Abstract
In Saccharomyces cerevisiae, the synthesis of chitin is temporally and spatially regulated through the transport of Chs3p (chitin synthase III) to the plasma membrane in the bud neck region. Traffic of Chs3p from the trans-Golgi network (TGN)/early endosome to the plasma membrane requires the function of Chs5p and Chs6p. Chs6p belongs to a family of four proteins that we have named ChAPs for Chs5p-Arf1p-binding Proteins. Here, we show that all ChAPs physically interact not only with Chs5p but also with the small GTPase Arf1p. A short sequence at the C-terminus of the ChAPs is required for protein function and the ability to bind to Chs5p. Simultaneous disruption of two members, Δbud7 and Δbch1, phenocopies a Δchs6 or Δchs5 deletion with respect to Chs3p transport. Moreover, the ChAPs interact with each other and can form complexes. In addition, they are all at least partially localized to the TGN in a Chs5p-dependent manner. Most importantly, several ChAPs can interact physically with Chs3p. We propose that the ChAPs facilitate export of cargo out of the Golgi.
Keywords: Arf1, ChAPs, Coat, Golgi, vesicular transport
Introduction
The trans-Golgi network (TGN) is the central sorting station in the cell (Gu et al, 2001). From the TGN, proteins are distributed to different compartments such as the lysosomes, endosomes or plasma membrane. To reach their specific destinations, cargo proteins interact with receptors that facilitate their inclusion into the appropriate transport vesicle. After cargo delivery, these cargo receptors are retrieved from the acceptor compartment to allow for another round of transport. The cargo receptors recognize distinct coat proteins, and might even help to recruit them or stabilize them on the membrane (Le Borgne et al, 1996). Multiple types of vesicles have been identified that transport cargo from the TGN to different organelles. The AP-1 complex and clathrin facilitate transport between the TGN and the early endosomes. AP-3 recruits clathrin to the TGN to produce vesicles targeted to the lysosome (or the vacuole in Saccharomyces cerevisiae). Finally, the clathrin-binding GGAs (Golgi-localized, Gamma-ear-containing, Arf-binding) are responsible for the exchange of proteins and lipids between the TGN and the late endosomes. Although these different pathways have been studied extensively, it is likely that there are other, as yet undefined vesicles transporting cargo to other specialized regions of the cell. For example, the identity of the coat protein essential for transport from the late endosome or from the TGN directly to the plasma membrane remains elusive. Although the FAPPs (4-phosphate adaptor proteins) have been recently implicated in post-Golgi carrier formation (Godi et al, 2004), they were not found on transport intermediates after fission. In addition, at least in S. cerevisiae, the major location of secretion changes during the cell cycle: normally, secretion occurs at the bud tip, which is the region where the bud grows; however, at the end of mitosis, secretion switches to the mother-bud junction. Although the exocyst and other factors necessary for consumption of vesicles at the plasma membrane also relocate from the bud tip to the mother-bud junction, it remains unclear whether the vesicles directed to the bud tip and into the bud neck carry the same coat. Furthermore, the number of cargo receptors identified at the TGN is clearly insufficient to distribute all the different cargo proteins to their various locations. In recent years, it has become clear that the vesicles directed to the mother-bud junction late in the cell cycle must carry specific cargoes, whereas soluble cargo like the yeast pheromone α-factor might be excluded from these vesicles.
One of the best-defined cargoes that cycle between the plasma membrane and internal stores/endosomes in a cell cycle-dependent manner is chitin synthase III, Chs3p. This enzyme serves as a valuable model to investigate regulated vesicular traffic in yeast (Valdivia and Schekman, 2003). Chs3p is polarized at the incipient bud site and the neck region between the mother and the small daughter. When the daughter reaches approximately the same size as the mother, Chs3p is internalized and sequestered into specialized endosomes called chitosomes (Ziman et al, 1998). Transport of Chs3p to the plasma membrane requires the function of two proteins called Chs5p and Chs6p. Chs5p is a peripheral late Golgi protein that colocalizes with the trans-Golgi marker Kex2p (Santos and Snyder, 1997). Chs6p is a cytosolic protein of unknown function (Ziman et al, 1998) in the transport of Chs3p to the plasma membrane. Mutants in CHS6 are resistant to calcofluor, which is characteristic of factors involved in chitin biosynthesis (Ziman et al, 1998). Furthermore, Chs3p is trapped in internal compartments in chs6 mutants. AP-1- and clathrin-coated vesicles (CCVs) have been implicated in retrieval of Chs3p from the early endosomes to the TGN (Valdivia et al, 2002). In yeast and mammals, the formation of CCVs that contain AP-1 is dependent on the small GTPase Arf1p (Le Borgne et al, 1996; Chen and Graham, 1998; Gaynor et al, 1998; Kirchhausen, 2000; Yahara et al, 2001). The interaction between AP-1 and the activated form of Arf1p is stabilized by cargo proteins that retain AP-1 at the membrane, leading to the assembly of the clathrin lattice and the formation of CCVs. Valdivia et al (2002) reported that ARF1 interacts genetically with CHS5 and CHS6, suggesting a link between ARF1 function and Chs3p traffic. However, Arf1p function was assumed to be connected to the formation of CCVs at the early endosomes. We wanted to explore the possibility that Arf1p and Chs5p and Chs6p might be more closely linked than was apparent from these data and to ask whether Chs5p and Chs6p might play a direct role in the formation of specialized vesicles at the TGN, which contain Chs3p as cargo.
In this report, we show that indeed Chs5p and Chs6p interact biochemically with Arf1p. Furthermore, we discovered that Chs6p is part of a family with three additional members: Bud7p, Ymr237p and Ykr027p, and we named this family ChAPs (Chs5p-Arf1p-binding Proteins). Although mutants in BUD7 display a bud-site selection phenotype, for none of these proteins had a cellular role been established (Zahner et al, 1996; Ni and Snyder, 2001). Surprisingly, all ChAPs bind to Arf1p. Furthermore, double deletion of BUD7 and BCH1 results in the same phenotype as deletion of CHS6 with respect to calcofluor resistance and Chs3p trafficking. The different ChAPs interact with each other and partially localize to the TGN, dependent on the presence of Chs5p. Moreover, the ChAPs interact directly with Arf1p, and most importantly, the ChAPs can be crosslinked to Chs3p, indicating a direct requirement of the ChAPs in export of certain cargo from the Golgi.
Results
Chs6p is part of an ancient eukaryotic family
Chs5p and Chs6p are involved in the traffic of Chs3p to the plasma membrane (Ziman et al, 1998; Valdivia et al, 2002). Yet, the role of Chs5p and Chs6p in this process is ill defined. We were wondering whether homologs of Chs5p or Chs6p with similar function might exist, which would give us a hint about the function of these proteins. We therefore searched the database for homologs of Chs5p and Chs6p in S. cerevisiae. Although there were no clear homologs of Chs5p, we found three homologs for Chs6p: Bud7p, and the uncharacterized proteins Ymr237w and Ykr027w. We named Ymr237w and Ykr027w, Bch1p and Bch2p, respectively, for Bud7p and Chs6p homologs 1 and 2. The entire family was referred to as ChAPs.
Bud7p was identified as a mutant in the bud-site selection process (Zahner et al, 1996). Homozygous diploid deletions in BUD7 result in a random budding pattern, but the role of Bud7p in this process is unknown. ChAPs were found to be present also in other fungi, like zygomycetes, basidiomycetes and nonbudding ascomycetes, and bioinformatic analysis revealed that Bch1p is most likely the most ancient member of this protein family (Supplementary Figure S1). A gene duplication event founded the Chs6p subbranch of the family. The duplication of the genome during development of the genus Saccharomyces resulted finally in Bud7p and Bch2p as closest homologs of Bch1p and Chs6p, respectively (Supplementary Figure S3; Wolfe and Shields, 1997). Therefore, it is not surprising that all members of this family are located on different chromosomes. BCH1, CHS6, BUD7 and BCH2 are located on chromosomes XIII, X, XV and XI, respectively. Outside of fungi, Bch1p was also identified in mycetozoa, red algae and ciliates, but not in the various completely sequenced genomes of metazoa and viridiplanta. Thus, the ChAPs represent an ancient eukaryotic protein family that ramified during development of ascomycotic yeasts. But what is the role of these proteins?
The ChAPs are involved in different processes
To study the ChAPs in a systematic manner, we created single deletions. The single deletion strains obtained were assayed for growth at various temperatures and on different nutrient sources. From all the ChAP deletion strains tested, only the Δchs6 strain was temperature-sensitive at 37°C and resistant to calcofluor (Figures 1A and 3A; Valdivia et al, 2002). Calcofluor is a toxic dye, which binds to chitin and thereby poisons yeasts containing chitin in their cell walls. Resistance to calcofluor is used as a diagnostic tool for Chs3p mislocalization. Remarkably, calcofluor-sensitivity was restored upon deletion of ARF1 in a Δchs6 strain (Figure 3A; Valdivia et al, 2002).
Figure 1.
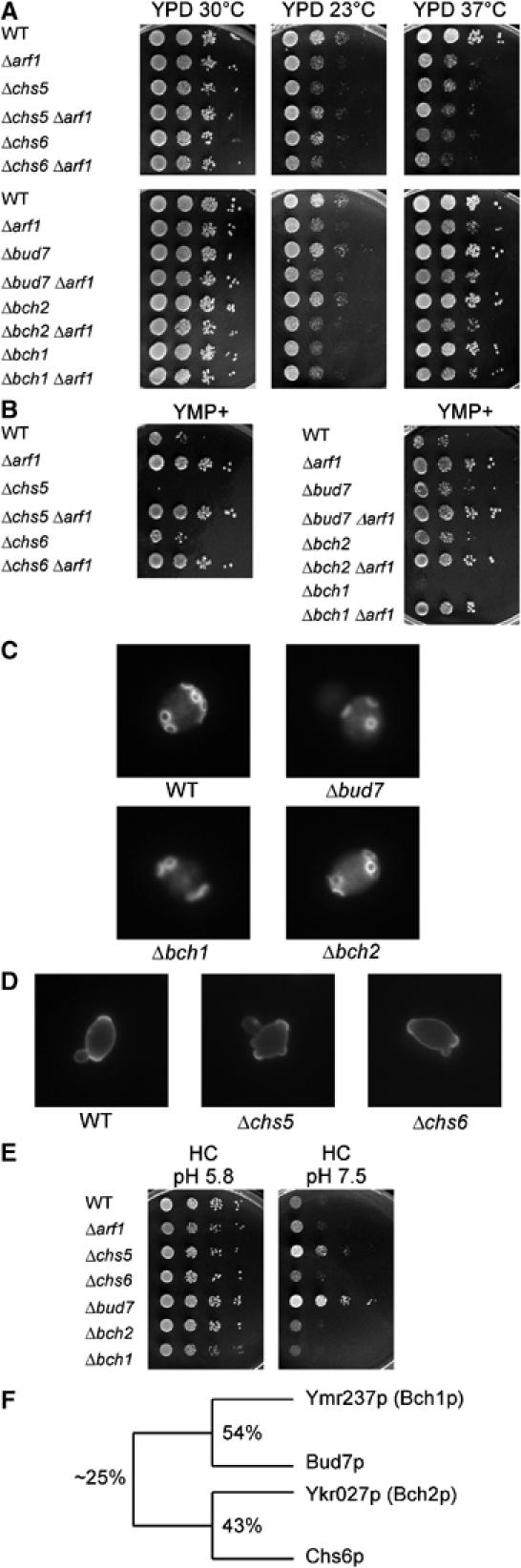
Analysis of single deletion mutants of the ChAPs together with mutants in CHS5 and ARF1. For the drop assays, haploid strains were grown overnight to logarithmic phase in rich medium. Serial dilutions (1:10) were dropped onto plates and incubated for 2 days at 30°C, unless indicated otherwise. (A) Growth at different temperatures. (B) Growth on YMP+ plates, which contain an elevated level of ammonium sulfate. (C) Analysis of the budding pattern. Diploid yeast strains were grown to logarithmic phase and stained with calcofluor. (D) Analysis of the budding pattern of mutants in CHS5 and CHS6. Diploid yeast strains were grown to logarithmic phase in YPD+0.7 M sorbitol. After fixation, cells were stained with FITC–concanavalin A. (E) Growth on minimal medium plates buffered at pH 5.8 and 7.5. (F) Scheme depicting the relationship of the members of the ChAP family. The percentage of protein sequence identity is given.
Figure 3.
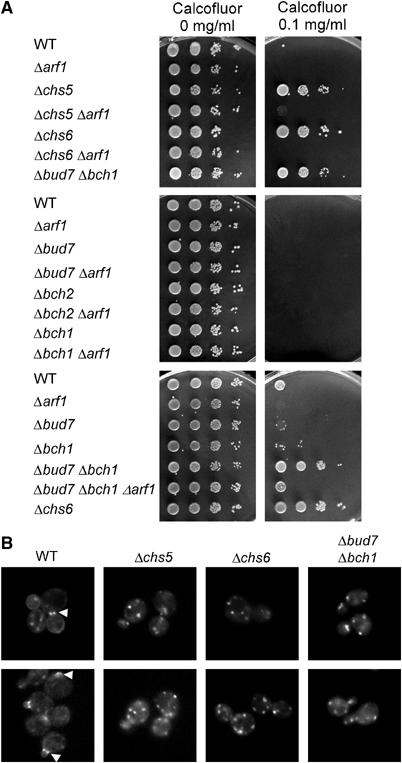
Chs5p and members of the ChAPs are involved in transport of chitin synthase Chs3p to the plasma membrane. (A) Haploid strains were grown to logarithmic phase in rich medium. Serial dilutions (1:10) were dropped onto plates containing the toxic dye calcofluor and incubated for 2 days. (B) Haploid yeast cells bearing a chromosomal CHS3-GFP were grown to logarithmic phase. Arrowheads point to the signal at the bud neck and the incipient bud site. Pictures were taken from freshly mounted cells and two images per strain are shown.
In contrast to the deletion phenotype of CHS6, a Δbch1 strain grew slowly at 23°C and was highly sensitive to growth on YMP+ plates (Figure 1A and B). YMP+ is a rich medium containing elevated levels of ammonium. Deletion of any of the other three members of the family was no more sensitive towards YMP+ than the wild type. For a Δbch2 strain, we did not observe any obvious defect under any of a large number of different growth conditions. As reported before, a homozygous diploid Δbud7 strain displayed a random budding pattern (Figure 1C). The bipolar budding pattern in Δbch1 and Δbch2 homozygous diploid deletions was indistinguishable from wild type. To assess the budding pattern of Δchs5 and Δchs6 homozygous diploid strains, which do not stain with calcofluor, we used concanavalin A coupled to FITC for the visualization of bud scars in Δchs5 and Δchs6 cells (Figure 1D). As reported before, deletion of CHS5 resulted in a random budding pattern (Santos et al, 1997). In contrast, no defect in bud-site selection was observed in Δchs6 cells. We also observed another previously undescribed phenotype for a Δbud7 deletion, which was the fast growth on plates buffered at pH 7.5, demonstrating also a role for Bud7p in haploid cells (Figure 1E). The fact that phenotypes observed for specific mutants were only associated with one member of the ChAPs indicates that the individual family members serve different functions in the cell and are not entirely redundant. However, they might still act in the same process.
Strikingly, however, the Δchs5 deletion strain exhibited all phenotypes observed for the deletion of different members of the ChAPs (Figures 1 and 3A), namely resistance to calcofluor, temperature- and cold-sensitivity, YMP+ sensitive growth, fast growth on plates at pH 7.5 and a random budding pattern in diploid strains. These data suggest that Chs5p acts in the same pathway either as an upstream regulator or a downstream convergence point in ChAPs family-related functions.
Chs5p and Bch1p interact with the activated form of Arf1p
Given the genetic interaction between CHS5, CHS6 and ARF1 (Figure 3A; Valdivia et al, 2002), we asked whether they also interact biochemically. Cytosolic extracts from a wild-type yeast strain were passed over Arf1p affinity columns preloaded with GTP or GDP, respectively (Trautwein et al, 2004). A conformational change was provoked on the Arf1 proteins by spontaneous nucleotide exchange to elute specific interactors. To test for the specificity of the affinity matrix, we assayed for the binding of known interactors of Arf1p. As expected from their biological functions, the coatomer complex and the ARF-GAP Glo3p bound preferentially to Arf1p-GTP, whereas the ARF-GEF Gea2p bound only to Arf1p-GDP (Figure 2A). This approach has been also used successfully to identify Pab1p as an Arf1p-binding protein, which uses COPI-coated vesicles for mRNA transport to the ER (Trautwein et al, 2004). We found that Chs5p bound specifically to the activated form of Arf1p (Figure 2B). We also identified Arf1p-GTP-associated proteins by mass spectrometry. Out of 10 protein bands analyzed, seven corresponded to already known interactors of Arf1p, thus again validating the approach and an eighth protein band corresponded to Chs5p, confirming the immunoblot assay. Interestingly, another band was identified as Bch1p, demonstrating that at least one of the ChAPs can associate, directly or indirectly, with Arf1p.
Figure 2.
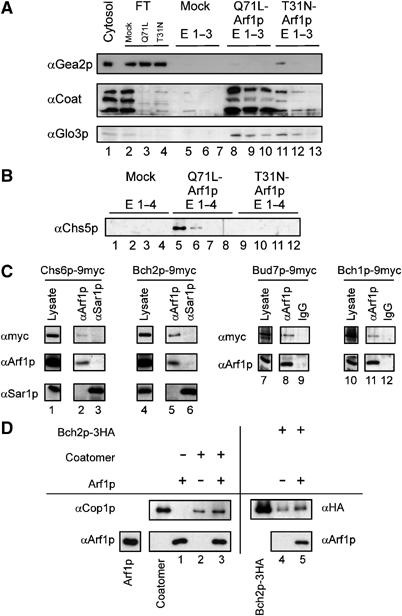
Chs5p binds to activated Arf1p. (A) Differential Arf1p affinity chromatography to evaluate binding to Arf1p. Yeast cytosol was incubated with either Arf1p-Q71L (Arf1p-GTP) or Arf1p-T31N (Arf1p-GDP) column material. After washing, spontaneous nucleotide exchange was elicited resulting in a conformational change on Arf1p and the release of conformation-specific bound proteins. The eluates (E1–E3) from an affinity chromatography experiment were analyzed by immunoblot. FT is the flow-through of unbound protein. Beads without Arf1p were mock-treated and served as negative control. The Arf1p-GEF Gea2p is enriched in the Arf1p-GDP column, whereas both the coatomer complex (three subunits are shown) and the Arf1p-GAP Glo3p bind predominately to Arf1p-GTP. (B) Chs5p binds to activated Arf1p. Eluates (E1–E4) from the Arf1p affinity chromatography were analyzed for the presence of Chs5p by immunoblot. (C) The ChAPs interact with Arf1p. Co-immunoprecipitation experiments were performed using strains in which the ChAPs were chromosomally tagged with 9myc. The lysates were treated with affinity-purified α-Arf1p-IgGs or α-Arf1p and α-Sar1p sera and Protein A–Sepharose. The precipitate was analyzed by immunoblot with antibodies directed against the myc epitope, Sar1p and Arf1p. Lanes 1 and 4 and lanes 7 and 10 represent 1.3 and 1.7% of the lysate, respectively. The lysates and corresponding precipitates were from the same immunoblot. Image processing was identical for lysate and corresponding precipitates. (D) Bch2p interacts directly with Arf1p. Liposomes were incubated with Bch2p in the presence or absence of Arf1p-GTP. The liposomes were floated through a sucrose cushion and the top layer was analyzed by immunoblot against Arf1p and Bch2p. The flotation of coatomer served as control and Cop1p was detected by immunoblot. In all, 10% of the input of Arf1p, coatomer and Bch2p were loaded for comparison.
To test whether the other ChAPs can also interact with Arf1p, we performed co-immunoprecipitations with α-Arf1p antibodies on lysates from strains where the ChAPs were appended chromosomally with a 9myc-tag. The proteins were expressed from their endogenous promotor and were fully functional. A signal for Bch1p-9myc, Bud7p-9myc, Chs6p-9myc and Bch2p-9myc was obtained in the precipitate (Figures 2C and 7A), indicating that all ChAPs can associate with Arf1p. This interaction seemed to be specific because another small GTPase, Sar1p, was unable to precipitate any of the ChAPs (Figure 2C and data not shown). However, the interaction between Arf1p and the ChAPs might be indirect. To clarify this point, we performed a liposome binding experiment. One of the ChAPs, Bch2p, was incubated with activated Arf1p and liposomes. The association of Bch2p with Arf1p and the liposomes was assessed by flotation. Bch2p binding to liposomes was increased in the presence of Arf1p (Figure 2D). The increase in binding occurred to a similar extent as in the positive control, when coatomer was present, indicating a specific Arf1p-dependent recruitment of Bch2p to liposomes. A similar result was obtained for Chs6p (data not shown). These data demonstrate that at least Bch2p and Chs6p can interact directly with Arf1p.
Figure 7.
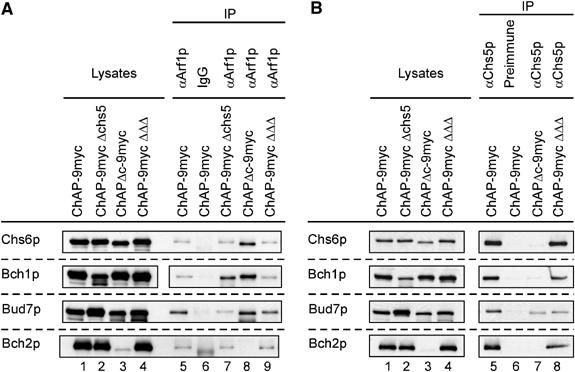
The ChAPs interact with Arf1p and Chs5p. Co-immunoprecipitation experiments were performed using strains in which one ChAP was individually chromosomally tagged with 9myc in either WT, in Δchs5 or in a strain in which the three remaining ChAPs were deleted. Furthermore, the experiment was also performed with lysates from ChAPΔc-9myc strain. The lysates were treated with either affinity-purified α-Arf1p (A) or α-Chs5p serum (B) and Protein A–Sepharose. The precipitate was analyzed with α-myc. In all, 1.7% of the lysates was loaded in lanes 1–4. The lysates in (A) and (B) are from the same experiment. The lysates and corresponding precipitates were from the same immunoblot.
Δbud7Δbch1 display the same phenotype as Δchs6
The ChAPs seemed to be linked to both Chs5p and Arf1p. The only established role for a ChAP in conjunction with Chs5p and Arf1p is the transport of Chs3p, which requires Chs6p. Our results raise the possibility that Chs5p acts not only together with Chs6p but also together with the other three family members to export Chs3p from the TGN. Alternatively, Chs5p may play different roles in conjunction with various members of the ChAPs.
To test the possibility that ChAPs interact with Chs5p to export Chs3p from the Golgi, we generated deletions of the ChAPs in various combinations. These deletion strains were tested on plates containing calcofluor. As described above, Δchs5 and Δchs6 strains were resistant to calcofluor and became sensitive again when ARF1 was disrupted (Figure 3A; Valdivia et al, 2002). However, more importantly, a double deletion of BUD7 and BCH1 resulted also in calcofluor resistance (Figure 3A). To test whether the calcofluor resistance of a Δbud7Δbch1 strain was owing to a defect in transport of Chs3p to the plasma membrane, we created a functional Chs3p-GFP, which is expressed from the CHS3 chromosomal location under the endogenous promotor. In WT cells, we observed a GFP signal primarily at the bud neck and the incipient bud site and also sometimes in structures resembling the TGN as well as a more dispersed signal, which probably corresponds to chitosomes (Figure 3B). This localization is somewhat different from the localization for Chs3p-GFP described by Valdivia et al (2002). However, they coexpressed Chs7p, an ER-resident export factor for Chs3p and Chs3p-GFP from plasmids, whereas we are observing the endogenous protein only. As expected, in Δchs5 and in Δchs6 cells, the GFP signal was exclusively restricted to structures corresponding to the TGN/chitosomes and almost never (<1%) observed in the bud neck (Figure 3B; Valdivia et al, 2002). Similarly, in a Δbud7Δbch1 strain Chs3p-GFP was present only in TGN/chitosomal structures (Figure 3B).
To confirm that the calcofluor resistance of a Δbud7Δbch1 strain was owing to reduced chitin levels at the plasma membrane, we stained the deletion strains with calcofluor. In both Δchs5 and Δchs6 cells, the cell wall was stained very poorly. This poor staining was rescued by an additional ARF1 deletion, because Chs3p reaches the plasma membrane by an alternative pathway (Valdivia et al, 2002, and data not shown). In case of the Δbud7Δbch1 double deletion, only weak staining of the cell wall was visible (data not shown). Taken together, these observations indicate that in the Δbud7Δbch1 strain, Chs3p transport to the plasma membrane was disturbed. The calcofluor-sensitivity was restored in the Δbud7Δbch1Δarf1 strain, indicating that Bud7p and Bch1p might be involved in the same pathway as Chs6p (Figure 3A). Moreover, the results strongly suggest that not only Chs6p but also other members of the ChAPs are required for efficient Chs3p transport from the TGN to the plasma membrane.
The ChAPs interact with each other
As more than one ChAP member is necessary for Chs3p transport to the plasma membrane, this raised the possibility that the ChAPs interact with each other. We constructed a quadruply tagged strain to be able to detect all ChAPs in the same strain and performed co-immunoprecipitation experiments.
Chs6p was fused to GFP, Bud7p was appended with 9myc, Bch1p carried a double AU5 epitope and Bch2p was tagged with 3HA. The resulting strain behaved like the corresponding wild-type strain, indicating that all chromosomal fusions were functional and expressed at endogenous levels. When polyclonal AU5 antibodies were used for precipitation, all the three other ChAPs were detected (Figure 4). Co-precipitation was specific, because no ChAPs were detected from control strains lacking the AU5 tag. A similar result was obtained for Bud7p-9myc that was precipitated with monoclonal myc antibodies. Again, Bch2p, Chs6p and Bch1p were detected when Bud7p-9myc was precipitated and were absent when control lysates lacking the myc epitope were used. Similar results were obtained for co-immunoprecipitations with monoclonal HA antibodies and polyclonal GFP antibodies (data not shown).
Figure 4.
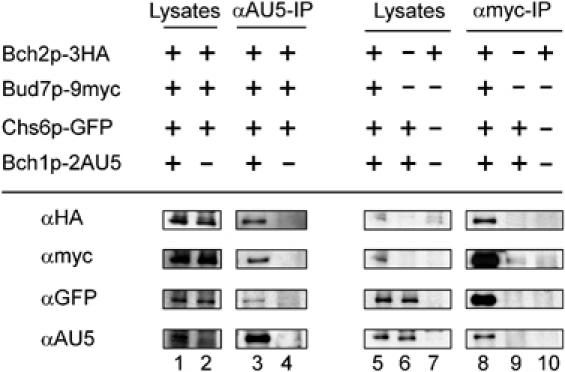
The ChAPs interact with each other. A quadruply chromosomally tagged strain along with control strains were subjected to immunoprecipitation. The precipitates were analyzed by SDS–PAGE and immunoblot with antibodies directed against all four different epitope tags. Precipitations with α-AU5 and with α-myc-IgGs are shown. In all, 1.7% of the lysates were loaded in lanes 1, 2 and 5–7. The lysates and corresponding precipitates were from the same immunoblots. Image processing was identical for lysates and corresponding precipitates.
These experiments demonstrate that the four ChAPs are able to interact with each other, either directly or indirectly. However, the intensities of the signals of the precipitations suggest that ChAPs form complexes with variable stoichiometries. We verified the presence of multiple complexes by blue native gel electrophoresis (data not shown). This variability suggests that a single ChAP might assemble individually into distinct complexes with other ChAPs.
The ChAPs localize at least partially to the TGN
Our results suggest that the ChAPs collaborate to transport Chs3p to the plasma membrane. But where does this transport step occur? We have shown that the ChAPs interact with Arf1p and it has been reported that activated Arf1p resides on Golgi membranes (Stearns et al, 1990). Furthermore, Chs5p, which might be implicated in the same trafficking step, is associated with the TGN (Santos and Snyder, 1997). Importantly, Δchs5, Δchs6 and Δbud7Δbch1 all accumulate Chs3p in the TGN. To investigate the localization of the members of the ChAPs, we performed immunofluorescence on strains in which the ChAPs were chromosomally tagged with 9myc. These strains also contained a chromosomal SEC7-GFP, which was used as a marker of the TGN. Bch2p was localized exclusively to the TGN (colocalization with Sec7p-GFP), whereas Chs6p and Bud7p each localized to the TGN and to other punctate structures in the cytoplasm (Figure 5). Bch1p exhibited a very diffuse staining throughout the cytoplasm, which overlapped partially with the TGN. Whereas Chs6p, Bud7p and Bch2p appeared to be expressed to about the same level, Bch1p was expressed at a noticeably higher level. This was evident from immunofluorescence (the exposure time was much shorter for Bch1p) as well as from immunoblots of cell lysates (Figure 9B). Taken together, all members of the ChAPs were at least partially localized to the TGN.
Figure 5.
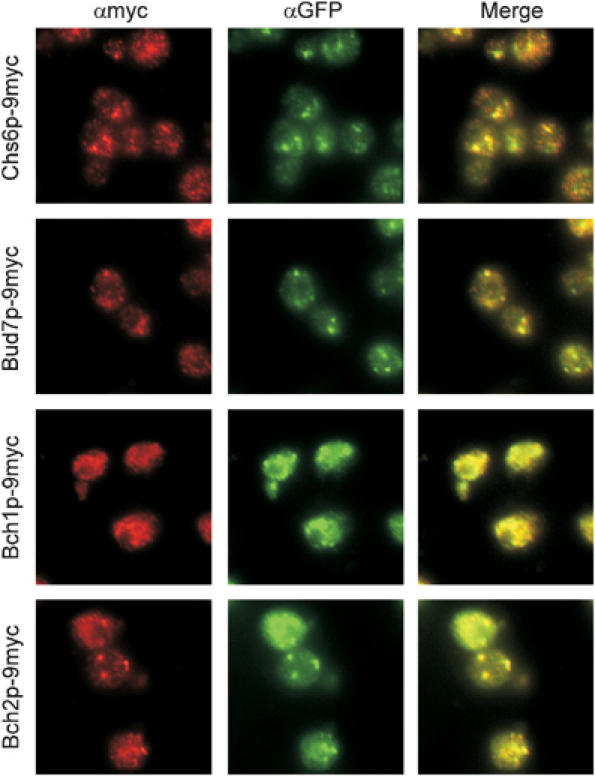
The ChAPs are at least partially localized to the TGN. The ChAPs were chromosomally tagged with 9myc and Sec7p with GFP. Strains were grown to logarithmic phase, fixed and stained with α-myc and α-GFP antibodies. α-Myc and α-GFP were detected with Cy3 and FITC, respectively.
Figure 9.
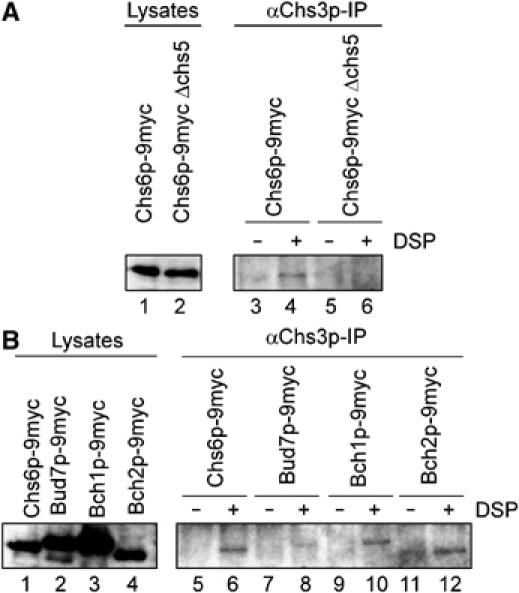
Chs3p interacts with the ChAPs. (A) Crosslinking experiments were performed using either WT or Δchs5 strains carrying Chs6p-9myc. The lysates were treated with either DMSO or DSP before Chs3p immunoprecipitation. The precipitate was analyzed by immunoblot with α-myc. In all, 8% of the lysates was loaded in lanes 1 and 2. (B) Crosslinking experiments were performed using strains in which the ChAPs were chromosomally tagged with 9myc as in (A). The lysates and corresponding precipitates were from the same immunoblot, and image processing was identical.
As activated Arf1p, Chs5p and the ChAPs all localize to the TGN, we were wondering whether the localization of Chs5p and the ChAPs might be dependent on the presence of activated Arf1p. Therefore, we treated yeast cells with the fungal metabolite brefeldin A (BFA), which is an inhibitor of ARF-GEF function. Already after 15 min of BFA treatment, Golgi localization of Chs5p-6HA and Bch2p-9myc was lost (Supplementary Figure S2), indicating that Chs5p and the ChAPs depend on Arf1p-GTP for their Golgi localization.
The C-terminus of the ChAPs is essential for function
None of the ChAPs is essential and even the quadruple knockout did not show a detrimental phenotype under normal growth conditions (data not shown). Therefore, to study further the molecular role of the different ChAPs, we generated small deletions in the different genes. Remarkably, when we removed the C-terminal 13–14 amino acids in Chs6p, Bud7p and Bch1p, the strains phenocopied the gene disruption (Figure 6). Similar to a Δchs6 strain, a Chs6pΔc-9myc strain was temperature-sensitive and resistant to calcofluor (Figure 6B). Furthermore, Chs3p-GFP was trapped in the TGN (data not shown). Likewise, Bud7pΔc-9myc homozygous diploid cells displayed a random budding pattern, indistinguishable from Δbud7 cells (Figure 6C) and Bch1pΔc-3HA was sensitive to YMP+ (Figure 6D). The observed phenotypes for these ChAPs were not owing to decreased stability or absence of the proteins in the cell, because we could detect similar levels by immunoblot (Figure 7). However, Bch2pΔc-9myc was degraded rapidly, so the role of the C-terminus could not be ascertained (Figure 6A). Nonetheless, because the other nonfunctional proteins were still expressed, the C-terminal-deleted strains served as valuable tools in the course of further experiments.
Figure 6.
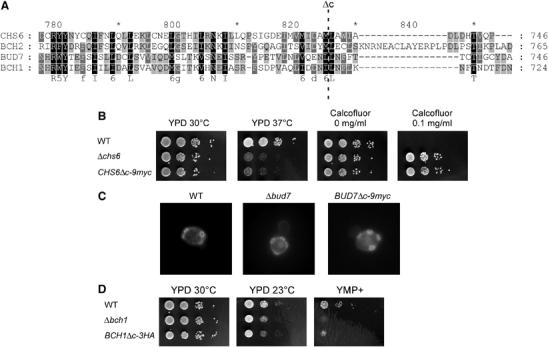
The C-terminus of the ChAPs is essential for their function. (A) Sequence alignment of the C-terminus of the ChAPs. The last 13 amino acids of Chs6p were deleted and replaced by 9myc. The C-termini of the other three family members were deleted and replaced by epitope tags according to this alignment. A dashed line indicates the site of deletion. (B) Growth of CHS6Δc-9myc as compared to WT and Δchs6 at different temperatures and on calcofluor plates. (C) Budding pattern of a homozygous diploid BUD7Δc-9myc strain. Diploid yeast strains were grown to logarithmic phase and stained with calcofluor. (D) Growth of BCH1Δc-3HA as compared to WT and Δbch1 at different temperatures and on YMP+ plates.
The C-terminus of the ChAPs is not required for the interaction with Arf1p
We have shown that the ChAPs are able to bind to Arf1p (Figure 2C and D). To test if the C-terminus of the ChAPs is required for this interaction, we performed co-immunoprecipitation experiments with lysates from strains in which the C-terminus of individual ChAPs was replaced by 9myc using affinity-purified α-Arf1p IgGs. In addition, the ChAPs were tagged individually in either wild type, Δchs5 or in a background where the other three ChAPs had been deleted and tested for the ability to associate with Arf1p under the different conditions. As shown in Figure 7A (lane 5), Chs6p-9myc, Bud7p-9myc, Bch1p-9myc and Bch2p-9myc were all co-precipitated with affinity-purified α-Arf1p-IgGs. Neither the deletion of CHS5 (Figure 7A, lane 7) nor the deletion of the three other ChAPs (Figure 7A, lane 9) altered the precipitation efficiencies. However, deletion of the C-terminus increased the amount of protein precipitated in the case of Chs6p, Bud7p and Bch1p (Figure 7A, lane 8). As mentioned above, Bch2pΔc-9myc was too unstable to be evaluated. In conclusion, all ChAPs interact independently with Arf1p and neither their complete C-termini nor Chs5p is required for this interaction.
The C-terminus of the ChAPs is required for the interaction with Chs5p
Mutations in CHS5 and CHS6 result in a similar phenotype and both proteins interact with Arf1p, raising the possibility that Chs6p might bind to Chs5p. To test if the other ChAPs would also bind to Chs5p, we performed precipitations with Chs5p antiserum on the same lysates that were used to detect the Arf1p interaction (Figure 7). All ChAPs co-immunoprecipitated with Chs5p (Figure 7B, lane 5). The deletion of three ChAPs did not alter significantly the precipitation efficiency of the remaining one (Figure 7B, lane 8). However, Chs6pΔc-9myc and Bch1pΔc-9myc were not co-precipitated with Chs5p (Figure 7B, lane 7). The amount of precipitated Bud7pΔc-9myc was slightly reduced. Most importantly, however, these experiments provide evidence that the ChAPs bind independently from each other to Chs5p and that their C-termini facilitate this interaction. Furthermore, the requirement for an intact C-terminus argues that ChAP functions are mediated through Chs5p.
Chs5p is essential for the TGN localization of the ChAPs
As the interaction of the members of the ChAP family with Chs5p appeared to be essential for protein function, we asked whether this function is related to the localization of the ChAPs. Chs5p localizes to the TGN (Santos and Snyder, 1997, and Figure 8). This localization is independent of the ChAPs, because we observed the same TGN staining in the absence of the entire ChAP family (Figure 8A). This result indicated that Chs5p might either recruit the ChAPs or stabilize the ChAPs at the TGN. If this were the case, one would expect that the TGN localization of the ChAPs would require Chs5p.
Figure 8.
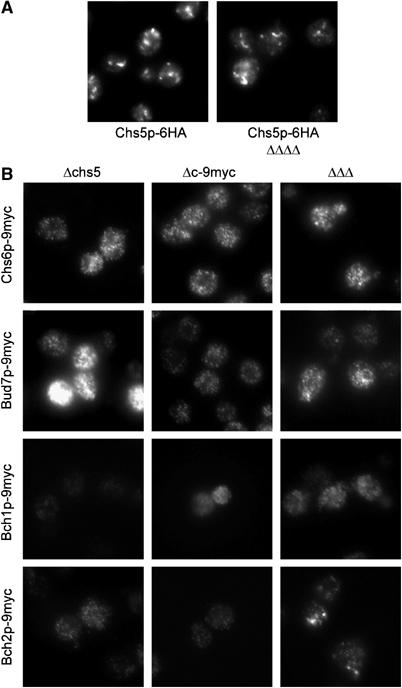
Chs5p is essential for the TGN localization of the ChAPs. (A) Chs5p-6HA staining in WT cells and in cells in which the whole ChAP family is deleted (ΔΔΔΔ). (B) Staining of ChAP-9myc in Δchs5 strains or strains in which the other three ChAPs were deleted (ΔΔΔ) or the ChAPΔc-9myc in a WT background. The same exposure time as in Figure 5 was used for each protein within one row.
The deletion of CHS5 had a dramatic impact on both expression levels as well as localization of the ChAPs. In Δchs5 strains, Bud7p is up- and Bch1p downregulated, both evident in immunofluorescence (Figure 8B) and immunoblots (Figure 7B, lanes 1 and 2). The same exposure time as in Figure 5 was used to allow a direct comparison. In addition, Sec7p-GFP staining in Δchs5 and in C-terminally deleted ChAP strains indicated a normal Golgi morphology (data not shown). Chs6p, Bud7p and Bch2p lost their TGN localization in a Δchs5 background and showed a diffuse staining (Figure 8B). Furthermore, the deletion of the C-terminus of the ChAPs had a similar effect on their TGN association. The impact on the localization of Bch1p was generally hard to score because of the already very diffuse staining of this protein in WT cells. These experiments provide further evidence that Chs5p as well as the C-terminus of the ChAPs is required for either their recruitment to or their stabilization at the TGN. Taken together, these results indicate that localization of the ChAPs to the TGN is a prerequisite for protein function. Because Chs5p and Chs6p have been shown to be required for export of Chs3p out of the TGN, the ChAPs might act together with Chs5p as cargo receptors/coat for specialized transport vesicles that bud from the TGN.
Next, we investigated the influence of the ChAPs on the localization of each other by performing immunofluorescence in triple deletion strains (Figure 8B). The most dramatic effect was observed for Chs6p and Bud7p, which showed a dispersed staining in the absence of the other three ChAPs, similar to what was observed in the Δchs5 strain. Bch2p retained TGN staining. Thus, the localization of Bch2p was less dependent on the other ChAPs than on Chs5p or its C-terminus. These results are corroborated by the finding that a Δchs5 strain displays all the phenotypes of the different ChAPs deletions. The data are in agreement with ChAPs assembly probably on membranes rather than the presence of a preformed complex.
Chs3p interacts with the ChAPs
If the interpretation of our data were correct, one would expect an interaction between Chs3p and at least a subset of the ChAPs. Because this interaction might be only transient, we chose a crosslinking approach to address this point. Cell lysates from different strains were treated with the cleavable crosslinker DSP, Chs3p was immunoprecipitated and the co-precipitating ChAPs were visualized by immunoblot. Chs6p-9myc was crosslinked to Chs3p (Figure 9A). This interaction was dependent on the presence of Chs5p. We could also detect an interaction of Chs3p with Bch1p-9myc and Bch2p-9myc (Figure 9B), indicating that they might also be involved in export of Chs3p from the TGN. Taken together, our results are consistent with a role of the ChAPs as cargo receptor for Chs3p or even a coat for Chs3p-containing vesicles.
Discussion
In this paper, we characterized and analyzed a new family of proteins, the ChAPs, which is involved in transport of chitin synthase Chs3p to the plasma membrane. We found that they interact with Chs5p, with the small GTPase Arf1p and with the cargo Chs3p. These interactions most likely take place at the trans-Golgi. The C-terminus of the ChAPs is necessary for the interaction with Chs5p and for localization to the TGN. We and others have established scoreable phenotypes for the individual deletion for three of the four ChAPs (Bulawa, 1992; Zahner et al, 1996). The deletion of CHS5 displayed all these phenotypes, indicating that Chs5p acts in the same pathway, either upstream of the ChAPs or as a convergence point. What could be the role of the ChAPs in Chs3p transport?
It is tempting to speculate that the ChAPs could form together with Chs5p a novel coat. This conclusion is supported by (1) their direct binding to Arf1p-GTP; (2) their ability to interact with each other; (3) their recruitment to the trans-Golgi in a Chs5p-dependent manner and (4) probably most importantly by their ability to bind to cargo. This idea is supported by the results from Santos and Snyder (1997), who suggested that Chs5p is present on transport vesicles. In addition, we could not detect any clathrin or coatomer in co-immunoprecipitates with the ChAPs (MT and AS, unpublished results), indicating that the ChAPs and Chs5p may act separately from the major coats.
Alternatively, the ChAPs could act as chaperones like the ER-resident Shr3p and Chs7p, which are required for inclusion of Gap1p and Chs3p, respectively, into COPII-coated vesicles, but do not enter the vesicle themselves (Kuehn et al, 1996, 1998; Trilla et al, 1999). However, it seems unlikely that ChAPs are chaperones, because they are soluble proteins that have to be recruited to the membrane in order to fulfill their function. This recruitment to membranes requires the presence of Chs5p on the late Golgi, because deletion of CHS5 abolished ChAPs' TGN localization. Yet, another possibility would be that Chs5p and the ChAPs control the generation of specialized transport carriers at the trans-Golgi, but are not part of a coat, similarly to the FAPPs, which are phosphoinositide-binding proteins on nascent carriers and which can also interact with Arf1 to control budding at the TGN (Godi et al, 2004). Although neither ChAPs nor Chs5p contain a PH domain, a different anchoring mechanism possibly through Chs5p could restrain the ChAPs at the late Golgi. However, Chs6p is not exclusively localized to the TGN. A significant portion is also found on chitosomes, which are specialized endosomes (Ziman et al, 1996). In addition, Bch1p was mostly cytoplasmic. This indicates that the association of the ChAPs with Golgi membranes might be more dynamic than that of the FAPPs.
In the case of Chs3p, Chs6p would recognize the cargo and therefore allow the inclusion into the polymerizing coat. It is tempting to speculate that each of the ChAPs recognizes a specific subset of cargo. Bud7p might be able to interact specifically with proteins involved in bud-site selection and polarity establishment, whereas Bch1p could be required to deliver amino-acid permeases or specific amino-acid transporters to the membrane, because of the YMP+ phenotype. What all these cargoes would have in common is their tightly controlled temporal and spatial localization to a subdomain of the plasma membrane. It is unlikely that Chs5p is a specific cargo receptor, because deletion of CHS5 accumulates not only all phenotypes of single ChAPs deletions but also additional phenotypes. Therefore, it is likely that Chs5p may play a much more general role in the vesicle formation at the TGN.
One specific member of the ChAPs is necessary but not sufficient for transport of a particular cargo. Chs3p requires the presence of Chs6p for export from the trans-Golgi; however, simultaneous deletion of BCH1 and BUD7 trapped Chs3p in the Golgi, despite the presence of Chs6p, clearly indicating a role for the other members of the ChAPs. Furthermore, in addition to Chs6p, Bch1p and Bch2p were also crosslinked to Chs3p. One possibility might be that Bch1p, Bch2p and Bud7p could help to form primers that would represent a starting point for vesicle formation (Springer et al, 1999). Alternatively, Bch1p, Bch2p and Bud7p could help to stabilize a Chs6p–Chs3p interaction until a transport vesicle could be generated. Both of these explanations would not require the presence of all ChAPs for a specific transport event, which is what we observe for Chs3p transport, bud-site selection and sensitivity to YMP+. However, more than one ChAP might be necessary for efficient cargo transport and thus the ChAPs might form facultative oligomers. This conclusion is supported by the apparent different stoichiometries in the co-immunoprecipitation experiments (Figure 4) and the various complexes detected by blue native gel electrophoresis (MT and AS, unpublished results).
Whatever the involvement of the ChAPs in the generation of vesicles at the trans-Golgi is, these vesicles are most likely not default secretory vesicles, rather than special ones, because they carry cargoes that are only required at a certain time in the cell cycle. These specialized vesicles are most likely targeted to the bud neck and the incipient bud site to deliver Chs3p and at least a polarity cue, which might help the cell to find the correct next budding site. However, we did not observe any change in localization of Chs5p and the ChAPs over the cell cycle, indicating that there might be an additional factor, which controls the vesicle generation temporally. The identification of this elusive factor might be difficult because neither the ChAPs nor Chs5p are essential under standard laboratory growth conditions. Therefore, generation of these specialized transport vesicles might not be essential either. In addition, multiple pathways lead out of the Golgi. Valdivia et al (2002) have provided evidence that there is an alternative trafficking route for Chs3p to the plasma membrane when ARF1 and CHS6 or CHS5 are deleted. Although the cells of these double deletions were rendered calcofluor-sensitive again, a similar combination of deletions with ARF1 and BUD7 did not rescue the bud-site selection defect of the Δbud7 strain (MT and AS, unpublished results), suggesting that the cargo for Bud7p might still reach the plasma membrane, but as the temporal and spatial control of the delivery of the polarity cue was lost, the random budding pattern could not be rescued.
Materials and methods
Yeast media
Standard yeast media were prepared as described (Sherman, 1991). Calcofluor plates were based on minimal medium containing additionally 0.1% yeast extract, 1% MES (pH 6.0) and 0.1 mg/ml calcofluor (Sigma). YMP+ plates contained 6.3 g/l yeast nitrogen base without amino acids and ammonium sulfate (Difco), 4.5 g/l yeast extract, 9 g/l peptone, 9 g/l succinic acid, 4.5 g/l NaOH and 25 g/l ammonium sulfate (Lillie and Pringle, 1980).
Yeast genetic methods
Standard genetic techniques were used throughout (Sherman, 1991). Chromosomal tagging and deletions were performed as described (Knop et al, 1999; Gueldener et al, 2002). For CHS6, BCH1 and BCH2, the whole ORF was deleted, whereas BUD7 was deleted only until 100 bp upstream the Stop codon leaving the overlapping ORF YOR300w intact. Strains and primers used are given in Table I and Supplementary Table S1. All PCR-based chromosomal manipulations were confirmed by analytical colony PCR.
Table 1.
Yeast strains used in this study
| Designation | Genotype | Reference |
|---|---|---|
| YPH499 | MAT a ade2 his3 leu2 lys2 trp1 ura3 | Sikorski and Hieter (1989) |
| YPH500 | MAT α ade2 his3 leu2 lys2 trp1 ura3 | Sikorski and Hieter (1989) |
| YPH501 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 | Sikorski and Hieter (1989) |
| YAS431 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS5∷LEU2 (Kluveromyces lactis) | This study |
| YAS571 | MAT a ade2 his3 leu2 lys2 trp1 ura3 ARF1∷HIS3MX6 | This study |
| YAS525 | MAT α ade2 his3 leu2 lys2 trp1 ura3 CHS6∷URA3 (K. lactis) BCH2∷KAN (Tn903) | This study |
| YAS430 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷LEU2 (K. lactis) BCH1∷HIS5 (Schizosaccharomyces pombe) | This study |
| YAS563-2a | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷URA3 (K. lactis) | This study |
| YAS563-3a | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷LEU2 (K. lactis) | This study |
| YAS563-4a | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷KAN (Tn903) | This study |
| YAS563-5a | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷HIS5 (S. pombe) | This study |
| YAS563-10a | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷LEU2 (K. lactis) BCH1∷HIS5 (S. pombe) | This study |
| YAS988 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 CHS5∷LEU2 (K. lactis)/CHS5∷TRP1 (K. lactis) | This study |
| YAS778 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 CHS6∷URA3 (K. lactis)/CHS6∷URA3 (K. lactis) | This study |
| YAS779 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 BUD7∷LEU2 (K. lactis)/BUD7∷URA3 (K. lactis) | This study |
| YAS780 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 BCH2∷KAN (Tn903)/BCH2∷KAN (Tn903) | This study |
| YAS781 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 BCH1∷HIS5 (S. pombe)/BCH1∷HIS5 (S. pombe) | This study |
| YAS793 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS5∷LEU2 (K. lactis) ARF1∷HIS3MX6 | This study |
| YAS794 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷URA3 (K. lactis) ARF1∷HIS3MX6 | This study |
| YAS795 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷LEU2 (K. lactis) ARF1∷HIS3MX6 | This study |
| YAS796 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷KAN (Tn903) ARF1∷HIS3MX6 | This study |
| YAS797 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷HIS5 (S. pombe) ARF1∷KANMX6 | This study |
| GPY60 | MAT α ura3–52 leu2,3–112 his4–579 pep4∷URA3 prb1 trp1 gal2 | R Schekman |
| YAS603 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-3HA-His3MX6 | This study |
| YAS839 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS5∷CHS5-6HA-TRP1 (K. lactis) CHS6∷URA3 (K. lactis) BUD7∷LEU2 (K. lactis) BCH2∷KAN (Tn903) BCH1∷HIS5 (S. pombe) | This study |
| YAS328 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6-9myc-TRP1 (K. lactis) | This study |
| YAS335 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) | This study |
| YAS576 | MAT α ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) | This study |
| YAS697 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis)/BUD7∷BUD7-9myc-TRP1 (K. lactis) | This study |
| YAS339 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1-9myc-TRP1 (K. lactis) | This study |
| YAS589 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-9myc-TRP1 (K lactis) | This study |
| YAS333 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) | This study |
| YAS379 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) | This study |
| YAS380 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) | This study |
| YAS582 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-9myc-TRP1 (K lactis) CHS5∷LEU2 (K. lactis) | This study |
| YAS653 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6-9myc-TRP1 (K. lactis) BUD7∷LEU2 (K. lactis) BCH2∷KAN (Tn903) BCH1∷HIS5 (S. pombe) | This study |
| YAS615 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) ura3 CHS6∷URA3 (K. lactis) BCH2∷KAN (Tn903) BCH1∷HIS5 (S. pombe) | This study |
| YAS614 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1-9myc-TRP1 (K. lactis) CHS6∷URA3 (K. lactis) BUD7∷LEU2 (K. lactis) BCH2∷KAN (Tn903) | This study |
| YAS654 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-9myc-TRP1 (K lactis) CHS6∷URA3 (K. lactis) BUD7∷LEU2 (K. lactis) BCH1∷HIS5 (S. pombe) | This study |
| YAS684 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6Δc-9myc-TRP1 (K. lactis) | This study |
| YAS798 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7Δc-9myc-TRP1 (K. lactis) | This study |
| YAS799 | MAT α ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7Δc-9myc-TRP1 (K. lactis) | This study |
| YAS802 | MAT a/α ade2/ade2 his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 BUD7∷BUD7Δc-9myc-TRP1 (K. lactis)/BUD7∷BUD7Δc-9myc-TRP1 (K. lactis) | This study |
| YAS698 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1Δc-9myc-TRP1 (K. lactis) | This study |
| YAS800 | MAT α ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1Δc-3HA-His3MX6 | This study |
| YAS801 | MAT α ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2Δc-9myc-TRP1 (K lactis) | This study |
| YAS792 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) CHS6∷CHS6-yEGFP-KanMX6 BCH2∷BCH2-3HA-HIS3MX6 BCH1∷BCH1-2AU5-LEU2 (K. lactis) | This study |
| YAS699 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) CHS6∷CHS6-yEGFP-KanMX6 BCH2∷BCH2-3HA-HIS3MX6 | This study |
| YAS700 | MAT α ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6-yEGFP-KanMX6 BCH1∷BCH1-2AU5-LEU2 (K. lactis) | This study |
| YAS604 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) BCH2∷BCH2-3HA-His3MX6 | This study |
| YAS381 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS5∷CHS5-6HA-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS382 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS385 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS386 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS655 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS384 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS387 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷BUD7-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS388 | MAT a ade2 his3 leu2 lys2 trp1ura3 BCH1∷BCH1-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS656 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-9myc-TRP1 (K. lactis) CHS5∷LEU2 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS835 | MAT a ade2 his3 leu2 lys2 trp1 ura3CHS6∷CHS6c-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS836 | MAT a ade2 his3 leu2 lys2 trp1 ura3BUD7∷BUD7c-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS837 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH1∷BCH1c-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS838 | MAT α ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2c-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 | This study |
| YAS906 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷LEU2 (K. lactis) BCH1∷HIS5 (S. pombe) ARF1∷KAN (Tn903) | This study |
| YAS947 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS3∷CHS3-yEGFP-KanMX6 | This study |
| YAS941 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS5∷LEU2 (K. lactis) CHS3∷CHS3-yEGFP-KanMX6 | This study |
| YAS942 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷URA3 (K. lactis) CHS3∷CHS3-yEGFP-KanMX6 | This study |
| YAS943 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BUD7∷LEU2 (K. lactis) BCH1∷HIS5 (S. pombe) CHS3∷CHS3-yEGFP-KanMX6 | This study |
| YAS947 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS6∷CHS6Δc-9myc-TRP1 (K. lactis) CHS3∷CHS3-yEGFP-KanMX6 | This study |
| YAS1057 | MAT a ade2 his3 leu2 lys2 trp1 ura3 CHS5∷CHS5-6HA-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 ERG6∷LEU2 (K. lactis) | This study |
| YAS1055 | MAT a ade2 his3 leu2 lys2 trp1 ura3 BCH2∷BCH2-9myc-TRP1 (K. lactis) SEC7∷SEC7-GFP-URA3 ERG6∷LEU2 (K. lactis) | This study |
Protein purifications
BCH2-3HA and CHS6-GFP were cloned into pTYB12 (NEB) and purified from Escherichia coli according to the manufacturer's instructions. Proteins were buffer exchanged to B88 (20 mM HEPES, pH 6.8, 5 mM Mg(Ac)2, 150 mM KAc, 250 mM sorbitol) using PD10 columns (Amersham). CHS5 was cloned into pET100/D-TOPO (Invitrogen) and purified from E. coli using standard procedures. Coatomer, Arf1p and Sar1p were purified as described (Spang and Schekman, 1998).
Differential affinity chromatography
The affinity chromatography was performed as described by Trautwein et al (2004). In brief, equal amounts of recombinant ΔN17-Arf1Q71Lp and ΔN17-Arf1T31Np were coupled to sepharose–resin and a nucleotide exchange was performed in NE buffer (25 mM HEPES (pH 7.5), 100 mM NaCl, 0.5 mM MgCl2, 1 mM EDTA, 1 mM DTT) for 1 h at 37°C (GTP for Arf1pQ71Lp, GDP for Arf1T31Np). Yeast cytosol was allowed to bind for 1 h at 4°C. Proteins were eluted with exchange buffer containing 500 μM GX′P (GDP for Arf1pQ71Lp, GTP for Arf1T31Np). The samples were separated on SDS–PAGE and immunoblotted.
Antibodies
Antisera against Gea2p (Spang et al, 2001), coatomer (Rexach et al, 1994), Glo3p (Poon et al, 1999) and Arf1p (Spang and Schekman, 1998) have been described. Polyclonal antibodies against Sar1p, Chs5p and the N-terminal fragments 1–171 of Chs3p were raised in rabbits.
Co-immunoprecipitation
Yeast lysates from equal amounts of cells were prepared by spheroplasting as described (Rexach et al, 1994). Spheroplasts were sedimented (2 min, 1000 g), lysed in 20 mM HEPES (pH 6.8), 150 mM KAc, 5 mM Mg(Ac)2, 1% Tween-20 with protease inhibitors and cleared by centrifugation (10 min, 16 000 g).
Immunoprecipitations were performed with 10 μg affinity-purified α-Arf1p antibodies or affinity-purified rabbit IgGs, 10 μl α-Arf1p, α-Sar1p, α-Chs5p or preimmune serum, 5 μl α-HA-11 (Eurogentec), 5 μl α-Myc (9E10; Sigma), 5 μl α-AU5 (Abcam) or 5 μl α-GFP (Torrey Pines) and 100 μl 20% Protein A–Sepharose per 1 ml cleared lysate for 1 h at 4°C. The beads were washed, resuspended in sample buffer and analyzed by immunoblot.
Liposome-binding experiments
Azolectin (20 mg; Sigma) was mixed with 6 ml diethylether and 1 ml 20 mM HEPES (pH 7.4). After sonication, the ether was removed by evaporation. Liposomes were extruded 12 times through 0.6 μm membranes (Avanti) and then incubated with 1.35 μM Arf1p and 625 μM GTPγS in NE buffer for 1 h at 30°C. Bch2p (110 nM) or coatomer (40 nM) were added to the reaction and incubated for 10 min at 4°C. The samples were adjusted to 1 M sucrose in B88, overlaid with 75 μl 0.75 M sucrose in B88 and 20 μl B88. The liposomes were floated for 3 h at 100 000 g (TLA100). The top 30 μl were analyzed by immunoblot.
Immunofluorescence and fluorescence microscopy
Immunofluorescence was performed as described (Pringle et al, 1991). Cells expressing chromosomally epitope-tagged proteins were grown to log phase. After fixation and spheroplasting, the cells were stained with α-HA-7 (Sigma) or α-myc (9E10; Sigma) antibodies. Cy3-coupled secondary antibodies were used for visualization (Jackson ImmunoResearch Laboratories). For comparison, all strains except the triple and quadruple deletion of the ChAPs also contained a chromosomal copy of SEC7-GFP to assess colocalization with the TGN. SEC7-GFP was integrated by transformation of a SpeI-linearized pUSE-URA3 integration plasmid containing SEC7-GFP (Seron et al, 1998). For the visualization of GFP after fixation, α-GFP and secondary antibodies coupled to FITC were used.
Analysis of the budding pattern was carried out as described (Lord et al, 2002). Briefly, cells grown for at least 16 h to log phase were stained after fixation in either 1 mg/ml calcofluor or 1 mg/ml FITC-ConA (Molecular probes). To determine the budding pattern, only cells were scored with no more than four bud scars.
Pictures were acquired with an Axiocam mounted on an Axioplan 2 fluorescence microscope (Zeiss) using Axiovision software. Image processing was performed using Adobe Photoshop 7.0.
Crosslinking experiments
For each sample, yeast lysate from six OD600 was prepared by glass bead lysis in 200 μl B88 buffer with protease inhibitors. The lysate was cleared by centrifugation at 25 000 g for 15 min at 4°C. A measure of 20 μl DMSO or 20 μl DSP (Pierce) dissolved in DMSO were added to 140 μl lysate. The crosslinking reaction took place 30 min at RT and was stopped with 7 μl 1 M Tris (pH 7.5) for 15 min. Then, 8 μl of 20% SDS were added and heated to 65°C for 15 min. A measure of 900 μl IP buffer (50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 0.1% SDS) were added and the sample was centrifuged for 10 min at 20 000 g. The supernatant was subjected to immunoprecipitation for 1 h at RT using 5 μg affinity-purified α-Chs3p antibodies crosslinked to Protein A–Speharose with DMP (Pierce). The precipitates were analyzed by immunoblot.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure Legends and Table S1
Acknowledgments
We thank M Mayer and F Seiler for technical assistance. We are grateful to JR Pringle and IG Macara for helpful discussions. We are indebted to IG Macara for critically reading the manuscript. We thank B Glick, J Hegemann and M Knop for plasmids. We acknowledge all members of the Spang laboratory for supportive discussions. This work was supported by the Max Planck Society. AS is an EMBO Young Investigator.
References
- Bulawa CE (1992) CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol 12: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Graham TR (1998) An arf1Delta synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics 150: 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Chen CY, Emr SD, Graham TR (1998) ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol Biol Cell 9: 653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA (2004) FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6: 393–404 [DOI] [PubMed] [Google Scholar]
- Gu F, Crump CM, Thomas G (2001) Trans-Golgi network sorting. Cell Mol Life Sci 58: 1067–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T (2000) Three ways to make a vesicle. Nat Rev Mol Cell Biol 1: 187–198 [DOI] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972 [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R (1998) COPII–cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 391: 187–190 [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Schekman R, Ljungdahl PO (1996) Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J Cell Biol 135: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Griffiths G, Hoflack B (1996) Mannose 6-phosphate receptors and ADP-ribosylation factors cooperate for high affinity interaction of the AP-1 Golgi assembly proteins with membranes. J Biol Chem 271: 2162–2170 [DOI] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR (1980) Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143: 1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Chen T, Fujita A, Chant J (2002) Analysis of budding patterns. Methods Enzymol 350: 131–141 [DOI] [PubMed] [Google Scholar]
- Ni L, Snyder M (2001) A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 12: 2147–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC (1999) Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK (1991) Immunofluorescence methods for yeast. Methods Enzymol 194: 565–602 [DOI] [PubMed] [Google Scholar]
- Rexach MF, Latterich M, Schekman RW (1994) Characteristics of endoplasmic reticulum-derived transport vesicles. J Cell Biol 126: 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Duran A, Valdivieso MH (1997) CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol 17: 2485–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M (1997) Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol 136: 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron K, Tieaho V, Prescianotto-Baschong C, Aust T, Blondel MO, Guillaud P, Devilliers G, Rossanese OW, Glick BS, Riezman H, Keranen S, Haguenauer-Tsapis R (1998) A yeast t-SNARE involved in endocytosis. Mol Biol Cell 9: 2873–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Herrmann JM, Hamamoto S, Schekman R (2001) The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol Biol Cell 12: 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Schekman R (1998) Reconstitution of retrograde transport from the Golgi to the ER in vitro. J Cell Biol 143: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S, Spang A, Schekman R (1999) A primer on vesicle budding. Cell 97: 145–148 [DOI] [PubMed] [Google Scholar]
- Stearns T, Willingham MC, Botstein D, Kahn RA (1990) ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci USA 87: 1238–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein M, Dengjel J, Schirle M, Spang A (2004) Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol Biol Cell 15: 5021–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trilla JA, Duran A, Roncero C (1999) Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J Cell Biol 145: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW (2002) The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell 2: 283–294 [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Schekman R (2003) The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc Natl Acad Sci USA 100: 10287–10292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713 [DOI] [PubMed] [Google Scholar]
- Yahara N, Ueda T, Sato K, Nakano A (2001) Multiple roles of Arf1 GTPase in the yeast exocytic and endocytic pathways. Mol Biol Cell 12: 221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR (1996) Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol 16: 1857–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Schekman RW (1996) Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol Biol Cell 7: 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R (1998) Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell 9: 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure Legends and Table S1


