equivalent to that in S51A MEFs by 16 h. These data provide further evidence that the inhibition of translation that occurs after acute and prolonged hypoxia is mechanistically distinct.
Disruption of the eIF4F complex during hypoxia
The assembly of the cap-binding complex eIF4F is a common control point for translation initiation and was thus a likely candidate for maintaining low rates of translation during prolonged hypoxia. We examined the levels of eIF4E and proteins that associate with it as an active complex (eIF4GI) or as an inactive complex (4E-BP1). Figure 3A shows that the levels of eIF4E do not change during hypoxia. In contrast, 4E-BP1 (Figure 3B) shows both a small induction at 8 h and a strong dephosphorylation after 16 h of hypoxia. This protein runs as different migrating bands representing different phosphorylation levels (Pause et al, 1994). The fastest migrating band is substantially increased after 16 h of hypoxia, and represents the hypophosphorylated 4E-BP1, which is known to have a higher affinity for eIF4E. A small decrease in the abundance of the scaffold protein eIF4GI (Figure 3C) was observed after 8 h, consistent with a decrease in its rate of synthesis measured in a microarray study using polysomal RNA (unpublished data). Overexposure of the blots indicated no reproducible changes in the cleavage of eIF4G. The influence of hypoxia on 4E-BP1 phosphorylation appears to be largely independent of eIF2α phosphorylation, as it is not differentially affected in the WT and S51A MEFs (unpublished data). However, until the relative contributions of various upstream signaling pathways to 4E-BP phosphorylation under hypoxia are better understood, it is premature to conclude that no connection between eIF2α and eIF4F exists.
Figure 3.
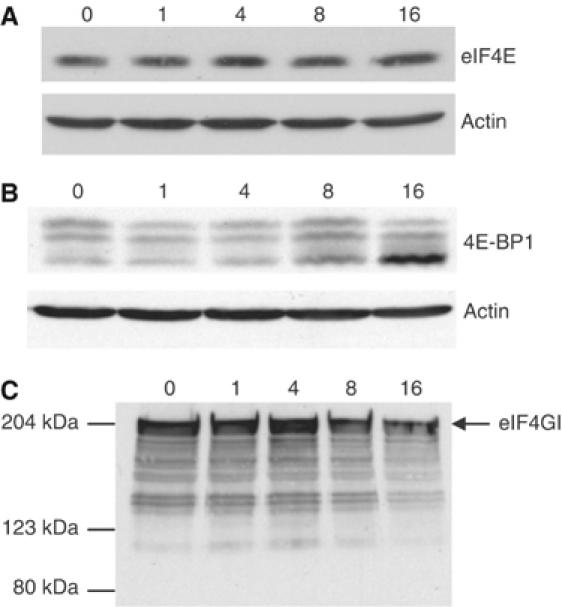
Expression of eIF4E, 4E-BP1 and eIF4GI during hypoxia. HeLa cells were exposed to 0.0% O2 for 0–16 h and cell lysates separated by SDS–PAGE. Immunoblots were performed using antibodies against actin, (A) eIF4E, (B) 4E-BP1 and (C) eIF4GI. The phosphorylation forms of 4E-BP1 have different electrophoretic mobilities and are represented by several bands on the immunoblot. Full-length eIF4GI run at about 220 kDa; the blot is overexposed to detect cleavage products.
To more strictly assess the influence of hypoxia on eIF4F, we investigated the association of eIF4E with eIF4GI and eIF4GII as well as with its inhibitor 4E-BP1 in HeLa cells. During aerobic conditions where translation is efficient, eIF4E is associated with large amounts of both eIF4GI and eIF4GII, and only a small amount of 4E-BP1 (cap lanes in Figure 4A and B). Cap-associated eIF4G migrated somewhat slower than the overall pool of eIF4G, suggesting a possible modification of this phospho-protein when bound to the cap. In contrast, after 4 or 16 h of hypoxia, there is a dramatic loss in binding to both eIF4GI and eIF4GII, indicating dissociation of the eIF4F complex. At 16 h, this dissociation correlates with a large increase in binding between eIF4E and 4E-BP1, consistent with the increase in the hypophosphorylated levels of 4E-BP1 at this time. It also correlated with decreased phosphorylation of eIF4E (Supplementary Figure S1) at 16 h, but the physiological significance of this remains unclear. However, although dissociation of eIF4G and eIF4E is complete after 4 h of hypoxia, a corresponding change in eIF4E phosphorylation or eIF4E/4E-BP1 association is not seen at this time point. This suggests that a mechanism distinct from 4E-BP1 dephosphorylation may also inhibit eIF4F during hypoxia.
Figure 4.
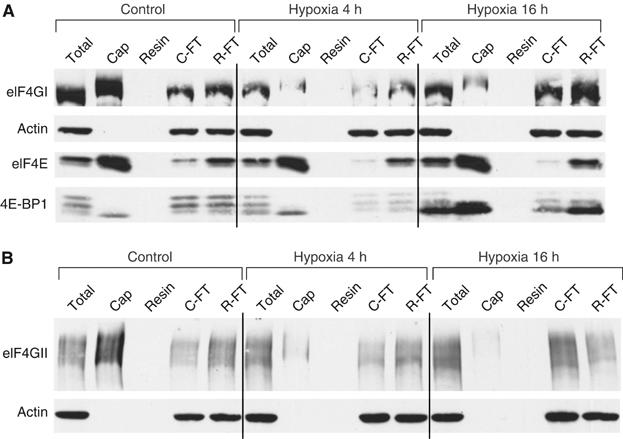
eIF4F is disrupted during prolonged hypoxia. HeLa cells were exposed to 0.0% O2 for 0–16 h and cell lysates probed for the presence of various eIF4E complexes. Lysates were incubated with an m7-cap analogue (‘Cap') or uncapped resin as a negative control. Immunoblots were performed with antibodies against actin, (A) eIF4GI, eIF4E, 4E-BP1 and (B) eIF4GII. ‘Cap': proteins bound to the capped resin; ‘Resin': proteins bound to the uncapped resin; ‘C-FT': unbound fraction after incubation with capped resin; ‘R-FT': unbound fraction after incubation with uncapped resin.
Translocation of eIF4E by 4E-T
A potential cause of eIF4F disruption that has not been well characterized is the translocation of eIF4E to the nucleus or to cytoplasmic bodies of mRNA processing (P-bodies). A 5–20% fraction of eIF4E is known to localize to the cell nucleus (Lejbkowicz et al, 1992). The shuttling protein 4E-T is the only known regulator of eIF4E localization and is capable of binding and transporting it to the cell nucleus (Dostie et al, 2000). eIF4E also colocalizes with 4E-T in P-bodies, where mRNA is degraded or stored (Andrei et al, 2005). Hypoxia caused a redistribution of both eIF4E and 4E-T from predominantly cytoplasmic staining under aerobic conditions to substantial nuclear staining during hypoxia (Figure 5A–C). This redistribution occurred progressively over time in hypoxic conditions, correlating with the gradual dephosphorylation of 4E-T (Figure 5D). In addition, hypoxic cells exhibit significant eIF4E and 4E-T staining in the perinuclear area, which may be associated with the nuclear envelope or the ER. Interestingly, hypoxia also increased the number of 4E-T speckles, which have been described as P-bodies (Ferraiuolo et al, 2005).
Figure 5.
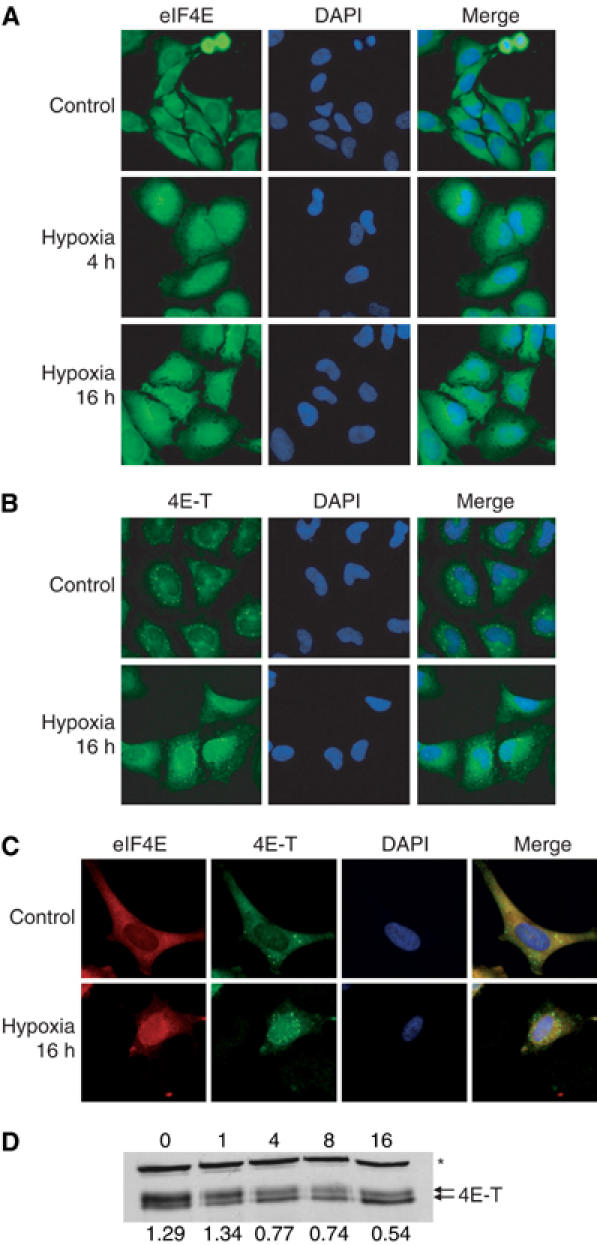
4E-T and eIF4E relocalize during hypoxia. HeLa cells were treated with 0.0% O2 for 0–16 h. Cells were stained with DAPI and (A) a polyclonal antibody against eIF4E, (B) a polyclonal antibody against 4E-T or (C) a monoclonal antibody against eIF4E and a polyclonal antibody against 4E-T. Cells were visualized by confocal microscopy and individual pictures merged to determine colocalization. (D) Cell lysates were separated by SDS–PAGE and immunoblots performed using antibodies against 4E-T. The ratio of the individual bands was quantified with optical densitometry. A crossreacting band is indicated (*).
Gene-specific regulation of translation
As translation efficiency is highly gene specific, we anticipated that individual genes would show different patterns of translation efficiency during acute and prolonged hypoxia. To investigate this, we fractionated polysomal mRNA and subsequently measured the mRNA abundance of individual genes by quantitative RT–PCR (Figure 6A). We first confirmed that concomitant with an increase in polysome association, the non/subpolysomal abundance decreased (Supplementary Figure S2). Subsequently, we quantified both the transcript recruitment and distribution within the polysomes (expressed as the relative fraction of translated transcripts and the average number of ribosomes per translated transcript, respectively).
Figure 6.
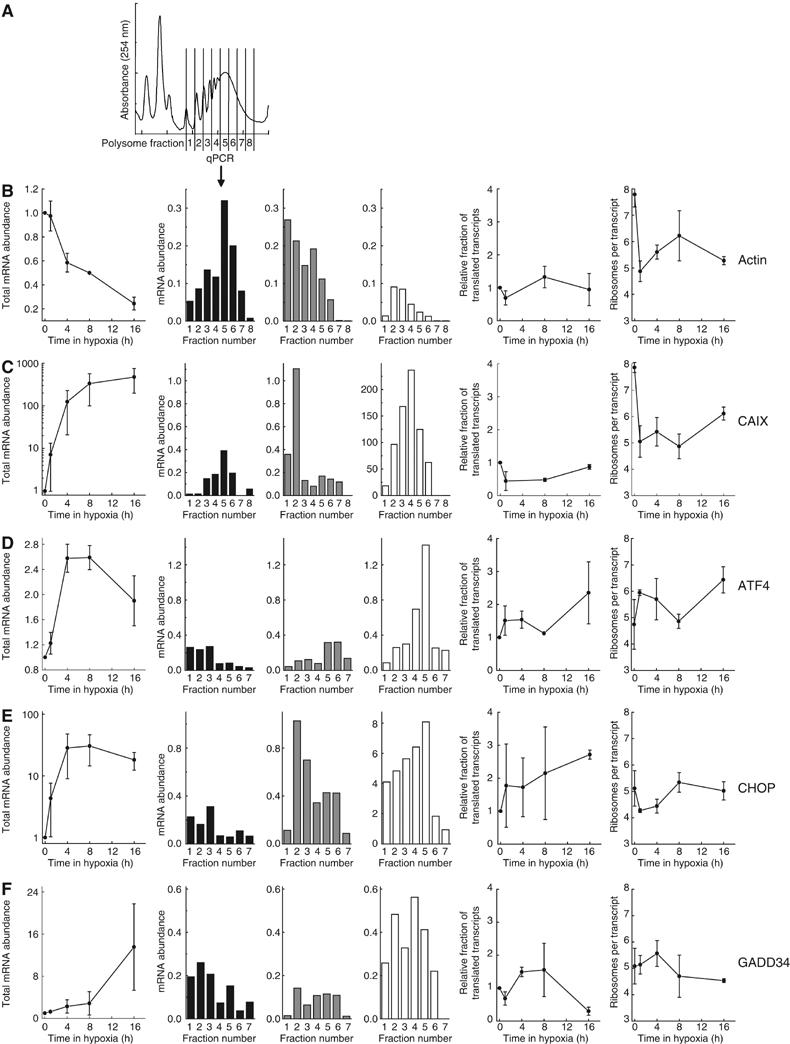
Gene-specific regulation of translation during hypoxia. HeLa cells were exposed to 0.0% O2 for 0–16 h and cell lysates were separated on sucrose gradients. (A) Fractions were collected as indicated, RNA was isolated and reverse transcribed. Thereafter, the total mRNA abundance of (B) β-actin, (C) CAIX, (D) ATF4, (E) CHOP and (F) GADD34 was determined using real-time quantitative PCR. The left panel shows total mRNA levels from unfractionated samples, normalized by 18S rRNA signal. The following three panels use black, gray and white bars to represent the gene abundance in polysome fractions following 0, 1 or 16 h hypoxia, respectively. The last two graphs show components of translation efficiency. This includes the relative fraction of transcripts in polysomes (i.e. corrected for total mRNA abundance) and the average number of ribosomes per mRNA. Graphs show the average from two independent experiments, and the histograms show the results from one representative experiment.
We first measured the translational profile of the housekeeping gene β-actin (Figure 6B). In aerobic cells, it is efficiently translated with a majority of the mRNA in polysome fractions 5 and 6. After 1 h of hypoxia, there is a marked reduction in translation, as evidenced by a shift toward the lower polysome fractions, which recovers considerably by 16 h. The drop in translation efficiency at 1 h is due to reductions in the relative fraction of translated mRNA and in the average number of ribosomes per translated transcript (Figure 6B). At later time points, only the average number of ribosomes per transcript remained low. The kinetic changes in translation efficiency for β-actin are similar to those observed for overall translation efficiency.
Many proteins are induced at the transcriptional level by hypoxia and we suspected that these genes might be preferentially translated during hypoxia. We investigated the translation of the HIF-1 target gene carbonic anhydrase IX (CAIX), which is important for tumor cell growth and survival during hypoxia (Robertson et al, 2004). Figure 6C shows an ∼500-fold transcriptional induction of CAIX during hypoxia. Polysome analysis indicates that, similar to β-actin, CAIX is initially efficiently translated but severely inhibited after 1 h of hypoxia. A significant restoration of the polysome distribution occurs after 16 h and thus ensures protein synthesis at this time where there is also significantly more cellular mRNA. As for β-actin, the initial inhibition of CAIX translation is due to a drop in the recruitment of the mRNA into polysomes and in the number of ribosomes per transcript. However, during prolonged hypoxia, the mRNA recruitment recovers and lower translation efficiency is attributed only to a small reduction in the average number of ribosomes per transcript.
ATF4 is a central transcription factor mediating the UPR following ER stress. Both thapsigargin (which causes ER stress) and 16 h of hypoxia result in eIF2α phosphorylation and translational induction of ATF4 in a PERK-dependent manner (Harding et al, 2000; Blais et al, 2004; Bi et al, 2005). Under normal conditions, the translation efficiency of this gene is low, with most of the mRNA found in fractions 1–3 (Figure 6D). In direct contrast to β-actin and CAIX, its translation is substantially increased during acute hypoxia, due to increased recruitment into the polysomes and an increase in the average number of ribosomes per transcript. In agreement with Blais et al (2004), we also observed a further increase in ATF4 translation efficiency during prolonged hypoxia.
An important transcriptional target of ATF4 is the C/EBP transcription factor CHOP (Fawcett et al, 1999), which induces cell cycle arrest and apoptosis during ER stress. Figure 6E shows that CHOP is regulated both transcriptionally and translationally by hypoxia. Translation is only moderately inhibited during acute hypoxia, as shown by a drop in the average number of ribosomes per transcript. However, this reduction is much smaller than average overall reduction (Figure 1C) and the reductions observed for both β-actin and CAIX. After 16 h of hypoxia, translation of CHOP is stimulated, as indicated by a recovery in the number of ribosomes per transcript and a marked increase in the fraction of translated mRNA.
Recovery from ER stress requires the GADD34 gene, which is induced in a PERK-dependent (Novoa et al, 2001) and CHOP-dependent (Marciniak et al, 2004) manner. GADD34 stimulates the activity of PP1c to dephosphorylate eIF2α. We found that, like CHOP, GADD34 is regulated both transcriptionally and translationally during hypoxia. Interestingly, its translation efficiency is highest after 4 h of hypoxia, which coincides with the start of recovery from eIF2α phosphorylation and overall translation inhibition (Figure 6F). In contrast, GADD34 mRNA is unable to completely bypass the translation inhibition after 16 h.
Gene-specific regulation of translation—dependence upon eIF2α
The gene-specific changes in translation noted above likely reflect the underlying eIF2α- and eIF4F-dependent mechanisms of translation control during hypoxia. We thus analyzed gene-specific translation in WT and S51A MEFs to establish the dependence of individual genes on eIF2α regulation (Figure 7). In contrast to WT MEFs, S51A MEFs show no loss in translation efficiency of β-actin or CAIX during acute hypoxia (Figure 7A and B). The loss in translation efficiency for these genes in the WT cells is similar to that observed in HeLa cells and is due primarily to a reduction in the average number of ribosomes per transcript. For both these genes, S51A MEFs show virtually no reduction in this parameter during the acute phase of hypoxia. However, in contrast to acute hypoxia, the translation efficiency during prolonged hypoxia is similar for these two genes in both cell lines.
Figure 7.
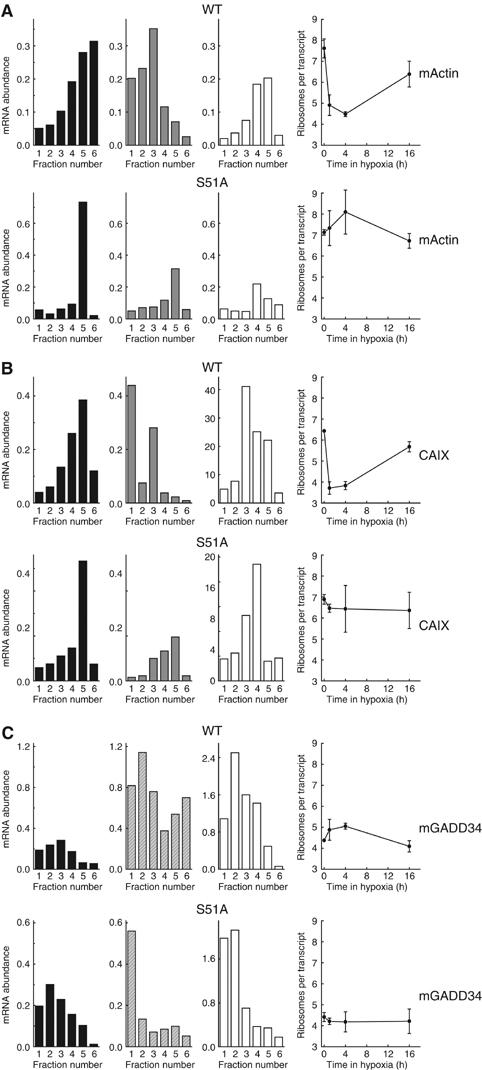
Dependence on eIF2α-P for gene-specific translational regulation. WT and S51A MEFs were exposed to 0.0% O2 for 0–16 h and cell lysates were separated on sucrose gradients. RNA was isolated from polysome fractions and reverse transcribed. Thereafter, the abundance of (A) β-actin, (B) CAIX and (C) GADD34 was determined using real-time quantitative PCR. The first three panels use black, solid gray, hatched grey and white bars to represent the gene abundance in polysome fractions following 0, 1, 4 or 16 h hypoxia, respectively. The following graph depicts one component of translation efficiency, that is, the average number of ribosomes per mRNA.
For ATF4 (Supplementary Figure S2), CHOP (data not shown) and GADD34 (Figure 7C), acute hypoxia causes a stimulation of translation in WT MEFs that is similar to HeLa cells. However, the translational induction is entirely absent in S51A MEFs. The increase in translation efficiency for GADD34 in WT MEFs during acute hypoxia results mainly from an increase in the average number of ribosomes per transcript (Figure 7C). Cells that are defective in eIF2α phosphorylation show impaired regulation of this parameter. Thus, for all genes examined, the observed changes in translation efficiency during acute hypoxia are dependent on eIF2α phosphorylation.
Discussion
Rapid and persistent downregulation of protein synthesis is thought to be a means of energy preservation and to protect against the lethal effects of hypoxia (Koumenis et al, 2002; Wouters et al, 2005). Here we show that the inhibition of global mRNA translation during hypoxia exhibits a biphasic response (Figure 8). The initial rapid inhibition (i.e. 15 min–4 h) is primarily dependent on eIF2α phosphorylation, whereas inhibition during prolonged hypoxia is independent of eIF2α. Phosphorylation of eIF2α under conditions of anoxia is extremely rapid, occurring almost as quickly as we can establish hypoxia in our system (15–30 min). We have previously shown that eIF2α is also phosphorylated under more moderate hypoxic conditions, although to a smaller degree and after longer times (Koumenis et al, 2002). We speculate that this rapid anoxic response may be especially important during the acute exposures to hypoxia/anoxia that frequently occur in tumors due to the transient opening and closing of blood vessels. This rapid response may explain the importance of eIF2α and ATF4 in the tolerance of cells to oxidative stress, which also occurs during hypoxia/reoxygenation cycles (Harding et al, 2003). This hypothesis is supported by a recent study by Bi et al (2005), who showed that activation of the PERK–eIF2α pathway during hypoxia contributes to overall tumor growth. Human tumor cells expressing a dominant-negative PERK allele as well as MEFs lacking PERK or expressing the S51A eIF2α produce smaller tumors with increased cell death in hypoxic areas than their WT counterparts (Bi et al, 2005). Thus, although activation of eIF2α phosphorylation in response to hypoxia is transient, this response appears critical for long-term cell survival within hypoxic regions of tumors.
Figure 8.
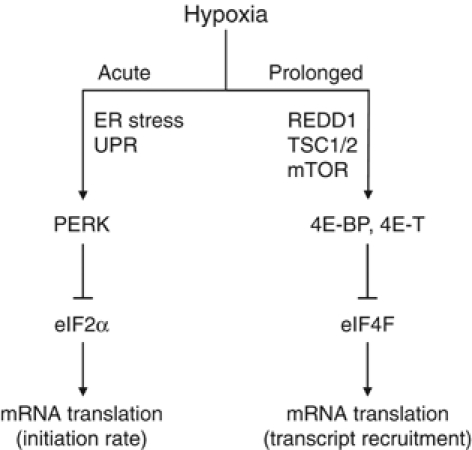
Model of the effects of hypoxia on overall mRNA translation. Acute hypoxia causes transient eIF2α phosphorylation due to PERK activation as a part of the UPR. This results in inhibition of the rate of translation initiation. Following prolonged hypoxic conditions, activation of 4E-BP and 4E-T causes disruption of eIF4F, which inhibits the recruitment of mRNA to polysomes. Both molecular mechanisms affect specific mRNAs to varying degrees, resulting in differential gene expression.
Inhibition of translation during prolonged anoxia is associated with disruption of the mRNA cap-binding complex eIF4F and sequestration of eIF4E by both dephosphorylated 4E-BP1 and 4E-T. To our knowledge, this report is the first to show a physiological stress-induced regulation of the localization of eIF4E and its transporter 4E-T. Accumulation of eIF4E in the cell nucleus or P-bodies renders it unavailable for cytoplasmic translation. Relocalization of eIF4E may have additional roles apart from reducing cap-dependent protein translation, including effects on mRNA processing, transport and degradation. Moderate hypoxia (1%) has also been shown to affect the eIF4F complex through inhibition of mTOR (Arsham et al, 2003). However, the kinetics and relative contribution of the eIF4F pathway on inhibition of global and gene-specific translation during more moderate hypoxia remain to be determined (see below).
Analysis of mRNA distribution within polysomes at different times also revealed interesting mechanistic differences during acute and prolonged hypoxia. Acute hypoxia caused a substantial drop in the average number of ribosomes per translated transcript. This presumably results from a reduction in the rate at which ribosomal subunits can be loaded onto mRNA, as each subunit requires a new nonphosphorylated eIF2α molecule. Inhibition of translation during prolonged hypoxia via eIF4F did not show this effect. Instead, translation was suppressed mainly by reducing the fraction of mRNA found within the polysomes. These results are consistent with a model in which the eIF4F cap-binding complex remains bound to the mRNA allowing sequential rounds of initiation by available eIF2α complexes. When translation is inhibited via eIF4F, many transcripts will lack this cap-binding complex and hence will not be competent to initiate translation. However, those transcripts that do contain eIF4F will be translated normally (Figure 8). A similar phenomenon has been observed for a subpopulation of mRNAs that contain 5′ terminal oligopyrimidine tracts (5′TOPs), which alternate between translationally repressed and active states in response to various stimuli (reviewed in Meyuhas and Hornstein, 2000). It is thus conceivable that the overall reduction in mRNA translation observed during prolonged hypoxia affects a subset of genes, such as those with 5′TOPs, to a greater degree than others.
An important issue that arises from our study is the nature of the oxygen-sensing pathways upstream of eIF2α and eIF4F. Substantial evidence suggests that the oxygen sensors are largely independent of the HIF oxygen-sensing pathway (Jaakkola et al, 2001; Koumenis et al, 2002). In the case of eIF2α, its phosphorylation occurs in an HIF-independent manner. Instead, it requires PERK activation (Koumenis et al, 2002) and is associated with activation of the UPR in response to ER stress (Romero-Ramirez et al, 2004; Bi et al, 2005). The upstream signaling that leads to eIF4F disruption is less clear, with perhaps both HIF-dependent and -independent components. Hypoxia has been shown to prevent insulin stimulation of mTOR and phosphorylation of its substrate 4E-BP1 during conditions of moderate hypoxia and serum starvation (Arsham et al, 2003). Similarly, Brugarolas et al (2004) showed that induction of REDD1 during hypoxia resulted in activation of the mTOR inhibitory complex TSC1/TSC2. As we also observe a decrease in the phosphorylation of 4E-BP1 after prolonged hypoxia, the eIF4F-dependent changes in translation reported here may also be due in part to inhibition of mTOR via REDD1 and TSC1/2. However, it is unlikely that this accounts entirely for eIF4F disruption and translation inhibition during hypoxia. REDD1 is an HIF-dependent gene and both mTOR inhibition and translation inhibition during hypoxia occur in HIF1α-knockout cells (Koumenis et al, 2002; Arsham et al, 2003). Furthermore, our data indicate that eIF4F disruption occurs before substantial binding of eIF4E to 4E-BP1. Here we have identified redistribution of eIF4E into the cell nucleus via 4E-T as an additional mechanism for eIF4F disruption during hypoxia. Further work will be needed to establish to what degree inhibition of translation is due to suppression of mTOR/4E-BP1 phosphorylation and 4E-T activation, as well as to the requirements of HIF in both of these pathways.
The fact that both eIF2α and eIF4F independently affect translation during hypoxia has important implications for the regulation of gene expression. mRNAs preferentially translated during acute hypoxia must be less dependent on eIF2α availability, whereas mRNAs that are actively translated during prolonged hypoxia must be less dependent on eIF4F. The translation of ATF4, which contains two upstream open reading frames (uORFs) in its 5′UTR, is perhaps the best example of a mammalian gene that displays this type of preferential translation (Harding et al, 2000). When eIF2α availability is high, translation begins at the 5′ most uORF and re-initiation occurs efficiently at the subsequent uORF, preventing translation from the correct start codon of ATF4. When eIF2α is phosphorylated, there is a higher probability of skipping the second uORF and re-initiating at the bona fide start codon (Lu et al, 2004). Here, we found that in addition to ATF4, the downstream genes CHOP and GADD34 are also translationally induced during acute hypoxia. The S51A MEFs, which are unable to phosphorylate eIF2α, are defective in this translational regulation. This result is consistent with a report showing that ER stress-induced expression of GADD34 protein can be prevented by keeping eIF2α dephosphorylated (Novoa et al, 2003). The bypass of translation inhibition may thus facilitate the ability of GADD34 to dephosphorylate eIF2α and promote recovery from ER stress.
Our results predict that during prolonged hypoxia, gene transcripts with lower dependency on eIF4F should be preferentially translated. The translation of ATF4 and CHOP was in fact stimulated in HeLa cells after 16 h of hypoxia when the eIF4F complex was disrupted. Preferential translation under conditions of limiting cap-binding complex activity can occur through a higher than average affinity for eIF4F (Lawson et al, 1988). Another group of mRNAs that can be translated independently of the eIF4F complex are those that contain an internal ribosomal entry site (IRES) in their 5′UTR (Carter et al, 2000; Holcik and Sonenberg, 2005). The presence of an IRES enables translation initiation under conditions where eIF4F-dependent translation is inhibited. Importantly, both mouse HIF-1α and VEGF have been shown to contain functional IRESs within their 5′UTR (Stein et al, 1998; Lang et al, 2002), although their biological importance is not yet firmly established. This provides a mechanism to ensure their translation during prolonged hypoxia where eIF4F is disrupted. The mechanism responsible for the selective translation of ATF4 and CHOP during prolonged hypoxia remains to be identified.
In conclusion, we have shown that mRNA translation is inhibited through multiple independent pathways with differing activation kinetics during hypoxia. These distinct modes of translational control influence the translation of individual genes to varying degrees and consequently can influence hypoxia-regulated protein expression in complex ways. An important finding is that inhibition of translation via eIF2α is transient, leading to dynamic changes in the translation efficiency of genes over the first 8 h of hypoxia. Our selected analysis of gene translation during hypoxia suggests that many genes may be differentially regulated by hypoxia. A complete survey of the genome for differentially translated genes during various exposures to hypoxia and their dependence of eIF2α is possible and will undoubtedly identify novel and important hypoxia-regulated proteins (Koritzinsky et al, 2005).
Materials and methods
Cell culture
Exponentially growing cervical carcinoma HeLa cells (American Type Culture Collection CCL-2), lung adenocarcinoma A549 cells, normal human fibroblasts (AG1522) or MEFs that were WT or had a homozygous knock-in mutation for eIF2α (S51A) (Scheuner et al, 2001) were grown on glass dishes or chamber slides in DMEM media supplemented with 10% fetal calf serum. The MEF media also contained MEM nonessential amino acids and 55 μM 2-mercaptoethanol (all Sigma-Aldrich). For preparation of extracts and viability assessments, see Supplementary data.
Hypoxic conditions
Cells were transferred to a hypoxic culture chamber (MACS VA500 microaerophilic workstation, Don Whitley Scientific). The composition of the atmosphere in the chamber consisted of 5% H2, 5% CO2, 0.0% O2 and residual N2.
m7GTP resin precipitation
A 1 mg portion of HeLa extract was incubated with 25 μl of m7GTP sepharose resin (Amersham Biosciences) for 3 h at 4°C. The resin was washed, boiled in Laemmli buffer and the polypeptides were resolved by SDS–PAGE.
Western blotting
Cell extracts were boiled in Laemmli buffer and polypeptides were resolved by SDS–PAGE and transferred onto 0.2 μm nitrocellulose membranes (Amersham Corp.). For primary antibodies, see Supplementary data. Detection of peroxidase-coupled secondary antibodies was performed with Enhanced Chemiluminescence (Amersham Corp.).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permeabilized in 4% paraformaldehyde and 0.1% Triton X-100. For antibodies, see Supplementary data. Cells were mounted in the ProLong™ Antifade Kit (Molecular Probes) and analyzed with a Zeiss inverted LSM 410 laser scan confocal microscope.
Polysomal fractionation and analysis
Polysomal fractionation and analysis were performed as described previously (Koritzinsky et al, 2005); see Supplementary data.
RNA isolation and reverse transcription
RNA isolation and reverse transcription were performed as described previously (Koritzinsky et al, 2005); see Supplementary data.
Quantitative PCR analysis
Real-time PCR was performed in either ABI 7700 or ABI 7500 (Applied Biosystems). For primers and probes, see Supplementary data. Unfractionated samples were normalized by 18S rRNA signal. Samples from polysome fractions were normalized by 18S rRNA measured by PCR divided by 18S rRNA measured by spectrometry during fractionation, corrected for loading. This facilitated correction for any differences in RNA isolation or reverse transcriptase efficiency between samples. The abundance of every gene was calculated relative to a master reference using standard curves.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary materials and methods
Acknowledgments
This work was financially supported by Netherlands Organization for Scientific Research (NWO), the Dutch Cancer Society (KWF Kankerbestrijding) and the Euroxy grant from the 6th framework of the EU to BGW, from the Norwegian Research Council to MK and from grant CA94214 from the National Institutes of Health (NIH) to CK.
References
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R (2005) A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsham AM, Howell JJ, Simon MC (2003) A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem 278: 29655–29660 [DOI] [PubMed] [Google Scholar]
- Bennewith KL, Durand RE (2004) Quantifying transient hypoxia in human tumor xenografts by flow cytometry. Cancer Res 64: 6183–6189 [DOI] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C (2005) ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, Wouters BG, Bell JC (2004) Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol 24: 7469–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr (2004) Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18: 2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas-Navia LI, Yu D, Braun RD, Brizel DM, Secomb TW, Dewhirst MW (2004) Tumor-dependent kinetics of partial pressure of oxygen fluctuations during air and oxygen breathing. Cancer Res 64: 6010–6017 [DOI] [PubMed] [Google Scholar]
- Carter MS, Kuhn KM, Sarnow P (2000) Cellular internal ribosome entry site elements and the use of cDNA microarrays in their investigation. In Translational Control of Gene Expression, Sonenberg N, Hershey JW, Mathews MB (eds) pp 615–636. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- DeGracia DJ, Kumar R, Owen CR, Krause GS, White BC (2002) Molecular pathways of protein synthesis inhibition during brain reperfusion: implications for neuronal survival or death. J Cereb Blood Flow Metab 22: 127–141 [DOI] [PubMed] [Google Scholar]
- Dostie J, Ferraiuolo M, Pause A, Adam SA, Sonenberg N (2000) A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J 19: 3142–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Cawthorne CJ, Williams KJ, Koritzinsky M, Wouters BG, Wilson C, Miller C, Demonacos C, Stratford IJ, Dive C (2004) Hypoxia-mediated down-regulation of Bid and Bax in tumors occurs via hypoxia-inducible factor 1-dependent and -independent mechanisms and contributes to drug resistance. Mol Cell Biol 24: 2875–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ (1999) Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J 339 (Part 1): 135–141 [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N (2005) A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol 170: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ (1996) Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379: 88–91 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Harris AL (2002) Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer 2: 38–47 [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N (2004) Upstream and downstream of mTOR. Genes Dev 18: 1926–1945 [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N (2005) Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Holland EC, Sonenberg N, Pandolfi PP, Thomas G (2004) Signaling control of mRNA translation in cancer pathogenesis. Oncogene 23: 3138–3144 [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ (2001) Targeting of HIF-alpha to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P (1999) Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA 96: 13118–13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Jefferson LS (2000) Regulation of translation initiation in mammalian cells by amino acids. In Translational Control of Gene Expression, Sonenberg N, Hershey JW, Mathews MB (eds) pp 561–579. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Koritzinsky M, Seigneuric R, Magagnin MG, Beucken T, Lambin P, Wouters BG (2005) The hypoxic proteome is influenced by gene-specific changes in mRNA translation. Radiother Oncol 76: 177–186 [DOI] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol 22: 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KJ, Kappel A, Goodall GJ (2002) Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 13: 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson TG, Cladaras MH, Ray BK, Lee KA, Abramson RD, Merrick WC, Thach RE (1988) Discriminatory interaction of purified eukaryotic initiation factors 4F plus 4A with the 5′ ends of reovirus messenger RNAs. J Biol Chem 263: 7266–7276 [PubMed] [Google Scholar]
- Leek RD, Stratford I, Harris AL (2005) The role of hypoxia-inducible factor-1 in three-dimensional tumor growth, apoptosis, and regulation by the insulin-signaling pathway. Cancer Res 65: 4147–4152 [DOI] [PubMed] [Google Scholar]
- Lefebvre VH, Van Steenbrugge M, Beckers V, Roberfroid M, Buc-Calderon P (1993) Adenine nucleotides and inhibition of protein synthesis in isolated hepatocytes incubated under different pO2 levels. Arch Biochem Biophys 304: 322–331 [DOI] [PubMed] [Google Scholar]
- Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sonenberg N (1992) A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci USA 89: 9612–9616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18: 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin de la Vega C, Burda J, Nemethova M, Quevedo C, Alcazar A, Martin ME, Danielisova V, Fando JL, Salinas M (2001) Possible mechanisms involved in the down-regulation of translation during transient global ischaemia in the rat brain. Biochem J 357: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ME, Munoz FM, Salinas M, Fando JL (2000) Ischaemia induces changes in the association of the binding protein 4E-BP1 and eukaryotic initiation factor (eIF) 4G to eIF4E in differentiated PC12 cells. Biochem J 351 (Part 2): 327–334 [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O, Hornstein E (2000) Translational control of TOP mRNAs. In Translational Control of Gene Expression, Sonenberg N, Hershey JW, Mathews MB (eds) pp 671–694. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Neumar RW, DeGracia DJ, Konkoly LL, Khoury JI, White BC, Krause GS (1998) Calpain mediates eukaryotic initiation factor 4G degradation during global brain ischemia. J Cereb Blood Flow Metab 18: 876–881 [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D (2003) Stress-induced gene expression requires programmed recovery from translational repression. EMBO J 22: 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC Jr, Sonenberg N (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371: 762–767 [DOI] [PubMed] [Google Scholar]
- Robertson N, Potter C, Harris AL (2004) Role of carbonic anhydrase IX in human tumor cell growth, survival, and invasion. Cancer Res 64: 6160–6165 [DOI] [PubMed] [Google Scholar]
- Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, Le QT, Koong AC (2004) XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res 64: 5943–5947 [DOI] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS (1998) HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J 17: 3005–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789 [DOI] [PubMed] [Google Scholar]
- Semenza GL (2003) Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3: 721–732 [DOI] [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol 18: 3112–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Wang L, Esko J, Giordano FJ, Huang Y, Gerber HP, Ferrara N, Johnson RS (2004) Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell 6: 485–495 [DOI] [PubMed] [Google Scholar]
- Tinton SA, Buc-Calderon PM (1999) Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett 446: 55–59 [DOI] [PubMed] [Google Scholar]
- Wouters BG, van den Beucken T, Magagnin MG, Koritzinsky M, Fels D, Koumenis C (2005) Control of the hypoxic response through regulation of mRNA translation. Semin Cell Dev Biol 16: 487–501 [DOI] [PubMed] [Google Scholar]
- Wouters BG, Weppler SA, Koritzinsky M, Landuyt W, Nuyts S, Theys J, Chiu RK, Lambin P (2002) Hypoxia as a target for combined modality treatments. Eur J Cancer 38: 240–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary materials and methods


