Abstract
Mutational analyses of the p12 Gag phosphoprotein of Moloney murine leukemia virus have demonstrated its participation in both virus assembly and the early stages of infection. The molecular mechanisms by which p12 functions in these events are still poorly understood. We performed studies to examine the significance of p12 phosphorylation in the viral life cycle. Alanine substitutions were introduced at the potential phosphorylation sites in p12, and the resulting mutants were tested for replication. Mutant viruses with changes at S61 and S78 were severely impaired, whereas the other mutant viruses were viable. S61 was shown to be required for normal levels of phosphorylation of p12 in vivo. These defective mutant viruses showed no apparent alteration to Gag protein processing or reduction in the yield of virions after transient transfection, but the mutants failed to form circular viral DNAs in acutely infected cells. Sequence analysis of revertant clones derived from S(61,65)A mutant virus revealed two classes: one group with a single mutation at a residue adjacent to S61 and another group with mutations introducing new positive charges surrounding S61. In vivo [32P]orthophosphate labeling indicated that the rescue of the S(61,65)A mutant virus did not result in a significant increase in the phosphorylation level of p12. Alanine substitutions of an arginine-rich stretch near S61 (at R-66, -68, -70, and -71) resulted in the same phenotype as the S(61,65)A mutant virus. The restored function of S(61,65)A mutant virus by second or third site mutations may result from a structural change or the addition of positively charged residues in the arginine-rich region.
The retroviral Gag protein plays an important role in the viral life cycle, involving not only virus assembly but also virion maturation after particle release and early postentry steps in virus replication (11, 43). In the case of the most thoroughly characterized of the gammaretroviruses, Moloney murine leukemia virus (MMLV), the Gag precursor protein mediates the assembly and release of virions during the late phase of virus infection. The Gag precursor protein is ultimately cleaved by the viral protease to yield four proteins: matrix or membrane associated (MA), p12, capsid (CA), and nucleocapsid (NC) (22). These separated Gag proteins then play roles in the early stages of virus infection and may facilitate virus entry, uncoating, and possibly nuclear entry of the viral DNA.
The p12 protein of the MMLV gag gene products has been shown to be involved in both virus assembly and early postentry steps (9, 47, 49). p12 is only 84 amino acid residues in length. Alanine substitution mutations of p12 have revealed that a PPPY motif is essential for the efficient release of virion particles. This proline-rich PPPY domain is interchangeable with the L-domains of the human immunodeficiency virus (HIV)-p6 protein (19, 30, 42), the Rous sarcoma virus (44), the Mason-Pfizer monkey virus (45), and equine infectious anemia virus (32). In various tests, the L-domain from one virus can function in place of the corresponding sequence of another virus, suggesting that the sequences utilize similar mechanisms and can act autonomously in many contexts (30, 44, 47). Although these late effects are limited to mutations in the PPPY domain, deletions of the N-terminal portion of p12 result in a block at an early stage of infection (9). Alanine substitution at both the N and the C termini of p12 also showed defects in early events (48, 49). These mutant viruses synthesized normal levels of linear viral DNA but no circular DNA. These results strongly suggest that p12 plays a role after reverse transcription and prior to DNA integration.
Phosphorylation of viral proteins often plays an important role in regulation of viral replication (4, 41). In HIV type 1, several viral proteins have been shown to be phosphorylated, including MA(p17) (13), CA(p24) (6), Vpr (50), Nef (7), Rev (15), Tat (26), and Vpu (31). In the case of the murine leukemia viruses, p12 is the only major phosphorylated protein (28, 29). The bulk of the phosphorylation of p12 has been shown to occur at serine and not at threonine or tyrosine residues (20, 27). p12 phosphorylation in vivo has been speculated to correlate with virion maturation (27, 46) and has been suggested to modulate the RNA-binding activity of p12 (35, 36). However, little is known about the effect of p12 phosphorylation on virus replication.
To determine the role of the murine leukemia virus p12 phosphorylation in the viral life cycle, we generated alanine substitution mutations at the serine residues of the p12 protein. The results suggest that the phosphorylation of p12 is important in the process of virus infection prior to integration and not in virus assembly or budding. We also demonstrate that S61 is required for nearly all p12 phosphorylation. The isolation of other mutants and revertants implies that an arginine-rich stretch in the C terminus of p12 is also essential for infection.
MATERIALS AND METHODS
Cell culture.
293 cells are human embryonic kidney (HEK) cells that express the E1 region of adenovirus 5. 293T cells are 293 cells that stably express simian virus 40 (SV40) large T-Ag. Rat2-2 cells are rat embryonic fibroblast cells. 293, 293T, and Rat2-2 cells were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum.
Plasmid and mutant construction.
Plasmid pNCA contains an infectious copy of MMLV proviral DNA (8); pNCS is a version of pNCA that carries an SV40 replication origin in the vector to allow high-level expression in 293T cells. Plasmids were constructed by introducing point mutations into plasmid vector pNCS. Point mutations were created by overlap extension PCR (18) with pNCS template DNA by using outside primers (forward primer in MA region, 5′-CTGGGTCAAGCCCTTTGTACACCCTAAGCC-3′; reverse primer in CA, 5′-GTCTGGGCGCTCGAGGGGAAAAGCG-3′). The sense strand primers utilized for creating mutations were as follows, and the antisense primers were their complement: S6/A, 5′-CCAGCCCTCACTCCTGCTCTAGGCGCCAAACCTAAA-3′; S(17,19)A, 5′-ACCTAAACCTCAAGTTCTTGCTGACGCTGGGGGGCCGCTCA-3′; S(61,65)A, 5′-AGGCACCGGACCCCGCCCCAATGGCAGCTCGCCTACGTGG-3′; S(78,81)A, 5′-CCCTGTGGCCGACGCCACTACCGCGCAGGCATTCCCCCTC-3′; S61A, 5′-GGAGAGGCACCGGACCCCGCCCCAATGGCATCTCGCCTA-3′; S65A, 5′-GACCCCTCCCCAATGGCAGCTCGCCTACGTGGGAGA-3′; S61D, 5′-AGAGGCACCGGACCCCGATCCAATGGCATCTCGCCTA-3′; S78A, 5′-CCCCCTGTGGCCGACGCCACTACCTCGCAGGCATTC-3′; S78D, 5′-CCCTGTGGCCGACGATACTACCTCGCAGGCATTCC-3′; S81A, 5′-GCCGACTCCACTACCGCGCAGGCATTCCCCGTC-3′; R(66,68)A, 5′-GGCATCTGCACTAGCAGGGAGACGGGAGCCCCCT-3′; R(66,68)K, 5′-GCATCTAAGCTAAAGGGGAGACGGGAGCCCCCT-3′; R(70,71)/A, 5′-CTACGTGGGGCAGCAGAGCCCCCTGTGGCCGAC-3′; R(70,71)K, 5′-CTACGTGGGAAGAAGGAGCCCCCTGTGGCCGAC-3′; R(66)/A, 5′-ATGGCATCTGCACTACGTGGGAGACGGGAGCCC-3′; R(66)/A/R, 5′-CCCACGTAGTGCAGATGCCATTGGGGA-3′; R(68)/A, 5′-ATGGCATCTCGCCTAGCAGGGAGACGGGAGCC-3′; R(70)/A, 5′-CTACGTGGGGCACGGGAGCCCCCTGTGGCCGAC-3′; R(71)/A, 5′-CTACGTGGGAGAGCAGAGCCCCCTGTGGCCGAC-3′; and P12/Flag, 5′-TACAAAGACGATGACGACAAGCCTGCGGGAGAGGCACCG-3′. All mutations were verified by DNA sequence analysis.
Mammalian cell transfection and viral infection.
To produce large amount of virus and to test the effects of mutations on viral assembly, 293T cells were transiently transfected by proviral DNAs containing the SV40 origin of replication, using the calcium phosphate method. The culture medium was changed at 48 h posttransfection, and the virus was harvested 72 h posttransfection and used for further analysis. Infection of Rat2-2 cells was carried out in the presence of Polybrene (8 μg/ml) for 2 h.
Virus purification and analysis of viral proteins.
To prepare virus stocks, 72 h after transfection of 293T cells with proviral DNAs, 9 ml of culture supernatant was collected from each plate (100 by 15 mm). Cell debris was removed by filtering the medium through a 0.45-μm (pore-size) filter, and HEPES buffer was added to a final concentration of 20 mM. Virions were first purified on a 25 to 45% sucrose-TNE step gradient for 2 h at 25,000 rpm and 4°C in a Beckman SW41 rotor. The interface was collected, and the virus was diluted with 4 ml of TNE (50 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA) and pelleted through a 25% sucrose cushion by centrifugation in the same rotor for 2 h at 25,000 rpm. The collected virions were suspended in TNE buffer and stored at −80°C.
For immunodetection of virion proteins, pelleted virus particles were lysed and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to nitrocellulose filters (Schleicher & Schuell) and probed by various antibodies, including goat anti-CA serum (NCI serum #79S-804). The membrane was stained by the ECL kit (Amersham Corp.) and exposed to X-ray film for detection. The filter could be stripped and reprobed with other antibodies.
In vivo labeling of virions and p12 immunoprecipitation.
293T cells were grown and transfected with wild-type or p12 mutant pNCS plasmids in 60-mm-diameter dishes. At 36 h posttransfection, the transfected cells were replaced with new DMEM without phosphate for 30 min. These cells were then labeled for 16 h with 1 mCi of 32P (NEN) per dish in 2 ml of phosphate-free DMEM (5% fetal calf serum). The 32P-labeled virions were harvested and purified by centrifugation through 25% (wt/vol) sucrose in phosphate-buffered saline buffer at 25,000 rpm for 2 h at 4°C in a Beckman SW41 rotor. The pellet was resuspended with radioimmunoprecipitation assay buffer (0.1% SDS, 1% NP-40, 0.1% sodium deoxycholate, 150 mM NaCl, 10 mM Tris-HCl; pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride and centrifuged in an Eppendorf model 5415C centrifuge at 13,000 rpm for 20 min. Supernatants were incubated with Flag antibody-conjugated agarose (M2 agarose; Sigma) for 1 h with constant shaking at 4°C. The beads were washed three times with radioimmunoprecipitation assay buffer, subjected to SDS-PAGE, and visualized with a PhosphorImager.
Analysis of viral DNA synthesized in vivo.
Preintegrative viral DNAs were isolated from Rat2-2 cells at 24 h postinfection (17) and analyzed by Southern blotting. PCR was used to detect circular viral DNA containing two long terminal repeats (LTRs). The primers used to amplify the LTR-LTR junction were MR5784 (5′-AGTCCTCCGATTGACTGAG-3′) and MR4091 (5′-CTCTTTTATTGAGCTCGGG-3) (39); the PCR conditions were 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, repeated for 30 cycles. PCR was also used to detect an internal region of linear viral DNA to serve as control. The primers used to amply the capsid region were 5′-CCCCTCCGCGCAGGAGGAAAC-3′ and 5′-ACAATAGCTTGCTCATCTCTCTATG-3′; the PCR conditions were 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, repeated for 24 cycles.
S(61,65)A revertant isolation.
Virus supernatants were harvested at the peak of reverse transcriptase (RT) activity from 293T cultures transfected with the S(61,65)A mutant. The virus stocks were used to infect fresh Rat2-2 cells. The infected cells were repassaged every 4 days until RT activity reached levels similar to those of the wild type. The emerging viruses were used to infect fresh Rat2-2 cells, and low-molecular-weight DNA was harvested (17). A 640-bp fragment spanning the MA-p12-CA coding region was PCR amplified from the DNA. The primers used were as follows: positive, 5′-CTGGGTCAAGCCCTTTGTACACCCTAAGCC-3′; and negative, 5′-ATCCCAGTCTGGGCGCTCGAGGGGAAAAGCG-3′. Thirty cycles of PCR were performed with ca. 10 ng of input DNA under the following conditions: 94°C for 45 s, 50°C for 30 s, and 72°C for 1 min. Amplified DNA was digested with BsrGI and XhoI. The BsrGI-XhoI fragments were cloned into the BsrGI and XhoI sites of a modified pUC18 vector and sequenced in their entirety. After sequencing, BsrGI-XhoI fragments from the pUC18 clones were exchanged for the BsrGI-XhoI fragment of pNCS to generate a series of pNCS-derived revertants.
RESULTS
Mutations of specific serine residues of p12 block virus replication.
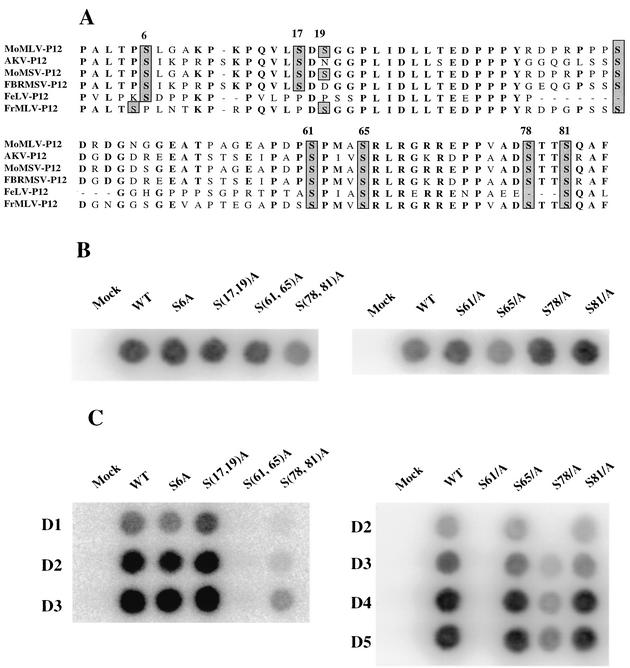
The Gag protein p12 of murine leukemia virus is the only virion protein that is phosphorylated at significant levels; the phosphate is present as phosphoserine (20, 46). To explore whether the serine phosphorylation of p12 plays an important role in the viral life cycle, we set out to generate a series of mutants. The p12 protein sequences from different gammaretroviruses were aligned to identify conserved serine residues as potential phosphorylation sites (Fig. 1A). To determine whether the phosphorylation of p12 protein affects virus replication, all serine residues of p12 except Ser-42, which has been previously shown to be dispensable in the viral cycle (49), were mutated to alanine residues by site-directed mutagenesis in a proviral clone. Single substitution mutants of Ser-6, Ser-61, Ser-65, Ser-78, and Ser-81 and double substitutions of Ser-(17,19), Ser-(61,65), and Ser-(78,81) were generated. The DNAs were then used to transfect 293T cells, and at 48 h posttransfection the virions produced from the transfected cells were harvested and assayed for RT activity (Fig. 1B). All of the mutants were able to induce the formation of virus particles. Next, the concentrations of the virions were normalized by RT levels, and the preparations were used to infect fresh Rat2-2 cells. The supernatants from infected Rat2-2 cells were collected every day, and the infected cells were split every 5 days. After 10 days of infection, the culture medium from the Rat2-2 cells infected by S6A, S(17,19)A, S65A, and S81A mutant viruses had RT activities similar to cells infected with wild-type virus (Fig. 1C). In contrast, substitution of S61 by alanine completely abolished virus spreading, and no detectable RT activity was found in the supernatant of infected Rat2-2 cells even 20 days postinfection. Only small amounts of RT activity were detected in the cells infected by S(78,81)A or S78A mutant viruses. We conclude that mutations at S61 blocked replication and that mutations at S78 strongly reduced replication.
FIG. 1.
Effect on virus replication of alanine substitution mutations in potentially phosphorylated serine residues of p12. (A) Sequences of retrovirus p12 genes. The sequences of p12 were aligned with the MacVector program. All conserved potentially phosphorylated serine residues are shadowed. (B) Transient transfection of 293T cells to assess viral assembly and particle release. The various mutations were introduced into a proviral construct, and the resulting DNAs were transfected into 293T cells. The virions collected from supernatant of transfected cells were analyzed for RT activity. (C) Virus spreading assay. The culture supernatants harvested from transfected 293T cells were normalized by RT activity and used to infect naive Rat2-2 cells. The culture supernatants from infected Rat2-2 cells were collected every day for up to 7 days after infection and assayed for RT activity. WT, wild type.
Mutant viruses assemble and release normal levels of mature virion particles.
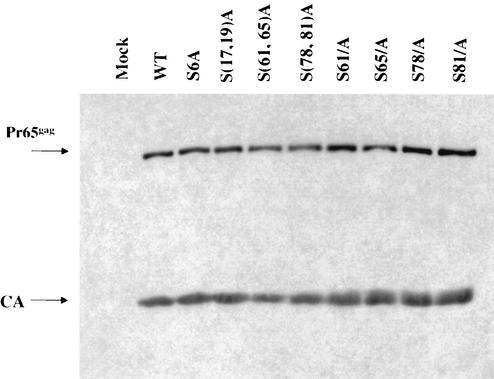
Studies were conducted to examine the assembly of the mutant viruses in more detail. The defective mutants and wild-type proviral DNAs were cotransfected with the pCMV-β-galactosidase plasmid into 293T cells. At 72 h after transfection of 293T cells, the culture supernatants containing virion particles were harvested from the transfected 293T cells, normalized by the enzymatic activity of β-galactosidase, and purified by ultracentrifugation through a 25% sucrose cushion. The purified virions were lysed and subjected to SDS-PAGE and Western blot with anti-CA antibody to measure the yield of virus (Fig. 2). No apparent difference was found in the viral protein profile between p12 wild-type and mutant virions. None of the mutations in the serine residues affected the late steps of the viral life cycle, protein processing, or virus assembly or release. These results are consistent with previous work (49) showing that alanine substitutions in the C terminus of p12 did not affect Gag protein processing or virus assembly.
FIG. 2.
Western blot analysis of gag gene products in the virions of p12 mutants. Virus particles were harvested from the culture supernatant of transfected 293T cells and lysed, and the capsid proteins were separated by electrophoresis, blotted, and probed with anti-CA antibodies. WT, wild type.
Mutant viruses fail to synthesize normal levels of circular viral DNA.
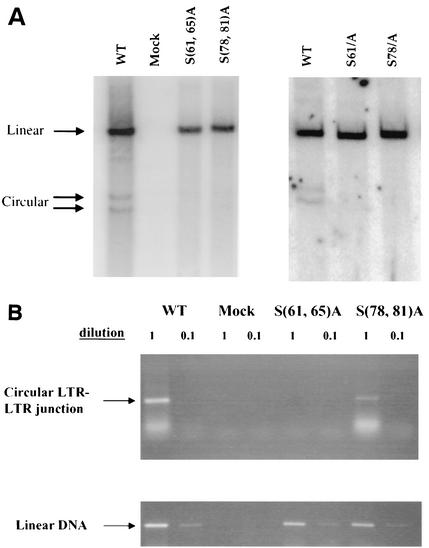
Since alanine substitution mutations at serine residues 61 and 78 did not affect virus assembly and Gag processing, studies were carried out to determine whether these substitution mutations affect early events. To monitor synthesis of viral DNA, virions of mutants S61A, S78A, S(61,65)A, and S(71,78)A were harvested from transfected 293T cells and used to infect fresh Rat2-2 cells. Low-molecular-weight DNAs were prepared from these infected Rat2-2 cells at 16 h postinfection and subjected to agarose gel electrophoresis for Southern blotting with a probe specific for the MMLV genome. Similar to the pattern observed for PM13 and PM14 mutants (49), the S61A and S(61,65)A mutants were also not able to synthesize detectable circular DNAs (Fig. 3A). The failure of S61A or S(61,65)A mutant to form circular viral DNAs is consistent with their impairment in virus spreading (Fig. 1C). Small but detectable amounts of circular DNAs were found in cells infected with the S78A or S(78,81)A mutants (Fig. 3A). The reduced level of viral circular DNAs in the infection with the S(78,81)A mutant virus was consistent with the delay in viral replication (Fig. 1C).
FIG. 3.
Southern blot and PCR analyses of linear and circular viral DNA synthesis after infection of Rat2-2 cells by p12 mutant viruses. Rat2-2 cells were infected with virus from transfected 293T cells, and low-molecular-weight DNA was isolated 16 h after infection. (A) Southern blotting was performed by using a radiolabeled viral DNA probe. The positions of linear and circular viral DNAs are indicated. The efficiency of recovery of the low-molecular weight DNAs was monitored by probing the blots for mitochondrial DNA. (B) PCR was performed on the same DNA samples with primers specific for the LTR-LTR junction of circular DNA or for an internal region of the linear DNA. Products were displayed by gel electrophoresis and visualized by ethidium bromide stain. WT, wild type.
To examine the ability of the mutants to direct the formation of circular DNAs with greater sensitivity, several of the same low-molecular weight DNAs were subjected to PCR with primers specific for the LTR-LTR junction region, which is present only in circularized viral DNAs (Fig. 3B). Although an amplified fragment of the expected size was readily detected in the wild-type infected cells, no such product could be detected in cells infected with mutant S(61,65)A. Reduced levels of the DNA were detected after infection with mutant S(78,81)A. To confirm that there was no effect on the formation of linear DNAs, PCR was performed with primers from an internal gag region present on all viral DNAs. Similar levels were detected in all of the infected cells (Fig. 3B).
S61 is required for p12 phosphorylation in vivo.
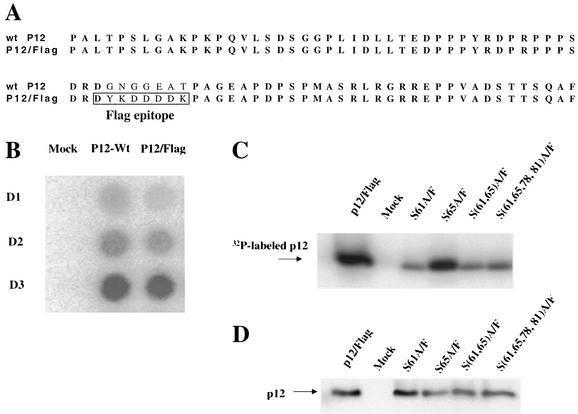
p12 is phosphorylated at serine residues (20, 27), and alanine substitutions of some of these series could affect its phosphorylation. To facilitate measurement of the phosphorylation status of p12, a Flag epitope was inserted into the central portion of p12 in the proviral DNA (Fig. 4A), a region that tolerates alanine substitution with no effect on virus replication (49). To test whether the Flag epitope had an effect on viral replication, the pNCS/Flag construct and a wild-type control were transfected into 293T cells, and the culture supernatants were used to infect Rat2-2 cells. The tagged variant replicated at a rate identical to that of the wild type (Fig. 4B). We expected that this epitope insertion would not change the phosphorylation pattern of p12 and used this variant as parent in the subsequent tests.
FIG. 4.
Phosphorylation of p12 in mutant virions. (A) The sequence alignment of wild-type p12 and p12/Flag. The Flag epitope was inserted in the central region of p12 in a proviral construct, a site known to be dispensable for virus replication. (B) Replication of p12/Flag virus. Wild-type or p12/Flag proviral constructs were transfected into 293T cells, and the released virus was used to infect Rat2-2 cells. Replication was monitored by RT assays. (C) Effect of alanine substitution mutations of potentially phosphorylated serine residues on the level of p12 phosphorylation. The Flag epitope was inserted into the p12 coding region of various mutants. 293T cells were cotransfected with p12/Flag wild type or the various mutants plus a pCMV-β-galactosidase DNA. One set of 293T cells was labeled with [32P]orthophosphate, and the 32P-labeled virions were harvested, purified by ultracentrifugation, and lysed. p12 was then isolated by immunoprecipitation with antisera against the Flag epitope. The 32P-labeled proteins were resolved on SDS-PAGE and exposed to a PhosphorImager screen. (D) A second set of 293T cells was transfected as before, and the virions were collected without 32P labeling. SDS-PAGE was performed to resolve the proteins, and the p12 levels were assessed by Western blotting carried out with anti-Flag antibodies.
Flag-tagged versions of the S61A, S78A, S(61,78)A, and S(61,65,78,81)A mutants were generated and used to transfect 293T cells. The transfected 293T cells were labeled with [32P]orthophosphate for 12 h, beginning 36 h posttransfection. The 32P-labeled virus particles from the transfected cells were harvested and lysed, and p12 protein was then immunoprecipitated with anti-Flag agarose beads. The purified p12 proteins were resolved by SDS-PAGE, and bands were quantified by using a PhosphorImager (Molecular Dynamics). The phosphorylation of the wild-type p12 was readily detected. The phosphorylation of the S61A mutant virus was dramatically reduced to ca. 15% that of the wild-type p12 (Fig. 4C). The S(61,78)A and S(61,65,78,81)A mutant virions showed similarly reduced levels of p12 phosphorylation. This suggests that S61 is the major phosphorylation site or is at least required for nearly all phosphorylation of p12. The additional changes of S65, S78, and S81 caused no further reductions in phosphorylation, suggesting that the residual phosphorylation occurred elsewhere. Changing S65 alone (Fig. 4C) showed fully wild-type levels of phosphorylation, indicating that this residue was not a major site of phosphorylation nor was it required for phosphorylation of other sites.
To confirm that the changes in the 32P labeling were not due to changes in the level of the protein in the virion particles, parallel transfections were performed, and the yield of virions from these transfected cells was quantified by RT assays and by estimates of the level of p12 in virions. The yield of particles was essentially equal, as judged by RT assays (data not shown), and the levels of p12 were constant at judged from Western blots of gels (Fig. 4C). Thus, the reductions in 32P labeling reflect changes in phosphorylation levels.
Aspartic acid cannot replace serine 61 or 78 to rescue viral replication.
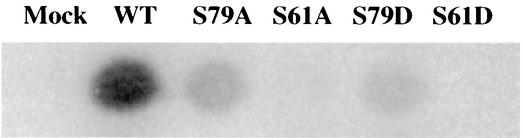
The in vivo 32P-labeling experiments suggested that the phosphorylation of S61 might be important for p12 function. To test whether an acidic amino acid could mimic the negative charge of phosphoserine, S61 and S78 were individually mutated to aspartic acid. The S61D and S78D mutant DNA constructs were introduced into 293T cells, and the virions from the transfected 293T cells were used to infect Rat 2 cells. These mutants were not replication competent. Thus, aspartic acid could not functionally substitute for the potentially phosphorylated serines at these positions in p12 (Fig. 5).
FIG. 5.
Replication of serine-to-aspartate substitution mutants. p12 mutant proviral DNAs in which various serine residues of p12 were replaced with aspartic acid were used to transfect 293T cells, the virus was collected and used to infect Rat2-2 cells, and the replication of the virus was assessed by assay for RT in the culture medium after 4 days. The results were similar to those observed for the corresponding serine-to-alanine substitutions, suggesting that the introduction of the negatively charged residue did not functionally replace the potentially phosphorylated serine residues. WT, wild type.
Isolation of revertants of a p12 serine substitution mutant.
To help define the structural features of p12 required for virus replication, revertant viruses were derived from the replication-defective mutant S(61,65)A virus. S(61,65)A virus particles were collected from transfected 293T cells and used to infect naive Rat2-2 cells. The infected cells were passaged repeatedly, and RT assays were carried out to monitor the emergence of revertants. After 12 weeks of culture, RT activities were found to reach wild-type levels in the supernatants from the infected Rat2-2 cells. Infections of fresh Rat2-2 cells with these revertant virus stocks showed that they replicated with nearly wild-type efficiencies.
To identify the mutations responsible for the reversion events, low-molecular-weight viral DNAs were prepared from cultures after acute infection with the putative revertant virus stock. PCR was carried out to amplify the MA-p12-CA portion of the viral DNA. The PCR products were cloned into the pUC18 plasmid vector, and the complete DNA sequences of a number of individual clones for each putative revertant were determined. Revertant clones 1, 6, 7, and 13 had the same mutation (Ala to Thr) at residue 65, changing one of the mutated residues back to threonine, a residue similar to the original serine (Fig. 6). Examination of the sequences of the rest of the putative revertants revealed that the original S(61,65)A mutation was maintained. Revertamt clones 2, 3, 8, 9, and 16 had acquired a common mutation (Met to Ile) at position 63. The remaining revertants had typically acquired additional positive charges by virtue of mutations in the nearby portions of p12. For example, Rev12 and Rev14 each had acquired two mutations, i.e., mutations G55R and G69R and mutations G49R and E50K, respectively. Rev5 and Rev10 each showed one similar mutation: Q83R and L66R, respectively.
FIG. 6.
Sequence analysis of S(61,65)A revertants. Revertant viruses were isolated from the medium of Rat2-2 cells infected with S(61,65)A mutant virus 3 months after infection. The revertant viruses were used to acutely infect naive Rat2-2 cells, and low-molecular-weight DNAs were isolated and used as a template for PCR to synthesize a DNA fragment spanning the region from MA to CA. The PCR products were subcloned and individual clones were sequenced.
Characterization of S(61,65)A revertant-derived clones.
To evaluate whether the mutations present in the various cloned DNAs were responsible and sufficient for the improved replication kinetics observed in the revertant virus stocks, these putative suppressor mutations were cloned back into the proviral construct pNCS. These pNCS-derived revertants were individually transfected into 293T cells, and the viruses produced were used to infect fresh Rat2-2 cells. The rate of spread of the virus in the infected cultures was monitored by RT assay. The replication kinetics of Rev1, Rev2, Rev12, and Rev14 were indistinguishable from that of the wild type, whereas Rev5 (Fig. 7A) and Rev15 (data not shown) both replicated with a moderate delay compared to the controls. In contrast, no replicating virus could be detected in cultures infected with the parental S(61,65)A virus, indicating that the compensatory mutations in the cloned DNAs were sufficient to fully or partially rescue the replication defect of the S(61,65)A mutant.
FIG. 7.
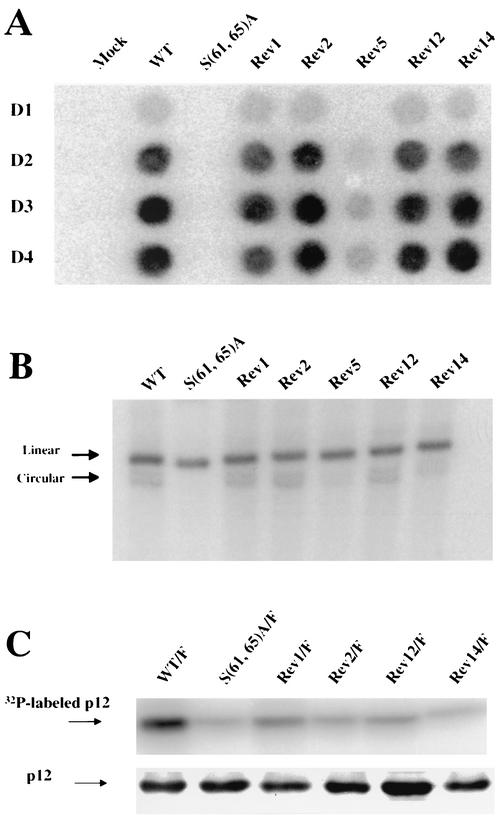
Phenotype of revertant viruses. (A) The replication of revertants in Rat2-2 cells. The p12 region of revertants was cloned back to a proviral DNA construct, used to generate virus, and the ability of the virus to replicate was assessed by RT assay. (B) Southern blot detection of linear and circular viral DNA synthesis after acute infection of Rat2-2 cells by wild type, S(61,65)A mutant, and revertant viruses. Rat2-2 cells were infected by the culture medium from transfected 293T cells with wild-type or revertant viruses as described in Fig. 1C. Low-molecular-weight DNA was purified 16 h after infection. Southern blotting was performed by using a radiolabeled viral DNA probe. The positions of viral linear and circular DNAs are indicated by arrows. The efficiency of recovery of the low-molecular-weight DNAs was monitored by probing the blots for mitochondrial DNA. (C) Phosphorylation of p12 by revertant viruses. Cultures expressing various mutants and revertants were labeled with [32P]phosphate, and virion particles were harvested from the medium and purified by centrifugation. The virions were disrupted, and viral proteins were analyzed by SDS-PAGE, followed by autoradiography (upper section). Virions from unlabeled cultures were isolated in parallel, and the levels of p12 protein were assessed by Western blot (lower section). WT, wild type.
The S(61,65)A mutation blocked the formation of viral circular DNAs but not of linear DNAs (Fig. 3) and blocked viral spread (Fig. 2C). To determine whether the specific defect of the S(61,65)A mutant in the formation of viral circular DNAs was corrected in the S(61,65)A revertants, Southern analysis was performed on DNAs from infected fresh Rat2-2 cells with revertant viruses, using labeled viral DNA as a probe (Fig. 7B). As shown in Fig. 7B, the defect in the formation of circular viral DNA was rescued to the level of wild type in Rev1, Rev2, Rev12, and Rev14, whereas only partial restoration of circular viral DNA synthesis was observed in Rev5. These results show that the suppressor mutations did act to correct the early block of the p12 parent.
No apparent change in the phosphorylation pattern of most revertants.
One mechanism by which the suppressor mutations of the various revertant viruses might restore function to the parental mutant would be to promote the needed phosphorylation of p12, perhaps at ectopic sites on the molecule. Alternatively, the mutations might somehow bypass the requirement for normal phosphorylation. To test which of these possibilities was correct, the phosphorylation levels of the various revertant viruses in vivo were assessed directly. The sequences needed to tag p12 with the Flag epitope were introduced into the genomes containing the revertants Rev1, Rev2, Rev12, and Rev14, and the mutant DNAs were used to transfect 293T cells. After 36 h, the cultures were labeled for 12 h with [32P]orthophosphate. The 32P-labeled virus particles were harvested and purified by ultracentrifugation through a 25% sucrose cushion. The virions were lysed, and the labeled Flag-tagged p12 protein was immunoprecipitated with α-Flag conjugated agarose, resolved by SDS-PAGE, and detected by autoradiography. The phosphorylation level of the p12 protein of Rev1, Rev2, Rev12, and Rev14 was very low and indistinguishable from that of the S61A mutant (Fig. 7C). This suggests that a restored phosphorylation of Rev1, Rev2, Rev12, and Rev14 is not responsible or required for the restoration of S(61,65)A function and virus replication.
An arginine-rich stretch in C terminus of p12 is crucial for the early stage of virus replication.
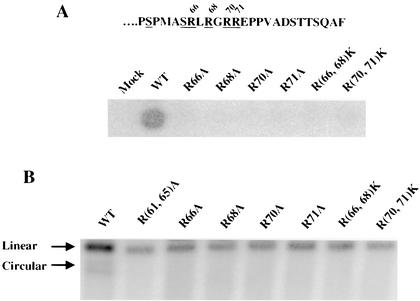
The studies of the S(61,65)A revertants described above revealed that the introduction of new positively charged residues, especially arginine residues, at several positions in p12 can suppress the defects of the S(61,65)A mutant parent (Fig. 7A). Previous studies have also shown that two other substitution mutants, PM14 and PM15, in the vicinity of Ser-61 are defective in the formation of viral circular DNAs (49). These two mutants have alanine substitution mutations in the arginine-rich stretch of p12. Experiments were therefore carried out to determine whether the nearby arginine residues—R66, R68, R70, and R71—play a critical role in the virus life cycle. The four arginine residues were substituted with either alanine or lysine to construct the single substitution mutants R66A, R68A, R70A, and R71A and the double mutants R(66,68)K and R(70,71)K. The mutant DNAs were transfected into 293T cells to produce virus particles. The mutant virus particles were then used to infect fresh Rat2-2 cells, and the ability of the viruses to spread in the culture was monitored by RT assays. The R66A, R68A, R70A, R71A, R(66,68)K, and R(70,71)K mutants all failed to produce infectious virus (Fig. 8A). Further, these mutant virions were poorly able to synthesize viral DNAs, especially circular forms, after acute infection (Fig. 8B). To confirm that there were no effects on the late stages of replication and to evaluate the pattern of Gag processing during virus production, Western analysis of virion proteins released by the 293T cells was performed. There were no apparent effects of the arginine mutations on Gag processing or virus assembly (data not shown). Taken together, these data indicated that the cluster of arginine residues is essential for the function of p12 in the early stage of virus infection.
FIG. 8.
Effects on virus replication of alanine or lysine substitution mutations replacing the arginine residues in the C terminus of p12. (A) Replication of arginine substitution mutants. The terminal amino acid sequences of p12 are shown, with the clustered arginine residues shown underlined. The arginine residues at positions 66, 68, 70, and 71 in the p12 region of a proviral DNA construct were mutated to alanine or lysine. The DNAs were used to transfect 293T cells, virus was collected and used to infect Rat2-2 cells, and virus replication of these mutants was assessed by RT assay of the culture media harvested at day 4 posttransfection. (B) Detection of viral DNA synthesis in cells infected with arginine substitution mutant viruses. Low-molecular-weight DNAs were isolated after acute infection of Rat2-2 cells with various arginine substitution mutants, and the viral DNAs were detected by Southern blotting. The positions of linear and circular viral DNAs in the wild-type control are indicated by arrows. The efficiency of recovery of the low-molecular-weight DNAs was monitored by probing the blots for mitochondrial DNA. WT, wild type.
DISCUSSION
The results presented here suggest that serine 61 of the MMLV p12 protein is crucial for the early phase of the viral life cycle and that serine 78 is also important, although to a lesser extent. Single alanine substitutions of these two residues and any combination of mutations that includes these changes resulted in drastic defects in virus replication. Infection by the mutant viruses was blocked at a specific stage: after completion of reverse transcription to form a linear double-stranded DNA but before the formation of double-stranded circular forms or the integrated provirus. This phenotype is identical to earlier mutants with larger mutations in either the N-terminal or C-terminal regions of p12 (48, 49) and suggests that these serines help mediate the normal function of p12.
It has been known for many years that p12 is a phosphoprotein, with the bulk of the phosphate present as phosphoserine (20, 27). The mutation of S61 caused a nearly complete loss of phosphorylation of p12, suggesting that this residue itself was likely to be the major site of phosphorylation. However, we cannot rule out the alternative possibility that this residue is somehow required for the phosphorylation of another position or positions in the protein. In contrast, the S78 mutation did not cause any detectable decrease in the overall level of p12 phosphorylation, suggesting that it is not a significant site of phosphorylation. Its effect on replication is most likely due to a direct effect of the substitution itself on protein conformation and function. We note that even when S61 was mutated, there was still a low level of phosphorylation of p12, which must occur elsewhere on the molecule. The low phosphorylation level of the multiply mutant S(61,65,78,81)A was similar to that of the single S61A mutant, implying that the residual phosphorylation can occur on other residues, such as S6, S17, S19, or S42. These residues were not important for virus replication, since alanine substitution at these positions had no effect on the virus. This result suggests that the residual phosphorylation of p12 was neither sufficient for good replication in the absence of S61 nor required when S61 was present.
The isolation and characterization of revertants of the p12 mutant S(61,65)A yielded some surprising suppressor mutations. Many of the revertants (Rev5, Rev12, and Rev14) acquired additional positive charges at nearby positions in p12, most often arginine residues. These additional charges presumably alter the conformation of the protein so as to override the need for the crucial S61 and its phosphorylation. It is possible that the charges enhance the binding of p12 to some interaction partner or to nucleic acids. This region has been noted as similar to sequences in histone H5 and has been suggested as responsible for the DNA- or RNA-binding activity of p12 (16). The importance of a cluster of arginine residues near the C terminus of p12 is highlighted by the effects of specific mutations introduced at these sites. Single alanine substitution of any one of these arginines, and even lysine substitutions, caused a strong replication defect. Thus, arginine residues in this region apparently play a specific role beyond the simple positive charge that they carry.
Other revertants did not compensate with added basic residues but instead contained specific changes very close to the crucial S61 position. Rev1 contained a very subtle change, M63I, in between the two alanine substitutions, whereas Rev2 changed the seemingly unimportant alanine substitution at S65 back to a threonine. The basis for the suppression by these changes is unclear, but presumably these mutations affect the conformation of the protein so as to compensate for the serine mutations. Significantly, neither the revertants with increased basic residues nor those with the more subtle changes showed a restoration in the level of phosphorylation of p12 back to the wild-type levels. Thus, these revertants somehow carried out normal virus replication without the normal levels of phosphorylation of p12. The result hints that although a residue such as S61 is crucial for normal replication it can be easily made irrelevant with small compensatory changes nearby and that the S61 phosphorylation per se is not obligatory. It may well be that even in the wild-type context the phosphorylation is not the crucial aspect of S61 function but that other contributions to the protein are the key features of this residue.
What might be the function of the p12 protein in early steps of infection? The serine substitutions impair the formation of viral circular DNAs (Fig. 3 and 7B), which has been regarded as the hallmark of entry of virus into the nucleus (3, 5, 12, 14). The failure of these mutants to synthesize viral circular DNAs could have several explanations. One possibility is that both the phosphorylation of S61 and the presence of arginine residues might be required for the entry of virus into the nucleus. A variation on this theme is that p12 serves as a nuclear localization signal (NLS) to carry the preintegration complex into nucleus. Many cases have been described in which phosphorylation of NLS elements regulate nuclear import of proteins. This is known for lamins (23), SV40 T antigen (33), protein kinase C (2), v-Jun (40), and NF-AT (38). In studies of nuclear entry of hepatitis B virus, the phosphorylated core proteins have been shown to preferentially bind to the nuclear pore complex over unphosphorylated ones in digitonin-permeabilized cells (21). It was suggested that phosphorylation of core proteins induces exposure of the NLS in the core protein that allows core binding to the nuclear pore complexes by the importin-mediated pathway and the access of viral genome into the nucleus.
In the case of HIV type 1 and other lentiviruses, the viral DNAs can enter the nucleus of nondividing cells, probably through an active import process (5, 10, 24, 25). In contrast, it has been shown that MMLV viral DNAs cannot gain access to the nucleus until the cells progress through mitosis, probably due to a requirement for nuclear envelope breakdown for the entry of the viral integration complex into the nucleus (34). These observations suggest that if p12 phosphorylation is responsible for virus entry into nucleus, the mechanism is probably not a conventional nuclear active import. A variant scenario is that p12 might act as a nuclear retention signal, which allows the efficient retention of viral preintegration complex by binding to nuclear proteins while the nuclear envelopes assemble after mitosis (49). Through this mechanism, the viral preintegration complex can get access to the nucleus and integrate into chromosome.
Another explanation for the requirement for p12 phosphorylation for the synthesis of circular viral DNA is that p12 may be a component of the preintegration complex by binding to viral DNA. Phosphorylation of p12 may be essential for the correct folding of functional preintegration complex, which in turn undergoes either integration or autointegration. This notion is supported by the observation that phosphorylated p12 protein can preferentially bind viral RNA molecules more efficiently than unphosphorylated p12 (35, 37). Given the RNA-binding activities of phosphorylated p12, it is conceivable that p12 may also have DNA-binding activity, and the phosphorylation of p12 may facilitate its binding to viral DNAs of preintegration complex.
The disposition of p12 in the preintegration complex and its folded structure would be enormously helpful information in understanding its function; unfortunately, little is known about the structure of p12. We utilized the Seg program (1) and Pepplot program (GCG) to try to predict its structure; the results were that p12 is predicted to be almost entirely nonglobular, without significant alpha-helical structures (data not shown). These predictions suggested that p12 is a proline-rich, disordered, and low-complexity protein. Direct biophysical studies of its structure might help confirm or dismiss these predictions. It is quite possible that p12 only takes on discrete structures in the presence of nucleic acids or other proteins. The isolation of p12 interacting proteins may shed light on its structure, its function, and the mechanism of entry of MMLV into the nucleus.
Acknowledgments
We are grateful to Valmik K. Vyas, Matthew J. Evans, and Juliana Leung for helpful discussions and reading of early drafts. We thank Bing Yuan, Gilda Tachedjian, Ariberto Fassati, Eran Bacharach, and Barbara Stoymers for technical support and advice.
This work was partially supported by Public Health Service grant CA30488 from the National Cancer Institute. A.Y. is an Associate and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Battiste, J. L., H. Y. Mao, N. S. Rao, R. Y. Tan, D. R. Muhandiram, L. E. Kay, A. D. Frankel, and J. R. Williamson. 1996. α-Helix-RNA major groove recognition in an HIV-1 Rev peptide RRE RNA complex. Science 273:1547-1551. [DOI] [PubMed] [Google Scholar]
- 2.Boulikas, T. 1995. Phosphorylation of transcription factors and control of the cell cycle. Crit. Rev. Eukaryot. Gene Expr. 5:1-77. [PubMed] [Google Scholar]
- 3.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 4.Bukrinskaia, A. G. 1996. Phosphorylation of proteins as a factor for regulating viral infection. Mol. Biol. 30:514-517. [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 pretintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartier, C., P. Sivard, C. Tranchat, D. Decimo, C. Desgranges, and V. Boyer. 1999. Identification of three major phosphorylation sites within HIV-1 capsid: role of phosphorylation during the early steps of infection. J. Biol. Chem. 274:19434-19440. [DOI] [PubMed] [Google Scholar]
- 7.Coates, K., S. J. Cooke, D. A. Mann, and M. P. G. Harris. 1997. Protein kinase C-mediated phosphorylation of HIV-I Nef in human cell lines. J. Biol. Chem. 272:12289-12294. [DOI] [PubMed] [Google Scholar]
- 8.Colicelli, J., and S. P. Goff. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47-59. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, S., and S. P. Goff. 1984. Mutations in gag proteins p12 and p15 of Moloney murine leukemia virus block early stages of infection. J. Virol. 49:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier, R. A., and M. H. Malim. 1999. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res. 52:275-299. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 12.Fritsch, E., and H. M. Temin. 1977. Formation and structure of infectious DNA of spleen necrosis virus. J. Virol. 21:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guntaka, R. V., O. C. Richards, P. R. Ahnkk, H.-J. Kung, N. Davidson, E. Fritsch, J. M. Bishop, and H. E. Varmus. 1976. Covalently closed circular DNA of avian sarcoma virus: purification from nuclei of infected quail tumor cells and measurement by electron microscopy and gel electrophoresis. J. Mol. Biol. 106:337-357. [DOI] [PubMed] [Google Scholar]
- 15.Hauber, J., M. Bouvier, M. H. Malim, and B. R. Cullen. 1988. Phosphorylation of the rev gene product of human immunodeficiency virus type 1. J. Virol. 62:4801-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, L. E., R. V. Gilden, and S. Oroszlan. 1979. Amino acid sequence homology between histone H5 and murine leukemia virus phosphoprotein p12. Science 203:1346-1348. [DOI] [PubMed] [Google Scholar]
- 17.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-371. [DOI] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikuta, K., and R. B. Luftig. 1988. Detection of phosphorylated forms of Moloney murine leukemia virus major capsid protein p30 by immunoprecipitation and two-dimensional gel electrophoresis. J. Virol. 62:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leis, J., D. Baltimore, J. M. Bishop, J. Coffin, E. Fleissner, S. P. Goff, S. Oroszlan, H. Robinson, A. M. Skalka, H. M. Temin, and V. Vogt. 1988. Standardized and simplified nomenclature for proteins common to all retroviruses. J. Virol. 62:1808-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leukel, M., and E. Jost. 1995. Two conserved serines in the nuclear localization signal flanking region are involved in the nuclear targeting of human lamin A. Eur. J. Cell Biol. 68:133-142. [PubMed] [Google Scholar]
- 24.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMillan, N. A., R. F. Chun, D. P. Siderovski, J. Galabru, W. M. Toone, C. E. Samuel, T. W. Mak, A. G. Hovanessian, K. T. Jeang, and B. R. Williams. 1995. HIV-1 Tat interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413-424. [DOI] [PubMed] [Google Scholar]
- 27.Naso, R. B., W. L. Karshin, Y. H. Wu, and R. B. Arlinghaus. 1979. Characterization of 40,000- and 25,000-dalton intermediate precursors to Rauscher murine leukemia virus gag gene products. J. Virol. 32:187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal, B. K., R. M. McAllister, M. B. Gardner, and P. Roy-Burman. 1975. Comparative studies on the structural phosphoproteins of mammalian type C viruses. J. Virol. 16:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal, B. K., and P. Roy-Burman. 1975. Phosphoproteins: structural components of oncornaviruses. J. Virol. 15:540-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul, M., and M. A. Jabbar. 1997. Phosphorylation of both phosphoacceptor sites in the HIV-1 Vpu cytoplasmic domain is essential for Vpu-mediated ER degradation of CD4. Virology 232:207-216. [DOI] [PubMed] [Google Scholar]
- 32.Puffer, B., L. Parent, J. Wills, and R. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rihs, H. P., and R. Peters. 1989. Nuclear transport kinetics depend on phosphorylation site-containing sequences flanking the karyophilic signal of the simian virus 40 T-antigen. EMBO J. 8:1479-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen, A., C. J. Sherr, and G. J. Todaro. 1977. Phosphorylation of murine type C viral p12 proteins regulates their extent of binding to the homologous RNA. Cell 10:489-496. [DOI] [PubMed] [Google Scholar]
- 36.Sen, A., C. J. Sherr, and G. J. Todaro. 1976. Specific binding of the type C viral core protein p12 with purified viral RNA. Cell 7:21-32. [DOI] [PubMed] [Google Scholar]
- 37.Sen, A., and G. J. Todaro. 1977. The genome-associated, specific RNA binding proteins of avian and mammalian type C viruses. Cell 10:91-99. [DOI] [PubMed] [Google Scholar]
- 38.Shibasaki, F., E. R. Price, D. Milan, and F. McKeon. 1996. Role of kinases and the phosphotase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 382:370-373. [DOI] [PubMed] [Google Scholar]
- 39.Smith, C. M., W. B. Potts III, J. S. Smith, and M. J. Roth. 1997. RNase H cleavage of tRNAPro mediated by M-MuLV and HIV-1 reverse transcriptases. Virology 229:437-446. [DOI] [PubMed] [Google Scholar]
- 40.Tagawa, T., T. Kuroki, P. K. Vogt, and K. Chida. 1995. The cell cycle-dependent nuclear import of v-Jun is regulated by phosphorylation of a serine adjacent to the nuclear localization. J. Cell Biol. 130:255-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittaker, G. R., and A. Helenius. 1998. Nuclear import and export of viruses and virus genomes. Virology 246:1-23. [DOI] [PubMed] [Google Scholar]
- 42.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 44.Xiang, Y., C. Cameron, J. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshinaka, Y., and R. B. Luftig. 1982. In vitro phosphorylation of murine leukemia virus proteins: specific phosphorylation of Pr65gag, the precursor of the internal core antigens. Virology 116:181-195. [DOI] [PubMed] [Google Scholar]
- 47.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan, B., A. Fassati, A. Yueh, and S. P. Goff. 2002. Characterization of Moloney murine leukemia virus p12 mutants blocked during early events of infection. J. Virol. 76:10801-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, Y., and L. Ratner. 2000. Phosphorylation of human immunodeficiency virus type 1 Vpr regulates cell cycle arrest. J. Virol. 74:6520-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]