Abstract
Telomeric DNA can fold into four-stranded structures known as G-quadruplexes. Here we investigate the ability of G-quadruplex DNA to serve as a substrate for recombinant Tetrahymena and native Euplotes telomerase. Inter- and intramolecular G-quadruplexes were gel-purified and their stability examined using native gel electrophoresis, circular dichroism (CD) and thermal denaturation. While intermolecular G-quadruplexes were highly stable, they were excellent substrates for both ciliate telomerases in primer extension assays. In contrast, intramolecular G-quadruplexes formed in K+ exhibited biphasic unfolding and were not extended by ciliate telomerases. Na+-stabilised intramolecular G-quadruplexes were extended by telomerase owing to their rapid rate of dissociation. The Tetrahymena telomerase protein component bound to inter- but not intramolecular K+-stabilised G-quadruplexes. This study provides evidence that parallel intermolecular G-quadruplexes can serve as substrates for telomerase in vitro, their extension being mediated through direct interactions between this higher-order structure and telomerase.
Keywords: ciliate, G-quadruplex, telomerase, telomere
Introduction
Telomeres are protein–DNA structures at the ends of eukaryotic chromosomes that protect chromosome ends from fusion and are vital in safeguarding genomic stability. The 3′ strand of telomeres is composed of tandem repeats of short G-rich sequences that protrude as a single-stranded DNA overhang. These repeats are T2AG3 in humans and T2G4 and T4G4, respectively, in the ciliates Tetrahymena and Euplotes (Blackburn and Gall, 1978; Klobutcher et al, 1981). Single strands of telomeric DNA can adopt higher order structures, known as G-quartets, under physiological conditions in vitro (Henderson et al, 1987; Sundquist and Klug, 1989). These structures are defined by the coordination of four guanine residues in a cyclic array, stabilised by Hoogsteen hydrogen-bonding and a centrally located cation (Figure 1A) (Williamson et al, 1989). Multiple layers of G-quartets stack to form G-quadruplexes, in which one or more strands are assembled together in either an intra- or intermolecular configuration (Figure 1). G-quadruplexes exhibit extensive structural polymorphism (reviewed in Simonsson, 2001). The DNA strand orientation may be either parallel (Figure 1C and D), antiparallel (Figure 1B) or in some cases both configurations (Figure 1E).
Figure 1.
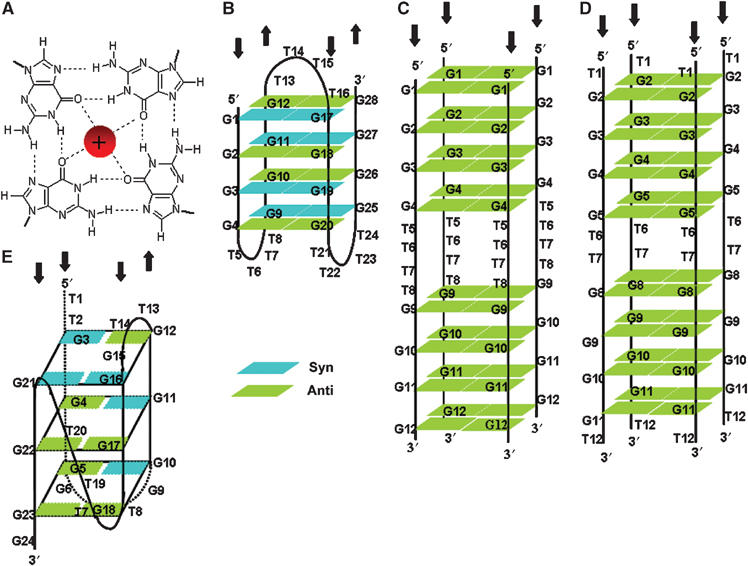
(A) Structure of a G-quartet. Hydrogen bonds between guanines and interaction with a monovalent cation, located centrally, are shown. (B) Solution structure of antiparallel Oxy 3.5 G-quadruplex in sodium (Wang and Patel, 1995) and potassium (Smith et al, 1995). (C, D) Predicted structure of parallel intermolecular Oxy 1.5 G-quadruplex in potassium (C) and 12GT in sodium (D) (this work). (E) Solution structure of antiparallel intramolecular 24GG G-quadruplex in sodium (Wang and Patel, 1994).
Evidence supporting the physiological relevance of G-quadruplex structures in vivo is rapidly mounting. Notably, several proteins have been isolated that drive intermolecular G-quadruplex assembly. For example, the β subunit of Oxytricha telomere binding protein has been demonstrated to accelerate formation of dimer and tetramer quartets (Fang and Cech, 1993). Similarly, RAP1, a major dsDNA telomeric binding protein from Saccharomyces cerevisiae, not only binds to but also promotes the formation of a parallel G-quadruplex in the presence of potassium (Giraldo and Rhodes, 1994; Giraldo et al, 1994). Furthermore, numerous factors that resolve G-quadruplex structure (Baran et al, 1997; Harrington et al, 1997; Sun et al, 1998, 1999; Enokizono et al, 2005) and some that specifically cleave G-quadruplex proximal DNA (Liu et al, 1993; Sun et al, 2001) have been identified. The most direct evidence for the existence of these secondary structures in vivo comes from the generation of antibodies against antiparallel G-quadruplex DNA that react with the ciliated protozoan Stylonychia lemnae macronuclei (Schaffitzel et al, 2001). The telomere binding proteins TEBPα and TEBPβ are both required for the formation of these G-quadruplexes at Stylonychia telomeres in vivo (Paeschke et al, 2005).
Telomerase is a ribonucleoprotein enzyme, which adds telomeric repeats to the chromosome end by reverse transcription of an integral RNA template (Greider and Blackburn, 1985). The observation that >85% of all cancers exhibit telomerase activity has attracted significant attention to this enzyme (Shay and Bacchetti, 1997). Initially, Zahler et al (1991) demonstrated that telomerase from the ciliate Oxytricha nova does not require primer folding into a G-quadruplex for elongation, and proposed that stabilisation of a primer's secondary structure may in fact inhibit telomere elongation in vivo. However, this seminal research used crude extracts containing telomerase in which G-quadruplex-interacting proteins may have been present, as well as crude mixtures of G-quadruplex DNA.
Using purified telomerase isolated from Euplotes aediculatus and purified recombinant Tetrahymena thermophila telomerase, we have re-evaluated the ability of telomerase to extend G-quadruplex DNA. We utilised telomeric oligonucleotides capable of forming both intramolecular and intermolecular conformations, stabilised by either K+ or Na+ ions. Owing to the heterogeneous nature of G-quadruplex conformations, we were careful to isolate particular conformations by gel purification and verify their identity. In the case of intramolecular antiparallel G-quadruplexes, our results validated the initial observation that increasing stabilisation of telomeric secondary structure inhibits telomerase-catalysed primer extension (Zahler et al, 1991). However, intermolecular parallel G-quadruplexes can be extended by both Euplotes and Tetrahymena telomerase, to a greater extent than would be predicted by their slow dissociation.
Results
Folding and characterisation of G-quadruplex DNA
To examine the ability of telomerase to extend G-quadruplex DNA, we prepared and isolated G-rich oligonucleotides folded in either K+ or Na+. Tetrahymena telomeric oligonucleotides (Table I) migrated according to their size on a native gel with no salt, indicating a lack of secondary structure (Figure 2A). In general, folding G-rich DNA gives rise to a heterogeneous population of structures. Indeed, in our hands, most telomeric oligonucleotides folded into heterogeneous mixtures when analysed by nondenaturing PAGE (Figures 2 and 3). In the presence of both Na+ and K+ salts, a large proportion of the 21 and 24-nucleotides (nt) Tetrahymena oligonucleotides migrated faster than expected, indicating that they have a compact structure, which is likely to be an intramolecular G-quadruplex, as has been observed previously (Figure 2B and C; Williamson et al, 1989).
Table 1.
Oligonucleotides used in this study
| Nomenclature | Sequence | Length |
|---|---|---|
| Tetrahymena oligonucleotides | ||
| 6TT | 5′-GGGGTT-3′ | 6 |
| 6GG | 5′-TTGGGG-3′ | 6 |
| 12GT | 5′-TGGGGTTGGGGT-3′ | 12 |
| 24TT | 5′-GGGGTTGGGGTTGGGGTTGGGGTT-3′ | 24 |
| 24GG | 5′-TTGGGGTTGGGGTTGGGGTTGGGG-3′ | 24 |
| 21GG | 5′-GGGGTTGGGGTTGGGGTTGGG-3′ | 21 |
| 48CC | 5′-(AACCCC)8-3′ | 48 |
| 48AA | 5′-(CCCCAA)8-3′ | 48 |
| 30AA | 5′-(CCCCAA)5-3′ (RNA) | 30 |
| Biot-24TT | 5′-Biotin-GGGGTTGGGGTTGGGGTTGGGGTT- 3′ | 24 |
| Biot-21GG | 5′-Biotin-GGGGTTGGGGTTGGGGTTGGG-3′ | 21 |
| Euplotes oligonucleotides | ||
| Oxy 1.5 | 5′-GGGGTTTTGGGG-3′ | 12 |
| Oxy 3.5 | 5′-GGGGTTTTGGGGTTTTGGGGTTTTGGGG-3′ | 28 |
| Ea23 | 5′-TTTTGGGGTTTTGGGGTTTTGGG-3′ | 23 |
| EaTR | 5′-CAAAACCCCAAAACC-3′ (RNA) | 15 |
| Non-telomeric oligonucleotides | ||
| Biot-PBR | 5′-Biotin-AGCCACTATCGACTACGCGATCAT- 3′ | 24 |
| T10 | 5′-TTTTTTTTTT-3′ | 10 |
| T15 | 5′-TTTTTTTTTTTTTTT-3′ | 15 |
| T20 | 5′-TTTTTTTTTTTTTTTTTTTT-3′ | 20 |
Figure 2.
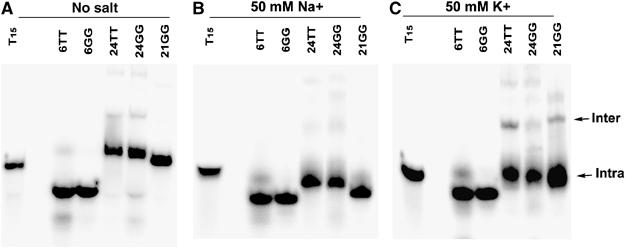
Native gel electrophoresis of the indicated oligonucleotides (1 μM) of Tetrahymena telomeric sequence in the absence of salt (A), in 50 mM NaGlu (B) or KGlu (C). T15 is an unstructured molecular weight (MW) marker.
Figure 3.
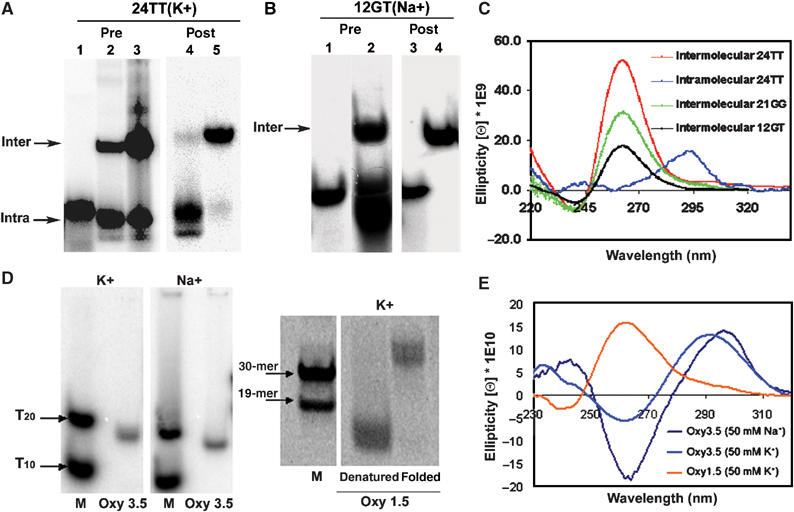
Characterisation of gel-purified G-quadruplexes. (A) Native gel electrophoresis of G-quadruplexes formed by Tetrahymena oligonucleotide 24TT in the presence of 50 mM KGlu. Lane 1: T15 unstructured MW marker. Lanes 2 and 3: 24TT oligonucleotide folded in the presence of 50 mM KGlu at 4 and 100 μM concentrations, respectively, prior to gel purification. The intra- and intermolecular bands were isolated from 4 and 100 μM 24TT G-quadruplex mixtures, respectively, and their post-purification profiles are shown in lanes 4 and 5. (B) Native gel electrophoresis of 12GT intermolecular G-quadruplex in 100 mM NaCl pre- (lane 2) and post-gel purification (lane 4). Lanes 1 and 3: T15 MW marker. (C) CD spectra of gel-purified intermolecular 24TT, 21GG, and intramolecular 24TT G-quadruplexes in 50 mM KGlu and intermolecular 12GT G-quadruplex in 100 mM NaCl. The DNA concentrations were 17 μM for intermolecular 24TT and 12GT, 10 μM for intermolecular 21GG and 2.6 μM for intramolecular 24TT. (D) Native gel electrophoresis of gel-purified G-quadruplexes formed by Euplotes oligonucleotides Oxy 3.5 (50 μM, in the presence of 50 mM K+ or NaGlu) and Oxy 1.5 (200 μM, in the presence of 50 mM KGlu). MW markers (M) are 5′-end labelled Poly-T (T10, T20) or non-telomeric DNA (19-mer and 30-mer) stained with Sybr Green. (E) CD spectra of gel-purified 5 μM Oxy 3.5 intramolecular G-quadruplexes (50 mM K+ or NaGlu) and 5 μM Oxy 1.5 intermolecular G-quadruplex (50 mM KGlu).
In the presence of K+, the 24TT and 21GG oligonucleotides also demonstrated slower mobility species, indicative of intermolecular G-quadruplexes consisting of two or more DNA strands (Figure 2C). The proportion of this band increased with increasing DNA concentrations, as would be expected for an intermolecular complex (data not shown). 24GG did not show a prominent intermolecular G-quadruplex band in K+ (Figure 2C), even though it has been reported that this oligonucleotide can form an intermolecular G-quadruplex in the presence of K+ (Hardin et al, 1991); the reason for this difference under our reaction conditions is unknown. The Tetrahymena telomeric oligonucleotide 12GT also demonstrated a mixture of bands in the presence of Na+, including a band of slower mobility than the T15 marker (Figure 3B). Crosslinking analysis of this band after gel purification indicates that it is a four-stranded intermolecular structure (Figure 1D, Supplementary Figure 1).
Since the pattern of G-quadruplexes formed by these oligonucleotides is heterogeneous, it was important to isolate individual conformations for further study. After gel purification, both inter- and intramolecular G-quadruplex conformations of 24TT in K+ were well preserved (Figure 3A, lanes 4 and 5; ∼95% purity), as was the intermolecular form of 12GT in Na+ (Figure 3B, lane 4, ∼99% purity). Structures formed from 21GG were less well-preserved after purification, yielding 60–85% purity (data not shown). The contaminating bands consisted of other G-quadruplexes, not linear DNA. All subsequent analyses were carried out using these gel-purified structures.
We confirmed that the observed bands represent G-quadruplexes using circular dichroism (CD). Parallel G-quadruplexes exhibit a positive CD peak at ∼265 nm and a negative CD peak at ∼240 nm, while antiparallel G-quadruplexes exhibit a positive CD peak at ∼295 nm and a negative CD peak at ∼260 nm (Williamson, 1994; Keniry, 2000). The gel-purified intermolecular conformations displayed strong positive and negative peaks at 260/240 nm, respectively, and negligible signals at 295 nm, supporting their assignment as parallel G-quadruplexes (Figure 3C). On the other hand, the isolated intramolecular 24TT G-quadruplex had a strong positive peak at 295 nm and no signal at 260 nm, indicative of an antiparallel conformation (Figure 3C). An intramolecular antiparallel structure has previously been observed by NMR and platination studies for 24GG in Na+ (Wang and Patel, 1994; Redon et al, 2001); the structures of the other Tetrahymena telomeric oligonucleotides used in this study have not been solved.
As substrates for Euplotes telomerase, we chose to study Oxy 1.5 and Oxy 3.5 (Table I), which form well-characterised G-quadruplexes. Oxy 3.5 folded in both K+ and Na+ demonstrated a major compact species that migrated below the T20 marker, supporting intramolecular G-quadruplex formation (Figure 3D). Oxy 1.5 folded in K+ showed a shift toward a slower moving species (∼48 nt), supporting a multimeric, possibly four-stranded, intermolecular quadruplex (Figure 3D).
G-quadruplex formation of these oligonucleotides was also verified by CD (Figure 3E). Oxy 3.5 folded in K+ or Na+ exhibited positive and negative CD peaks at 291/262 nm and 296/264 nm, respectively. These results verify the formation of antiparallel, intramolecular G-quadruplexes as previously characterised (Smith et al, 1995; Wang and Patel, 1995; Petraccone et al, 2004). Oxy 1.5 folded in K+ showed positive and negative CD peaks at 262 and 238 nm, suggesting the formation of a parallel G-quadruplex. The major compact or multimeric species from each oligomer was isolated by gel purification and used for further characterisations.
Stability of Tetrahymena G-quadruplexes
The stability of gel-purified G-quadruplexes was measured by examining the melting temperature (Tm) of the folded structures and by employing a complementary strand ‘trap' assay (Raghuraman and Cech, 1990). In the ‘trap' assay, an excess of a C-rich complementary strand is added to the G-quadruplex and samples are analysed over time by nondenaturing electrophoresis. Provided that the concentration of the C-strand is high enough to trap any unfolded G-strand molecules, the rate of Watson-Crick (WC) duplex formation is indicative of the rate of G-quadruplex unfolding. The rates we observed (below) were independent of the concentration of C-strand oligonucleotide, providing evidence that this strand is not actively invading the quadruplex (data not shown).
Upon addition of a 10-fold excess of C-rich (48CC) strand, ∼98 and 100% of 24TT and 21GG Na+-stabilised intramolecular G-quadruplexes, respectively, hybridised to the complementary strand within 1.5 min (Supplementary Figure 2A and D). This suggests that G-quadruplexes formed from both sequences in Na+ are highly unstable and unfold rapidly. Both G-quadruplexes exhibited a monophasic mode of unfolding with a half-life (t1/2) of <1.5 min.
The intramolecular 21GG G-quadruplex stabilised in K+ displayed biphasic unfolding kinetics. Approximately 40% of this G-quadruplex hybridised to the complementary strand within 1 min of the reaction; over the following 60 min a further 30% of this G-quadruplex slowly unfolded (Figure 4A and D). The unfolding profile of this G-quadruplex fitted to a double exponential equation, yielding an unfolding rate of 0.95±0.13 h−1 and a t1/2 of 0.7±0.1 h for the slower unfolding population. The population that was immediately accessible to the C-rich strand had a t1/2 of <1 min.
Figure 4.
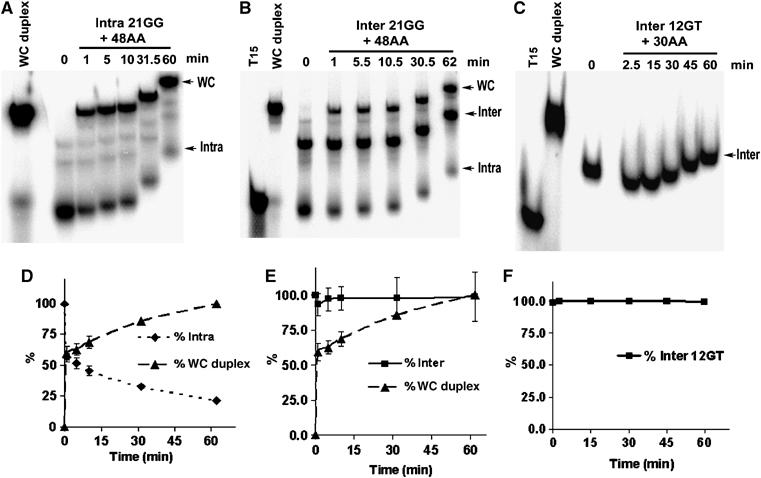
Complementary strand trap method for determining the rate of unfolding of gel-purified Tetrahymena G-quadruplexes. The indicated G-quadruplexes were incubated with 10- to 20-fold excess of complementary single-stranded DNA (48AA) or RNA (30AA) and at the indicated time intervals aliquots were loaded onto a native 12% polyacrylamide gel containing 50 mM KGlu (A, B) or 100 mM NaCl (C). T15: MW marker; WC: preannealed Watson–Crick duplex. Note that later time points have less time to migrate into the gel. Plots D–F show the quantitation of the corresponding gels; error bars represent standard deviation from an average of at least two experiments. (A) Unfolding of 32P-labelled intramolecular K+-stabilised 21GG G-quadruplex at 0.54 μM (85% purity). (B) Unfolding of 32P-labelled intermolecular K+-stabilised 21GG G-quadruplex at 3 μM (63% purity). (C) Unfolding of 32P-labelled intermolecular Na+-stabilised 12GT G-quadruplex at 11 μM (99% purity).
The intramolecular 24TT K+-stabilised G-quadruplex also demonstrated biphasic unfolding with an initial burst (∼32%) of hybridisation 1.5 min after addition of the C-rich strand followed by slower unfolding (Supplementary Figure 2B and E). Fitting this data to a double exponential equation yielded an unfolding rate of 0.15±0.02 h−1 for the slower unfolding population, which corresponds to a t1/2 of 4.8±0.8 h. The rapidly hybridisable population had a t1/2 of <1 min. Thus, for both oligonucleotides, there are two distinct species: one that is readily hybridisable by the complementary strand and one that is not.
Intermolecular G-quadruplexes formed from 21GG, 24TT or 12GT sequences followed a monophasic dissociation trend. Within the 62 min of the hybridisation reaction <6% of the total intermolecular 21GG G-quadruplex contributed to WC duplex formation (Figure 4B and E). The majority of the duplex formation can be attributed to the contaminating intramolecular counterpart of this G-quadruplex. Similarly, <7% of the gel-purified intermolecular 24TT G-quadruplex unfolded within 63 min of incubation with its complementary strand (Supplementary Figure 2C and F). Notably, in unfolding assays using the intermolecular 12GT quadruplex, which is much more pure than the other two, no WC duplex formation is detectable even after 60 min (Figure 4C and F). For all three intermolecular G-quadruplexes, the unfolding rate was too slow to fit to a single exponential equation.
Observations from ‘trap' assays described above were confirmed by measuring melting temperatures of the intermolecular 21GG and 24TT G-quadruplexes. Both G-quadruplexes displayed high Tm values that increased with increasing G-quadruplex concentration, indicative of intermolecular complexes (Table II). Overall, it is apparent that the parallel intermolecular G-quadruplexes are highly stable.
Table 2.
Tm values for Tetrahymena and Euplotes gel-purified G-quadruplexesa
| G-quadruplex | Cation | Strand orientation | Strand stoichiometry | Tm (°C)b | Tm (°C)c |
|---|---|---|---|---|---|
| Oxy 3.5 | Na+ | Antiparallel | Intramolecular | 60 | — |
| Oxy 3.5 | K+ | Antiparallel | Intramolecular | 85 | — |
| Oxy 1.5 | K+ | Parallel | Intermolecular | 88 | — |
| 21GG | K+ | Parallel | Intermolecular | 82 | 88 |
| 24TT | K+ | Parallel | Intermolecular | 77 | 90 |
| aUV melting curves were monitored at 295 nm in the appropriate folding buffer. Heating and cooling curves were superimposable with only slight hysteresis. | |||||
| b10 μM strand concentration. | |||||
| c20 μM strand concentration. | |||||
Stability of Euplotes G-quadruplexes
Similar measurements of Tm and complementary strand trap assays were carried out to determine the stability of the Euplotes intramolecular G-quadruplexes. In these trap assays, however, an RNA oligonucleotide consisting of the 15-nt Euplotes telomerase RNA template sequence (EaTR) was used as the trap, in order to mimic a telomerase assay.
The K+-stabilised Oxy 3.5 intramolecular G-quadruplex was incubated in the presence of increasing EaTR template in K+ reaction buffer (Figure 5A). Lone G-quadruplex exhibited a single band while pre-annealed Oxy 3.5-EaTR showed multiple band shifts, indicating that several higher-order species had formed. The presence of multiple species is likely due to several different binding modes of the RNA template to the DNA primer in 1:1 and 2:1 stoichiometries. These higher-order interactions were not characterised individually, but were treated as contributors to the total shifted species.
Figure 5.
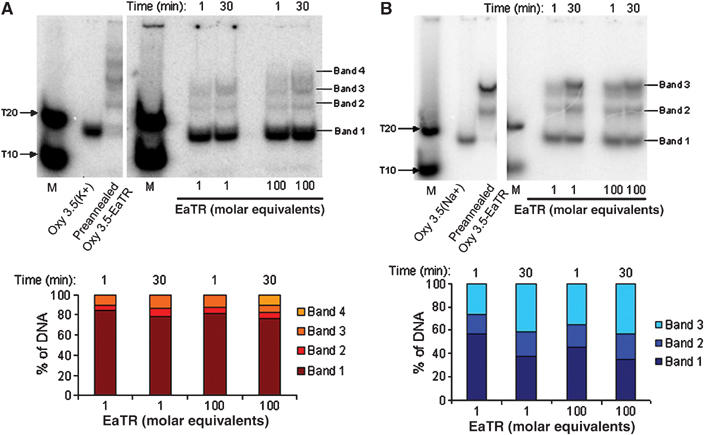
Euplotes complementary strand ‘trap' assays used to determine G-quadruplex stability in the presence of telomerase RNA template (EaTR). 5′ end-labelled 10 μM Oxy 3.5 intramolecular G-quadruplexes in 50 mM KGlu (A) or NaGlu (B) were incubated with 1 or 100 equivalents of unlabelled EaTR template for 1 or 30 min at 25°C and electrophoresed on 20% nondenaturing polyacrylamide gels (in 50 mM K+ (A) or NaGlu (B)). Lane 1: 5′ end-labelled unstructured poly-T markers (M). Lane 2: lone G-quadruplex. Lane 3: preannealed Oxy 3.5-EaTR polymorphic species.
To determine if the amount of shifted species changed over time, time course incubations of 1 and 100 molar equivalents of EaTR template at 1 and 30 min time points were conducted (Figure 5A). At 1 min with 100 equivalents of EaTR template, 78% of the K+-stabilised Oxy 3.5 G-quadruplex was conserved, while at 30 min with 100 equivalents of template, 76% of the quadruplex was conserved. This limited gain in shifted species over 29 min indicates that a small population (∼22%) of G-quadruplex was available to rapidly bind to the template, followed by a slower unwinding of the remaining hybridisable population. Thus, like the K+-stabilised Tetrahymena intramolecular G-quadruplex, this structure exhibits biphasic unfolding kinetics.
The Na+-stabilised Oxy 3.5 G-quadruplex time course revealed that with 100 equivalents of EaTR template at 1 and 30 min of hybridisation, 45 and 35% of the quadruplex was conserved, respectively (Figure 5B). Thus, like the Tetrahymena intramolecular G-quadruplexes, the Na+-stabilised Oxy 3.5 G-quadruplex is less stable than the K+ form. ‘Trap' assays with the Oxy 1.5 intermolecular G-quadruplex were unsuccessful due the radioactive end label interfering with intermolecular G-quadruplex assembly (Uddin et al, 2004); however, the stability of this Euplotes G-quadruplex proved to be very high as determined from Tm measurements (Table II).
Extension of folded G-quadruplexes by Tetrahymena telomerase
Telomerase activity assays were performed to determine if the G-quadruplexes characterised in the previous sections act as substrates for their respective telomerase enzymes. Na+-stabilised intramolecular G-quadruplexes formed from either 21GG or 24TT were readily extendable by recombinant Tetrahymena telomerase (Figure 6A, Supplementary Figure 3A). This was also the case for 24GG (data not shown). Extension of these G-quadruplexes is likely due to the fact that they are in rapid equilibrium with their linear forms. These G-quadruplexes exhibited higher Km values and equivalent or lower relative Vmax values than their linear counterparts (Table III). This indicates a lower relative specificity (kcat/Km) of the enzyme for these G-quadruplex substrates. This supports the idea that if a Na+-stabilised intramolecular G-quadruplex is in constant equilibrium with its unfolded state, then less linear DNA is available for binding and extension by telomerase.
Figure 6.
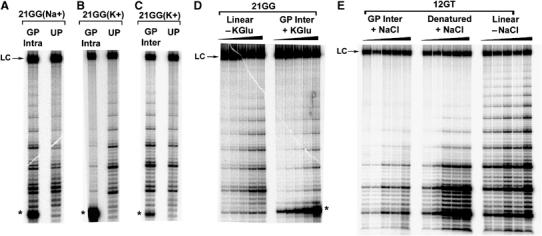
Telomerase activity assays using in vitro reconstituted recombinant Tetrahymena telomerase and G-quadruplexes formed from Tetrahymena oligonucleotides. For all panels, LC=loading control (32P-labelled 100-mer), * represents the unextended 5′ 32P-labelled gel-purified G-quadruplex, GP and UP refer to gel-purified and unpurified G-quadruplexes, respectively. All assays were conducted at 25°C for 60 min (A–C), 10 min (D) or 15 min (E) using ∼2 nM enzyme. (A) Telomerase extension of 32P-labelled intramolecular 21GG (‘GP Intra'; 1.8 μM, 85% purity) and unlabelled 21GG (1.8 μM) in 50 mM NaGlu. (B) Telomerase extension of 32P-labelled intramolecular 21GG (1.1 μM, 78% purity) and unlabelled 21GG (1.1 μM) in 150 mM KGlu. (C) Telomerase extension of 32P-labelled intermolecular 21GG (0.7 μM, 66% purity) and unlabelled 21GG (0.7 μM) in 150 mM KGlu. (D) Telomerase extension of 32P-labelled intermolecular 21GG (0.09–1.5 μM, 63% purity) in 50 mM KGlu. The control is unlabelled linear 21GG over the same concentration range, in the absence of KGlu. (E) Telomerase extension of intermolecular 12GT (0.06–8 μM, 99% purity) in 100 mM NaCl. Linear 12GT (−NaCl) and denatured 12GT (+NaCl) G-quadruplex over the same concentration range were used as controls.
Table 3.
Km values and relative Vmax ratios (G-quadruplex/linear DNA) for gel-purified intramolecular Tetrahymena G-quadruplexes stabilised in 50 mM NaGlu and their linear counterparts
| Primer | Conformation | Km±s.d. (nM) | Relative Vmax (folded/linear) |
|---|---|---|---|
| 24TT | Intramolecular | 1240±750 | 0.34±0.09 |
| 24TT | Linear | 560±60 | |
| 24GG | Intramolecular | 880±420 | 1.19±0.4 |
| 24GG | Linear | 160±50 | |
K+-stabilised gel-purified intramolecular G-quadruplexes were not extended by telomerase (Figure 6B, Supplementary Figure 3B). Some very faint extension products were discernable, which could be attributed to the presence of contaminating intermolecular G-quadruplex. This result was unexpected given the immediate hybridisation of 32–40% of this G-quadruplex to its complementary strand followed by a much slower dissociation of the remainder of the population (see Figure 4A and D, Supplementary Figure 2B and E). It is improbable that the absence of accessory protein(s) in the in vitro reconstituted preparation of Tetrahymena telomerase resulted in an inability of telomerase to extend K+-stabilised intramolecular G-quadruplexes, since partially purified native Tetrahymena telomerase also failed to utilise the K+-stabilised intramolecular 24GG G-quadruplex as a primer (data not shown).
Intermolecular G-quadruplexes, unlike their intramolecular counterparts, were excellent substrates for Tetrahymena telomerase (Figure 6C, D and E, Supplementary Figure 3B and C). Their extension is not likely due to spontaneous dissociation of the G-quadruplex into its linear state, since it is evident from the unfolding analyses that only a very small proportion (at most ∼6%) of the G-quadruplex unfolds within the first 10 min of the reaction (Figure 4E and F, Supplementary Figure 2F). If the extension of 21GG intermolecular G-quadruplex that is seen in Figure 6D is due to this unfolding then it should be equivalent to the level of extension of linear DNA of the same sequence at 6% of a given concentration of G-quadruplex. However, the extension of linear DNA at 0.09 μM, for example, is 3.8-fold lower than the extension of G-quadruplex at 1.5 μM. The same holds true for the 24TT intermolecular G-quadruplex (Supplementary Figure 3C). This argument is even stronger for the 12GT intermolecular G-quadruplex, since it is extended by telomerase (Figure 6E) in the absence of any detectable unfolding (Figure 4C).
To further characterise intermolecular G-quadruplexes as candidate substrates for telomerase, we measured the affinity (Km) and rate constants (kcat) of gel-purified intermolecular 24TT stabilised in K+ and its linear control. Telomerase has a reduced affinity for the intermolecular G-quadruplex, with a Km of 3160±1100 nM, as compared to that of the linear control at 450±230 nM. The kcat values for the G-quadruplex and its linear counterpart measure 0.4±0.1 and 0.18±0.01 min−1, respectively. Despite a higher kcat for the G-quadruplex, its kcat/Km remains lower than that of the linear primer (2.1±0.9 × 103 versus 6.7±2.5 × 103 s−1 M−1). Thus, while the G-quadruplex is a good substrate, its linear counterpart is favoured by telomerase.
Extension of folded G-quadruplexes by Euplotes telomerase
G-quadruplexes folded from Oxy 3.5 and Oxy 1.5 were tested for their ability to act as primers for native Euplotes telomerase. All telomerase assays included heat denatured control primer Ea23 and corresponding Oxy 1.5 or 3.5 sequences (Figure 7). The K+-stabilised Oxy 3.5 intramolecular G-quadruplex was a poor telomerase primer. The presence of a stable secondary structure in this primer directly correlates with an increase in Km and decrease in relative kcat/Km (Table IV). By contrast, Na+-stabilised Oxy 3.5 intramolecular G-quadruplex allowed greater overall activity. The lower Km value supports the trend that Na+ stabilises G-quadruplexes less effectively than K+, resulting in a more accessible primer.
Figure 7.

Euplotes primer extension assays used to determine if G-quadruplexes can be extended by telomerase. Denatured control primers (Ea23, Oxy 1.5 and Oxy 3.5) and G-quadruplexes from 25 nM–2 μM were incubated in 50 mM KGlu (Oxy 1.5) or 50 mM K+ or NaGlu (Oxy 3.5) at 25°C for 60 min in the presence of 1.4 ng of purified telomerase. LC=loading control.
Table 4.
Km values and relative Vmax ratios (folded/linear Ea23 control) for gel-purified Euplotes G-quadruplexes stabilised in 50 mM monovalent cation and their linear counterparts
| Primer | Stabilising cation | Conformation | Km±s.d. (nM) | Relative Vmax (folded/linear) |
|---|---|---|---|---|
| Ea23 | K+ | Linear/denatured | 90±20a | 1.00±0.06 |
| Oxy 3.5 | K+ | Linear/denatured | 100±30 | 0.56±0.07 |
| Oxy 3.5 | K+ | Intramolecular | 460±120 | 0.30±0.03 |
| Ea23 | Na+ | Linear/denatured | 40±3 | 1.00±0.02 |
| Oxy 3.5 | Na+ | Linear/denatured | 47±12 | 0.65±0.03 |
| Oxy 3.5 | Na+ | Intramolecular | 27±9 | 0.56±0.03 |
| Ea23 | K+ | Linear/denatured | 110±40 | 1.00±0.14 |
| Oxy 1.5 | K+ | Linear/denatured | 40±7 | 0.67±0.03 |
| Oxy 1.5 | K+ | Intermolecular | 74±16 | 0.57±0.04 |
| Km's of Ea23 and G-quadruplexes were determined simultaneously to account for inter-day variation. | ||||
The K+-stabilised Oxy 1.5 intermolecular G-quadruplex exhibited reduced affinity and specificity compared to the control primer Ea23. Interestingly, despite a higher Km and lower kcat/Km, the overall activity was as high as denatured Oxy 1.5, suggesting that this G-quadruplex is extended by telomerase to a greater extent than would be expected from its stability.
As further evidence that the extension of the Tetrahymena and Euplotes G-quadruplexes was not due to equilibrium of these structures with their linear counterparts, we subjected the G-quadruplex DNA to treatment with enzymes known to act on single-stranded DNA. Snake venom phosphodiesterase 1 (SVP1), which cleaves single-stranded DNA exonucleolytically from the 3′ end, digested a majority of linear control DNA (Supplementary Figure 4). However, close to 100% of intermolecular quadruplexes 24TT, 12GT and Oxy 1.5 were resistant to treatment with SVP1. In fact, the intramolecular quadruplexes were digested to a greater extent than the intermolecular ones, consistent with their faster rates of unfolding and in contrast to their lower extension by telomerase. Approximately 13% of intermolecular 21GG was digested, but this quadruplex contains ∼35% contamination with intramolecular 21GG. As expected, the terminal two 3′ residues were digested from 24TT, but none of the other G-quadruplexes showed digestion of terminal nucleotides, indicating that these structures do not contain single-stranded overhangs.
Terminal deoxytransferase (TdT) adds nucleotides to 3′ overhangs or double-stranded DNA. While TdT efficiently extended linear controls, intermolecular Oxy 1.5 and 12GT G-quadruplexes showed minimal extension by this enzyme (Supplementary Figure 5).
Together these data demonstrate that the intermolecular G-quadruplexes are not efficient substrates for two nucleic acid processing enzymes, SVP1 and TdT. This makes it unlikely that telomerase-catalysed extension is due to the presence of linear DNA.
Direct binding of Tetrahymena telomerase protein component (TERT) to G-quadruplex DNA
To establish whether the extension of K+-stabilised G-quadruplexes correlates with their ability to directly interact with recombinant Tetrahymena telomerase, we conducted DNA-telomerase binding assays. Biotinylated gel-purified inter- and intramolecular G-quadruplexes formed from 24TT and 21GG, in parallel with their linear biotinylated counterparts, were individually preincubated with in vitro reconstituted, 35S-labelled immunopurified Tetrahymena telomerase. The G-quadruplex bound telomerase was then recovered using Neutravidin coated beads and the amount of recovered protein visualised on an SDS–PAGE gel.
The Biot-24TT and Biot-21GG intermolecular G-quadruplexes pulled down comparable levels of TERT protein to their linear counterparts (Figure 8). Both G-quadruplexes bind to TERT with higher Kd values than their linear forms, reflecting the trend that is seen for the Km values (see above). The intramolecular G-quadruplexes folded from the same biotinylated sequences displayed negligible interaction with TERT. The intramolecular Biot-21GG G-quadruplex appears to bind to TERT to a greater extent than the Biot-24TT G-quadruplex, but the former contains an intermolecular G-quadruplex contaminant of up to 26% of the total DNA.
Figure 8.
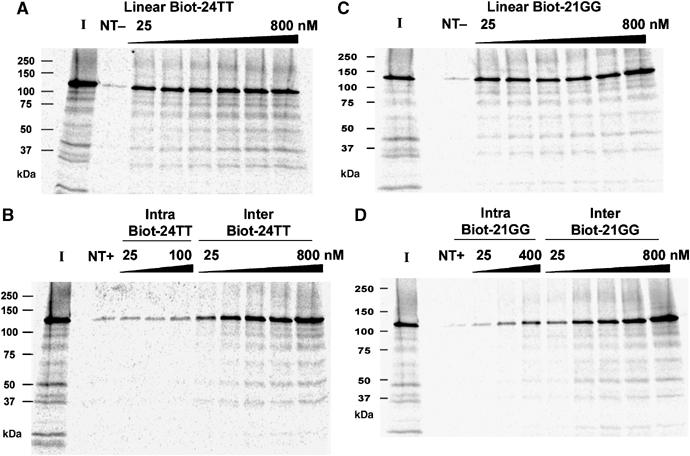
Primer pull-down assays of in vitro reconstituted immunopurified recombinant Tetrahymena telomerase using biotinylated G-quadruplexes stabilised in 50 mM KGlu. Biot-PBR, a non-telomeric (NT) control oligonucleotide (800 nM), in the presence (NT+) or absence (NT−) of KGlu. ‘I' represents input telomerase (25%). Gel-purified intra- and intermolecular Biot-24TT G-quadruplexes (B), or a linear Biot-24TT control (A), were used in pull-down assays at the indicated concentrations, in the presence or absence of 50 mM KGlu, respectively. Gel-purified intra- and intermolecular Biot-21GG G-quadruplexes (D), or a linear Biot-21GG control (C), were used in pull-down assays at the indicated concentrations, in the presence or absence of 50 mM KGlu, respectively.
Jointly these data suggest that, while telomerase has a strong binding affinity for K+-stabilised intermolecular G-quadruplexes, it has very poor ability to bind to intramolecular G-quadruplexes stabilised by K+.
Discussion
Ciliate telomerase is able to extend intermolecular parallel G-quadruplexes
The current consensus in the literature is that G-quadruplex DNA is sequestered from telomerase, and thus cannot serve as its substrate. Here we have presented evidence that at least a subclass of G-quadruplexes can be extended by telomerase. We have demonstrated that intermolecular parallel G-quadruplexes are robust substrates for both Tetrahymena and Euplotes telomerase, to an extent exceeding that predicted from their stability. Since at least partial resolution of the G-quadruplex structure is presumably required for base-pairing with the telomerase RNA template, we propose that telomerase itself possesses G-quadruplex resolvase activity, specific for this particular subclass of G-quadruplexes.
To discount the argument that intermolecular parallel G-quadruplexes are unstable and spontaneously dissociate, which could then facilitate their extension by telomerase, we examined their stability by measuring thermal stability and by using the WC complementary strand as a ‘trap' for dissociated DNA. The intermolecular G-quadruplexes were exceptionally stable with Tm values >77°C. Furthermore, in the trap assay, we found that 0–7% of the Tetrahymena intermolecular G-quadruplexes hybridised over 60 min, insufficient to account for the observed telomerase activity. Intramolecular K+-stabilised G-quadruplexes were actually faster-unfolding than their intermolecular counterparts, and yet were barely extended by telomerase, providing additional evidence that the amount of G-quadruplex unfolding occurring over these time frames is insufficient for high levels of telomerase extension. The intermolecular G-quadruplexes were also highly resistant to digestion with a single-strand specific nuclease or extension by TdT, indicating that there is insufficient linear DNA present to account for their much more robust extension by telomerase.
In addition, the Tetrahymena intermolecular parallel G-quadruplex was able to directly interact with the protein component (TERT) of recombinant Tetrahymena telomerase, which further reinforces the competence of this higher-order structure as a telomerase substrate. At present we cannot discriminate whether it is the intermolecular or parallel property of these G-quadruplexes that allows for binding and extension by the enzyme. Further investigation is in progress to address this issue. Presently, the solution structures for parallel intermolecular G-quadruplexes assembled from 21GG, 24TT or 12GT Tetrahymena telomeric sequences have not been solved; similarly, that of the Euplotes Oxy 1.5 parallel intermolecular G-quadruplex structure is unknown. Crosslinking experiments support a four-stranded structure for 12GT. The migration of the remaining G-quadruplexes on native gels suggests that Oxy 1.5 is a tetramer, while 21GG and 24TT are dimers. The latter quadruplexes must have some kind of ‘propeller-like' fold-out structure (Parkinson et al, 2002) in order to be parallel-stranded. However, at this stage, we also cannot rule out a four-stranded configuration.
Since native Euplotes telomerase and recombinant Tetrahymena telomerase are able to utilise intermolecular parallel G-quadruplexes as substrates, we postulate that the core telomerase complex, consisting of TERT protein and telomerase RNA, possesses a G-quadruplex resolvase capacity. Although we cannot rule out contribution from contaminating protein(s) that may assist telomerase in dealing with a structured primer, co-purification of such protein(s) in two different systems is unlikely.
Extension of intramolecular antiparallel G-quadruplexes correlates with their stability
While an unpurified heterogeneous mixture of G-quadruplexes in either Na+ or K+ was extended successfully by ciliate telomerase (Figure 6; Supplementary Figure 3B), the isolated antiparallel intramolecular G-quadruplex stabilised in K+ could no longer be extended. This finding was in accord with previous observations made by Zahler et al (1991) and is also consistent with a recent report by Zaug et al (2005), which demonstrated that recombinant human telomerase is unable to extend an intramolecular K+- stabilised G-quadruplex.
On examining the stability of K+-stabilised, intramolecular, antiparallel G-quadruplexes, two distinct phases of unfolding, a rapid phase and a slow phase, were observed, as previously reported by Raghuraman and Cech (1990). This implies that these G-quadruplexes contain two subpopulations, one that is readily hybridisable with a complementary strand with a half-life of <1 min and the second that is responsible for the slow phase of dissociation, with a half-life of several hours. Paradoxically, neither subpopulation is extendable by telomerase nor interacts with the telomerase protein component.
In contrast, Na+-stabilised G-quadruplexes of similar antiparallel intramolecular configuration were readily extended by telomerase. These quadruplexes, however, have very fast dissociation rates and are most likely in rapid equilibrium with their linear forms, which provides an explanation for their ability to be extended by telomerase. This is consistent with other reports in the literature, whereby lower melting points and faster folding-unfolding kinetics have been observed for Na+-stabilised G-quadruplexes compared to their K+-stabilised counterparts (Raghuraman and Cech, 1990; Hardin et al, 1991; Balagurumoorthy and Brahmachari, 1994; Risitano and Fox, 2003; Dominick and Jarstfer, 2004).
Therefore, the ability of ciliate telomerases to utilise intramolecular antiparallel G-quadruplexes as substrates is directly influenced by the relative stability of the G-quadruplex, which in turn is dictated by the identity of the stabilising cation.
In vivo relevance
Although parallel intermolecular G-quadruplexes have not been directly observed in ciliated protozoa in vivo, the potential for their formation exists. Indeed, the majority of 3′ telomeric overhangs in T. thermophila are 14–15 or 20–21 nt long (Jacob et al, 2001) and thus unable to form intramolecular G-quadruplexes. Similarly, the 3′ overhang in E. aediculatus is exactly 14 nt long (Klobutcher et al, 1981). In both organisms the predominant G-quadruplex is thus likely to be an antiparallel dimer G-quadruplex or a parallel 4-stranded structure. There is evidence for the in vivo existence of antiparallel dimeric G-quadruplexes in another ciliate, Stylonychia lemnae (Schaffitzel et al, 2001; Paeschke et al, 2005), but this does not rule out the potential existence of parallel quadruplexes in other organisms or at other stages of the life cycle.
In vitro, [Na+]/[K+] ratios affect the balance between antiparallel and parallel G-quadruplexes, with K+ favouring parallel conformations (Sen and Gilbert, 1990; Miura et al, 1995). Ca2+ has also been reported to induce a transition from antiparallel to parallel G-quadruplexes (Miyoshi et al, 2003). It is therefore conceivable that under different intracellular ionic conditions in vivo, one or the other conformation may be favoured, which would then influence the ability of telomerase to interact with the G-quadruplex. A switch from antiparallel to parallel G-quadruplexes has also been observed for the Tetrahymena and Oxytricha, but not human telomeric G-quadruplexes under molecular crowding conditions, which may more closely mimic the in vivo situation (Miyoshi et al, 2004, 2005).
The formation of intermolecular G-quadruplexes involving telomeres from adjacent chromosomes may facilitate telomere–telomere interactions. This idea was first introduced by Sen and Gilbert (1988), who suggested that parallel strand association in telomeric and other G-rich sequences could be involved in the alignment of four sister chromatids during meiosis. The clustering of telomeres in a meiotic bouquet arrangement has been observed in almost all organisms, including ciliates (Harper et al, 2004; Loidl and Scherthan, 2004). Furthermore, a component of the meiosis-specific synaptonemal complex in S. cerevisiae, Hop1, was demonstrated to promote pairing of double-stranded DNA helices via G-quartet formation, implicating intermolecular G-quadruplexes as the vehicles of chromosomal synapsis during meiotic prophase (Anuradha and Muniyappa, 2004). G-quadruplex mediated intermolecular assemblies could be stabilised by proteins such as TGP1 (Tetrahymena G-DNA binding protein 1) that has been demonstrated to directly bind intermolecular G-quadruplex structures formed from the (T2G4)4 Tetrahymena telomeric sequence (Lu et al, 1998). The resolvase capacity of telomerase could then allow for the unwinding of such interchromosomal associations and subsequent telomeric DNA synthesis.
Materials and methods
Oligonucleotide preparation
DNA oligonucleotides (Table I) were purchased from Sigma Genosys (Tetrahymena) or Integrated DNA Technologies (Euplotes) in desalted form. All oligonucleotides with the exception of 12GT were purified by electrophoresis on denaturing 20% polyacrylamide/8 M urea gels in 1 × TBE buffer (89 mM Tris, 89 mM Borate, 2 mM EDTA) and the major band was excised, eluted by crushing and soaking in TEN (10 mM Tris–HCl pH 7.5–8.0, 1 mM EDTA, 100–250 mM NaCl) and ethanol precipitated. In some experiments, the oligonucleotides were 5′ end-labelled with [γ-32P]ATP (see Supplementary data). Purified labelled and unlabelled Tetrahymena oligonucleotides were dialysed for 12–16 h at 4°C against Milli Q water to further desalt them.
The RNA oligonucleotides EaTR and 30AA (Table I) were purchased from Dharmacon (Lafayette, CO), stored in their protected forms, and deprotected following the protocol described by the manufacturer.
G-quadruplex formation, electrophoresis and purification
Tetrahymena oligonucleotides. Oligonucleotides of Tetrahymena telomeric sequence spiked with 5000–10 000 c.p.m. of radiolabelled oligonucleotide of the same sequence were heat denatured in 1 × Tetrahymena Telomerase buffer (50 mM Tris–HCl pH 8.3, 1.25 mM MgCl2, 5 mM dithiothreitol (DTT)) and either 50 mM sodium glutamate (NaGlu) or 50 or 150 mM potassium glutamate (KGlu) at 95°C for 5–10 min, then allowed to equilibrate at 23°C for 30 min. The folded DNA was added to 6 × native gel loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol and 30% glycerol) and electrophoresed on a nondenaturing 12% polyacrylamide gel for 3.5–5 h at 10 W at 23°C. Both buffer and gel contained the same constituents as those in which the DNA was folded with the exception of DTT. The gel was dried for 60 min at 80°C and exposed to a PhosphorImager screen (Molecular Dynamics) for 12–16 h.
To gel purify particular G-quadruplex conformations from 21GG, 24GG and 24TT, DNA was folded as described above in either 0.1 ml of relevant salt buffer at 100 μM final concentration or 1 ml of buffer at 2–4 μM concentration for inter- and intramolecular conformations, respectively. To isolate an intermolecular form of 12GT, 1 mM DNA was heat denatured as above and incubated at 25–37°C for 2–3 days in the presence of 1 × Telomerase buffer and 100 mM NaCl. The folded DNA was electrophoresed on a native gel as described above. The gel was exposed to BioMax Kodak film for 1 h to determine the location of the radiolabelled oligonucleotide. Comigration of this band with the bulk of the unlabelled DNA was confirmed by UV shadowing. The band of interest was excised and crushed in 250 mM KGlu/NaGlu or NaCl, 10 mM Tris–HCl pH 8.3, 1 mM EDTA and incubated for 12–16 h at 23°C with rotation. The supernatant was filtered with a 0.22 μm filter and the DNA precipitated with two volumes of absolute ethanol for 4–16 h at −20°C. The precipitated product was resuspended in the original folding buffer. DNA concentrations were determined by UV absorbance at 260 nm (see Supplementary Table I for extinction coefficients). An aliquot of the gel purified G-quadruplex was electrophoresed on a second native gel in the presence of the relevant salt to determine its purity using ImageQuant software. The remainder of the gel purified G-quadruplex was stored at 4°C in its folding buffer.
Euplotes oligonucleotides. 200 μM Oxy 1.5 and 1 μM Oxy 3.5 in 1 × Euplotes folding buffer (20 mM TrisOAc pH 7.5, 10 mM MgCl2, 1 mM DTT, and 50 mM K+ or NaGlu) were heated to 95°C for 5 min and allowed to slowly cool (∼2°C/min) to 25°C. Folded DNA was electrophoresed on 20% nondenaturing gels (50 mM K+ or NaOAc) for 3.5 h at 100 V at 4°C using a running buffer of 50 mM K+ or NaOAc in 1 × TBE. DNA was visualised by phosphorimaging (32P-labelled DNA) or UV shadowing (unlabelled DNA), isolated by a crush and soak method into 1 × TEK (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 100 mM KCl) or TEN buffer, and concentrated by ethanol precipitation. Samples were resuspended in the corresponding 1 × Euplotes reaction buffers, and quantified by scintillation counting (labelled DNA) or UV absorbance at 260 nm (unlabelled DNA; see Supplementary Table I for extinction coefficients). The integrity of the recovered structures was confirmed by native gel electrophoresis. Radiolabelled structures were used for solution hybridisation studies, whereas unlabelled G-quadruplexes were used for telomerase assays and CD analysis.
Biotinylated primer pull-down assays (binding studies)
Binding studies were conducted by immobilising biotinylated Tetrahymena G-quadruplex DNA on NeutrAvidin beads (Pierce Biotechnology) in the presence of radiolabelled recombinant Tetrahymena telomerase. Nonradioactive biotinylated oligonucleotides 24TT and 21GG were folded and gel purified as described above. Each biotin-conjugated G-quadruplex (10 μl) or its linear counterpart was incubated with 10 μl of 35S-labelled recombinant telomerase in 1 × Tetrahymena Telomerase buffer (see above) with or without 50 mM KGlu at 25°C for 10 min. NeutrAvidin beads (10 μl per reaction) were washed four times with 1 × Telomerase buffer with 10% glycerol. The beads were blocked twice for 15 min at 4°C in the same buffer with the addition of 0.75 mg/ml BSA, 0.15 mg/ml glycogen and 0.15 mg/ml yeast RNA. NeutrAvidin beads that were to be used for G-quadruplex samples included 50 mM KGlu in the wash and blocking steps to preserve the folded conformation. Blocked beads were resuspended in one volume of blocking buffer. Blocked bead slurry (20 μl) was added to the DNA-telomerase reaction mix and the samples were rotated at 4°C for 15 min. The beads were washed four times with 1 × Telomerase buffer with 10% glycerol and 300 mM LiOAC for linear DNA or 300 mM KGlu for G-quadruplex DNA. The beads were resuspended in one volume of the corresponding salt buffer. Half of each reaction was added to an equal volume of 2 × Laemmli's loading buffer (125 mM Tris–HCl pH 6.8, 4% SDS, 0.005% bromophenol blue, 20% glycerol and 0.72 M β-mercaptoethanol) plus 300 mM KGlu, denatured at 95°C for 3 min, loaded onto a 4–20% SDS–PAGE gel (Novex) and electrophoresed for 1.5 h at 120 V. The gel was fixed in 25% isopropanol and 10% acetic acid for 20 min, dried at 80°C for 1 h then exposed to a PhosphorImager screen for 12–16 h.
Circular dichroism, stability studies, preparation of telomerase and activity assays
DNA structural analysis by CD, complementary strand trap assays, the preparation of recombinant and native Tetrahymena and Euplotes telomerase, telomerase activity assays and treatment with SVP1 and TdT were carried out using published procedures. Further details are given in the Supplementary data online.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Table 1
Acknowledgments
We are grateful to Dr Paul Curmi and the members of his laboratory at the University of New South Wales, Australia for access to their circular dichroism spectrophotometer. This work was funded by the National Science Foundation (MBJ, MCB-0446019), the Wellcome Trust (TMB, Senior Research Fellowship GR066727MA) and the Dora Lush Biomedical Postgraduate Research Scholarship from the National Health and Medical Research Council of Australia (LO).
References
- Anuradha S, Muniyappa K (2004) Meiosis-specific yeast Hop1 protein promotes synapsis of double-stranded DNA helices via the formation of guanine quartets. Nucleic Acids Res 32: 2378–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagurumoorthy P, Brahmachari SK (1994) Structure and stability of human telomeric sequence. J Biol Chem 269: 21858–21869 [PubMed] [Google Scholar]
- Baran N, Pucshansky L, Marco Y, Benjamin S, Manor H (1997) The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res 25: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG (1978) A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120: 33–53 [DOI] [PubMed] [Google Scholar]
- Dominick PK, Jarstfer MB (2004) A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J Am Chem Soc 126: 5050–5051 [DOI] [PubMed] [Google Scholar]
- Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M (2005) Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by hnRNP D. J Biol Chem 280: 18862–18870 [DOI] [PubMed] [Google Scholar]
- Fang G, Cech TR (1993) The β subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell 74: 875–885 [DOI] [PubMed] [Google Scholar]
- Giraldo R, Rhodes D (1994) The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J 13: 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo R, Suzuki M, Chapman L, Rhodes D (1994) Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: A circular dichroism study. Proc Natl Acad Sci USA 91: 7658–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Hardin CC, Henderson E, Watson T, Prosser JK (1991) Monovalent cation induced structural transitions in telomeric DNAs: G-DNA folding intermediates. Biochemistry 30: 4460–4472 [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ (2004) A bouquet of chromosomes. J Cell Sci 117: 4025–4032 [DOI] [PubMed] [Google Scholar]
- Harrington C, Lan Y, Akman SA (1997) The identification and characterization of a G4-DNA resolvase activity. J Biol Chem 272: 24631–24636 [DOI] [PubMed] [Google Scholar]
- Henderson E, Hardin CC, Walk SK, Tinoco I Jr, Blackburn EH (1987) Telomeric DNA oligonucleotides form intramolecular structures containing guanine–guanine base pairs. Cell 51: 899–908 [DOI] [PubMed] [Google Scholar]
- Jacob NK, Skopp R, Price CM (2001) G-overhang dynamics at Tetrahymena telomeres. EMBO J 20: 4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry MA (2000) Quadruplex structures in nucleic acids. Biopolymers 56: 123–146 [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM (1981) All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci USA 78: 3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Frantz JD, Gilbert W, Tye BK (1993) Identification and characterization of a nuclease activity specific for G4 tetrastranded DNA. Proc Natl Acad Sci USA 90: 3157–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Scherthan H (2004) Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci 117: 5791–5801 [DOI] [PubMed] [Google Scholar]
- Lu Q, Schierer T, Kang SG, Henderson E (1998) Purification, characterization and molecular cloning of TGP1, a novel G-DNA binding protein from Tetrahymena thermophila. Nucleic Acids Res 26: 1613–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Benevides JM, Thomas GJ Jr (1995) A phase diagram for sodium and potassium ion control of polymorphism in telomeric DNA. J Mol Biol 248: 233–238 [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Karimata H, Sugimoto N (2005) Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew Chem Int Ed Engl 44: 2–5 [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Matsumura S, Nakano S, Sugimoto N (2004) Duplex dissociation of telomere DNAs induced by molecular crowding. J Am Chem Soc 126: 165–169 [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Nakao A, Sugimoto N (2003) Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+. Nucleic Acids Res 31: 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ (2005) Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol 12: 847–854 [DOI] [PubMed] [Google Scholar]
- Parkinson GN, Lee MP, Neidle S (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417: 876–880 [DOI] [PubMed] [Google Scholar]
- Petraccone L, Erra E, Esposito V, Randazzo A, Mayol L, Nasti L, Barone G, Giancola C (2004) Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry 43: 4877–4884 [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Cech TR (1990) Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res 18: 4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC (2001) Platination of the (T2G4)4 telomeric sequence: a structural and cross-linking study. Biochemistry 40: 8463–8470 [DOI] [PubMed] [Google Scholar]
- Risitano A, Fox KR (2003) Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry 42: 6507–6513 [DOI] [PubMed] [Google Scholar]
- Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA 98: 8572–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Gilbert W (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334: 364–366 [DOI] [PubMed] [Google Scholar]
- Sen D, Gilbert W (1990) A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 344: 410–414 [DOI] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33: 787–791 [DOI] [PubMed] [Google Scholar]
- Simonsson T (2001) G-quadruplex DNA structures—variations on a theme. Biol Chem 382: 621–628 [DOI] [PubMed] [Google Scholar]
- Smith FW, Schultze P, Feigon J (1995) Solution structures of unimolecular quadruplexes formed by oligonucleotides containing Oxytricha telomere repeats. Structure 3: 997–1008 [DOI] [PubMed] [Google Scholar]
- Sun H, Bennett RJ, Maizels N (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res 27: 1978–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels N (1998) The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem 273: 27587–27592 [DOI] [PubMed] [Google Scholar]
- Sun H, Yabuki A, Maizels N (2001) A human nuclease specific for G4 DNA. Proc Natl Acad Sci USA 98: 12444–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Klug A (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 342: 825–829 [DOI] [PubMed] [Google Scholar]
- Uddin MK, Kato Y, Takagi Y, Mikuma T, Taira K (2004) Phosphorylation at 5′ end of guanosine stretches inhibits dimerization of G-quadruplexes and formation of a G-quadruplex interferes with the enzymatic activities of DNA enzymes. Nucleic Acids Res 32: 4618–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Patel DJ (1994) Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure 2: 1141–1156 [DOI] [PubMed] [Google Scholar]
- Wang Y, Patel DJ (1995) Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J Mol Biol 251: 76–94 [DOI] [PubMed] [Google Scholar]
- Williamson JR (1994) G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct 23: 703–730 [DOI] [PubMed] [Google Scholar]
- Williamson JR, Raghuraman MK, Cech TR (1989) Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59: 871–880 [DOI] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, Cech TR, Prescott DM (1991) Inhibition of telomerase by G-quartet DNA structures. Nature 350: 718–720 [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR (2005) Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA 102: 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Table 1


