Abstract
Voltage-gated ion channels open and close in response to voltage changes across electrically excitable cell membranes1. Voltage-gated potassium (Kv) channels are homotetramers with each subunit constructed from 6 transmembrane segments, S1-S62. The voltage-sensing domain (segments S1-S4) contains charged arginines on S4 that move across the membrane electric field2,3, modulating channel open probability. Understanding the physical movements of this voltage-sensor is of fundamental importance and is the subject of current controversy. Recently, the crystal structure of the KvAP4 channel motivated an unconventional “paddle model” of S4 charge movement, suggesting that the segments S3b and S4 move as a unit through the lipid bilayer with a large (15-20 Å) transmembrane displacement5. We have tested the movement of these segments in functional Shaker K+ channels using luminescence resonance energy transfer to measure distances between the voltage-sensors and a pore-bound scorpion toxin. Our results are consistent with a 2 Å vertical displacement of S4, not the large excursion predicted by the paddle model. Such small movement supports an alternative model in which the protein shapes the electric field profile, focusing it across a narrow region of S46. We conclude that the voltage-sensor segments do not undergo significant transmembrane translation.
Conformational changes in proteins can be studied in great detail by using fluorescence energy transfer as a spectroscopic ruler7,8. Luminescence resonance energy transfer (LRET) is a modified version that employs a lanthanide donor complex with a long excited state lifetime9,10. This unconventional probe can donate energy to a conventional fluorescent acceptor in the standard distance dependent manner of Förster theory10 and energy transfer efficiency and distances are calculated from the time constants of acceptor fluorescence emission (Methods). LRET is capable of accurately measuring distances on Shaker channels in vivo since only donor-acceptor pairs produce sensitized acceptor emission that is measured after a brief time-gate rejects fast background fluorescence. Further advantages arise from the minimal spectral overlap of donor and acceptor, the zero intrinsic anisotropy of the donor lanthanide11, and the accuracy with which donor quantum yield10 and Ro can be estimated.
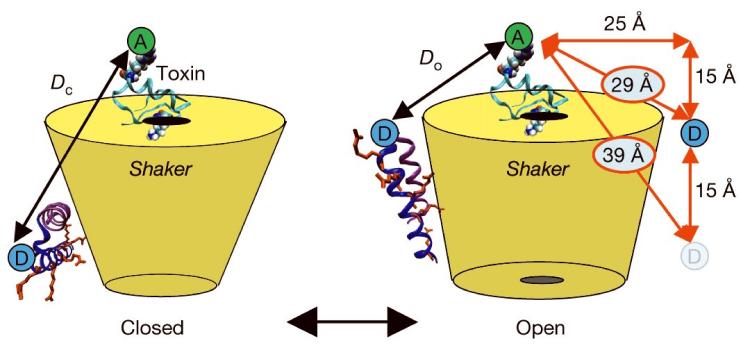
Gating-driven protein movements have previously been measured on the Shaker channel using LRET12 and conventional fluorescence resonance energy transfer13 with both donor and acceptor labelled to sites on the voltage-sensing domains. This configuration measured distance changes parallel to the membrane between S4 helices in the same tetrameric channel but did not directly measure transmembrane movements. Here we have attached the acceptor dye to the channel via a scorpion toxin that binds to the pore from the external solution14,15. Toxin binding is insensitive to the channel's open or closed status and does not alter the movement of the voltage-sensor15,16. Lanthanide donors were attached to several sites on S4, S3b, and the S3-S4 linker region near S4 in order to test directly in vivo whether the voltage-sensing segments undergo a large transmembrane movement (Fig. 1). Any paddle-type mechanism by definition requires transmembrane movement of 15-20 Å5, equivalent to a change in S4-to-toxin distance of ∼10 Å in the configuration used here, as estimated using conservative structural assumptions (Fig. 1). Testing the vertical translation of S3b-S4 is of central importance in evaluating the validity of the paddle mechanism, as the model's other unusual feature, the location of S4 at the lipid interface, has been shown experimentally to be plausible17,18.
Figure 1.
Cartoon representation of the paddle model. LRET measures distances from donor labelled sites (blue circles) on the S3b-S4 paddle (structure taken from the isolated voltage-sensor4). The voltage-sensing arginines are shown in red. The energy transfer acceptors (green circles) are attached to the top of the channel with a scorpion toxin. The paddle model predicts a change in distance, Dc - Do, of 10 Å, estimated by a conservative geometric calculation assuming a 15 Å vertical translation (red arrows).
For LRET, ionic currents of Shaker expressed in Xenopus oocytes were blocked with 100 nM fluorescent charybdotoxin (CTX) or agitoxin-2 (AgTX) such that almost all channels were blocked and residual unblocked current was limited to 10-30 μA to minimize voltage-clamping errors19. The charge-voltage relations for donor-labelled channels were measured separately from cells blocked with a saturating level of unlabelled toxin (Supplementary Data). Lanthanide-chelate donors (terbium) were attached to site-directed cysteines on the voltage-sensor domain (Methods). The labelling of voltage-sensing segments with fluorescent probes does not disrupt the movement of gating charge. The effect of fluorescein (and rhodamine) was tested, but not terbium-chelate (Supplementary Data). Toxin binding brought acceptor fluorophores in proximity to the labelled donor sites on the channel, and the distances were calculated as a function of voltage from the measured LRET time constants. Acceptor sensitized emission data from E333C on S3b and background controls (Methods) are shown in Fig. 2. LRET signals were fit well to two time constants that reflect the asymmetry of the toxin-acceptor position with respect to the central axis of the channel (Fig. 4). Distances from both time constants were calculated as well as a population-weighted average distance vs. voltage (Methods). These distances are shown for E333C on S3b and L361C on S4 (Fig. 2). The results clearly show that sites homologous to the KvAP voltage-sensor paddle move less than 1 Å with respect to the toxin when going from the closed to the open channel positions. If the S4 segment moves in a purely vertical direction, a change in LRET distance of 1 Å corresponds to a 2 Å vertical displacement, as estimated by a conservative geometric calculation similar to that shown in Fig. 1.
Figure 2.
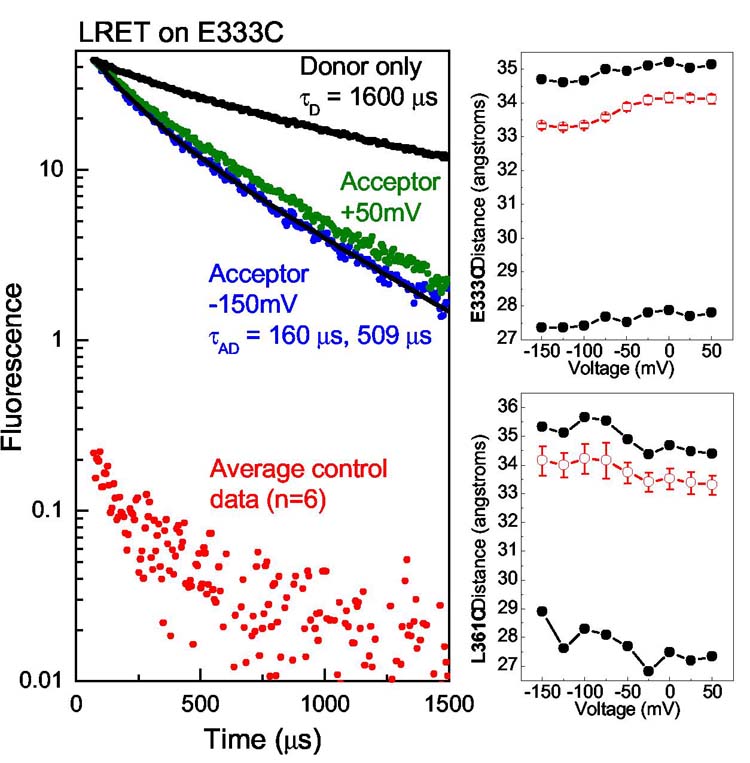
LRET raw data and distance calculations. Acceptor sensitized emission at two extreme voltages are shown (left) for the E333C mutant near the top of the paddle. The time constants displayed a small voltage dependence corresponding to a small movement 0.8 Å away from the toxin (top-right). The distances calculated from the two lifetime components and the weighted-average (Methods) are shown. L361C showed voltage dependent movement of 0.8 Å towards the toxin (bottom-right). Error bars for the average distance represent standard error of the mean (n = 13 for E333C, n = 8 for L361C).
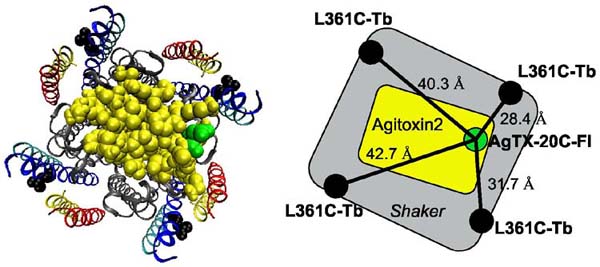
Figure 4.
A model of Shaker with docked agitoxin predicts four distances for each LRET experiment. The coordinates (left) provide an opportunity to compare our measurements with a picture of Shaker that has S3 and S4 placed against the pore domain. Distances for L361C on S4 are shown measured from alpha-carbons (right).
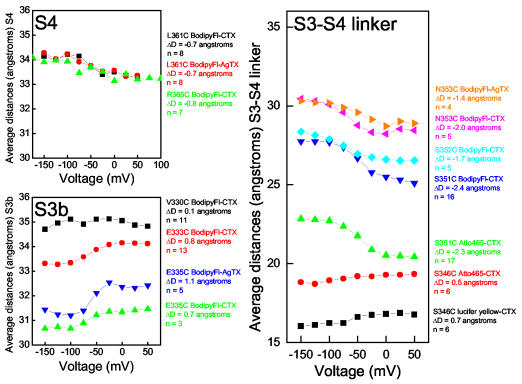
Small but unambiguous voltage-dependent movements were seen at many sites (Fig. 3) with S3 moving ∼1 Å away from the toxin, S4 moving ∼1 Å towards the toxin, and the sites in the linker moving up to 2.5 Å in a manner consistent with a change in linker tilt12. We note that S3b and S4 move in opposite directions, instead of translating together as a rigid unit. For three sites, N353C, E335C, and L361C, both AgTX and CTX gave similar calculated distances. Two sites on the S3-S4 linker were studied using two different acceptors, CTX-Lucifer Yellow and CTX-Atto465, useful for measuring distances as short as 15 Å (Methods). For S346C, the calculated distances differed by only 2.5 Å, which may be attributed to differences in dye size and linker lengths. For S351C the distances obtained using CTX-Atto465 vs. CTX-Bodipy Fl differed by 5 Å, but the gating-induced change in distance was unaffected by the choice of acceptor. Thus, the absolute distances are slightly uncertain, but the changes in distance are very reproducible. As a further control, we switched the donor and acceptor for one experiment, labelling E333C with fluorescein acceptors and attaching a CTX-Tb donor to the top of the channel. The resulting distance measurements were nearly identical to those in Fig. 2 (Supplementary Data). In every experiment, minimal voltage-dependent changes in LRET amplitudes as well as time constants demonstrate that only small changes in energy transfer occurred. These small changes refute the most central feature of the paddle model: substantial physical movement of gating charge transverse to the membrane plane.
Figure 3.
Average distances for many Shaker sites. The S4 and S3b sites are homologous to sites on the KvAP voltage-sensing paddle. The distances for S4 change just 0.8 Å, consistent with an approximately 2 Å vertical translation. S3b moves in the direction opposite to S4, moving just 0.8 Å away from the toxin. Sites in the S3-S4 linker are clearly closer to the toxin than the transmembrane segments, as expected, and move no more than a few angstroms.
We are confident that this LRET technique estimates distances faithfully, on proteins in general and K+ channels in particular. Beyond the technique's agreement with known structures in soluble proteins10, distances measured here agree well with independent estimates of distances from the Shaker voltage-sensors to the pore using tethered tetraethylammonium blockers20. For example, in the tether experiments, Q348C, D349C, and K350C were found to be 17-18 Å from the pore in the open state, very similar to our measurements of 17-19 Å and 21-25 Å for S3-S4 linker residues S346C and S351C, respectively. Likewise, E334C and E335C were found to be ∼30 Å from the pore in the tether experiments, close to our measurements for these same residues at the end of S3b, 32-34 Å. Although tethered blocker data and LRET measure distances to two different points near the central pore, the close agreement between the approaches demonstrate their power for constraining structural distances on the Shaker channel. Furthermore, tethered blockers measure distances only for the open state whereas LRET has the advantage of probing both open and closed states.
LRET measures absolute distances with less systematic error than traditional energy transfer techniques8,10, and can therefore be used to evaluate and constrain structural models. Recently, a structural model was proposed for the Shaker open state based on a combination of experimental data and molecular dynamics21,22. This model was supplemented with a computationally docked agitoxin23 so that theoretical distances from the toxin labelling site to sites on the Shaker voltage sensor could be compared directly with our LRET measurements (Fig. 4). The model predicts four theoretical distances and we have used simulations to test how well LRET experiments can measure the average distance for situations of such geometric complexity (Table 1, Methods and Supplementary Data). The simulations reproduce average distances in close agreement to model values, with the exception of S351 using CTX-Atto465 where the small Ro caused an underestimation. The LRET experimental results for two sites on S4 demonstrate very close agreement between model and data. However, the LRET measurements may systematically underestimate distances slightly because the position of the probes can wobble around their linker attachment points, weighting the measurement towards the distance of closest approach. The model prediction for S3b was unique in that it predicted a shorter distance (∼ 4 Å) than the distance measured experimentally. Nevertheless, the distance values obtained with LRET are consistent with the general structural view that S3 and S4 are transmembrane segments at all voltages.
Table 1.
Comparison of LRET distance analysis with distances from a model of the Shaker open state21
| Donor site, Acceptor used, Avg. model D Å*, Avg. simulation D Å**, LRET Avg. D | ||||
|---|---|---|---|---|
| (S4) L361, | BodipyFl-CTX, | 35.8, | 33.4, | 33.3 |
| (S4) R365, | BodipyFl-CTX, | 39.8, | 35.5, | 33.2 |
| (S3b) V330, | BodipyFl-CTX, | 30.9, | 30.2, | 34.8 |
| (S3-S4 Linker) S351, | BodipyFl-CTX, | 30.2, | 30.5, | 25.5 |
| (S3-S4 Linker) S351, | Atto465-CTX, | 30.2, | 25.0, | 21.0 |
The mean of four distances measured from the Cα of D20 on a docked AgTX to the Cα of specified sites on each subunit (Figure 4).
See methods and supplementary data.
The small vertical S4 movements presented here supplement the even smaller lateral movements between voltage-sensors previously obtained12 and indicate that the conformational changes that underlie gating charge movement are subtle rather than substantial. The paddle model could be altered to account for our data by allowing the paddle segments to swing laterally outward while undergoing vertical movement such that distances to the toxin remain constant. However, this kind of movement would be flatly inconsistent with the small lateral displacements observed in previous LRET measurements12. The small physical movements of voltage-sensing segments suggest that the membrane electric field must be focused over a very tight region of the voltage-sensor, as if aqueous crevices penetrate the protein and thereby shape the field profile6,24. Small S4 movements relative to these crevices and voltage-induced changes in crevice shape can produce the large gating charge that must traverse the field to account for the steep voltage dependence of voltage-gated channels.
Methods
Distance calculations. The lifetime of acceptor sensitized emission was used to calculate energy transfer efficiency using the relation E = 1 – τAD/τD, where τD is the lifetime of the donor in the absence of acceptor. τD was measured on channel sites in the absence of acceptors. On a few sites τD displayed a slight voltage dependence (S346C and S351C < 10%, E335C < 5%) and these changes were included in the analysis (distances changed < 1 Å). Sensitized emission data were fit to two exponentials using four parameters; A1, τ1, A2, τ2. Multiple time constants indicate that the acceptor molecule is not an equal distance from all four donors labelled to the voltage-sensing domains. An average lifetime was calculated by normalizing the sensitized emission lifetimes by the rate of energy transfer to obtain a ‘population average’25; where . Distances from τ1, τ2 and were calculated by finding E (above) and using where Ro is the characteristic distance of 50% energy transfer. Most data were taken using Bodipy Fl-maleimide acceptors (Molecular Probes) for which Ro = 39 Å. Other data were taken using Atto465-maleimide (Atto-Tec), Ro = 27 Å, and Lucifer Yellow-iodoacetamide (Molecular Probes), Ro = 23 Å.
Toxin biochemistry, Shaker expression and block. Charybdotoxin-R19C and agitoxin-2-D20C were prepared, labelled, and purified as previously described26. The mass of each fluorescent-toxin was verified with mass spectrometry and high affinity block with Shaker was evaluated qualitatively by examining the slow rate of toxin dissociation. The channel construct was the fast inactivation-removed, conducting Shaker H4IR, with the mutations F425G, K427D that increase the toxin binding to subnM affinities27. Xenopus ooctye preparation, channel mutagenesis (Stratagene), and mRNA synthesis (Ambion) were performed using standard procedures. Experiments were performed typically 3-5 days after micro-injection of 20 ng of Shaker mRNA. Voltage clamping was performed with a two-electrode setup (CA-1B, Dagan). Charge-voltage relations were measured using saturating wild-type CTX block (2 μM), and LRET measurements were recorded with nearly complete block using 100 nM fluorescent-toxin (Supplementary Data).
LRET protocols and controls. The optical setup consisted of an Olympus inverted IX-70 microscope with a 40x quartz objective (numerical aperture 0.8, Partec). The lanthanide was excited with a pulsed 337 nm nitrogen laser source (Oriel), reflected by a 400DCLP dichroic (Chroma). Donor and acceptor fluorescence were collected simultaneously with D490/10 and HQ520/20 filters, respectively (Chroma). Fluorescence was detected with two water-cooled R943-02 photomultiplier tubes (Hamamatsu) operated at −1760 V. Prompt fluorescence was rejected using an electronic gate (Products for Research) with a dead-time of 70 μs. The detector current was converted to voltage (106 V/A, Hamamatsu) and filtered at 50 kHz (8-pole Bessel filter, Dagan). The laser pulse was given exactly 40 ms after the initiation of a voltage step to ensure the channels had reached conformational equilibrium before measuring the LRET signals.
Background cysteines on oocytes were generally pre-labelled with β-maleimidopropionic acid (Sigma) for 1 hour after 2-3 day incubation at 12 degrees in order to increase specificity of donor labelling28. Oocytes were then incubated for 24-30 hours at 18 degrees to allow surface expression of Shaker. Cells were placed in depolarizing solution for 30 minutes with 100 μM DTT to reduce cysteine thiols for reaction with maleimide. DTT was washed away before the cells were placed in depolarizing solution containing 80 μM maleimide-lanthanide chelate. LRET signal to background was estimated for every LRET experiment by recording acceptor sensitized emission signals from oocytes that were expressing high levels of the background Shaker construct minus the experimental cysteine mutation. Controls were labelled identically to the LRET experiments.
LRET simulations using a Shaker model. Coordinates for the Shaker open-state model with docked agitoxin21,23 provide predictions for four different distances between the agitoxin-D20 alpha carbon and the four alpha carbons of selected sites on the voltage-sensors. Assuming these four distances, LRET signals were simulated assuming a bi-exponential donor with dominant component, 75% at 1600 μs, and a minor component, 25% at 300 μs. The minor component adds a systematic error that slightly underestimates distances (< 5%). The multiple components can be well described by fitting to two exponentials (Supplementary Data) as was the experimental data. These calculations show how the complicated geometry of the model can be reduced to distance estimations in close agreement with actual LRET measurements (Table 1).
Supplementary Material
Acknowledgements and the Competing Interests statement
We gratefully acknowledge Benoit Roux for putting together coordinates for a combined model of the AgTX-Shaker complex23 with the model for the Shaker open state21, L. Kolmakova-Partensky and Tatyana Lawrecki for technical assistance. This work was supported by grants from NSF MCB99-84841, the Carver Foundation, and the Cottrell funds of the Research Corp (to P.R.S.), NIH grant GM30376 (to F.P.), and HHMI (to C.M.). P.R.S. also thanks Jacquelyn Ackland, Jerome Stenehjem, and the other members of the Sharp Rehabilitation Center of San Diego, CA, for their great care, which made this science possible.
Footnotes
Supplementary Information accompanies the paper on Nature's website (http://www.nature.com).
References
- 1.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952;117:500–44. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–92. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–8. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 6.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–53. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 7.Clegg RM. Fluorescence Resonance Energy Transfer. Current Opinions in Biotechnology. 1995;6:103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 8.Selvin PR. The Renaissance in Fluorescence Resonance Energy Transfer. Nature Structural Biology. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 9.Selvin PR, Rana TM, Hearst JE. Luminescence resonance energy transfer. J. Amer. Chem. Soc. 1994;116:6029–6030. [Google Scholar]
- 10.Selvin PR. Principles and Biophysical Applications of Luminescent Lanthanide Probes. Annual Review of Biophysics and Biomolecular Structure. 2002;31:275–302. doi: 10.1146/annurev.biophys.31.101101.140927. [DOI] [PubMed] [Google Scholar]
- 11.Reifenberger JG, Snyder GE, Baym G, Selvin PR. Emission polarization of europium and terbium chelates. Journal of Physical Chemistry B. 2003;107:12862–12873. [Google Scholar]
- 12.Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 13.Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 14.MacKinnon R, Miller C. Mechanism of charybdotoxin block of the high-conductance, Ca2+-activated K+ channel. J Gen Physiol. 1988;91:335–49. doi: 10.1085/jgp.91.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein SA, Miller C. Mechanism of charybdotoxin block of a voltage-gated K+ channel. Biophys J. 1993;65:1613–9. doi: 10.1016/S0006-3495(93)81200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 17.Hessa T, White SH, von Heijne G. Membrane Insertion of a Potassium-Channel Voltage Sensor. Science. 2005 doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 18.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–5. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 19.Baumgartner W, Islas L, Sigworth FJ. Two-microelectrode voltage clamp of Xenopus oocytes: voltage errors and compensation for local current flow. Biophys J. 1999;77:1980–91. doi: 10.1016/S0006-3495(99)77039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaustein RO, Cole PA, Williams C, Miller C. Tethered blockers as molecular ‘tape measures’ for a voltage-gated K+ channel. Nat Struct Biol. 2000;7:309–11. doi: 10.1038/74076. [DOI] [PubMed] [Google Scholar]
- 21.Laine M, et al. Atomic proximity between S4 segment and pore domain in Shaker potassium channels. Neuron. 2003;39:467–81. doi: 10.1016/s0896-6273(03)00468-9. [DOI] [PubMed] [Google Scholar]
- 22.Laine M, Papazian DM, Roux B. Critical assessment of a proposed model of Shaker. FEBS Lett. 2004;564:257–63. doi: 10.1016/S0014-5793(04)00273-X. [DOI] [PubMed] [Google Scholar]
- 23.Eriksson MA, Roux B. Modeling the structure of agitoxin in complex with the Shaker K+ channel: a computational approach based on experimental distance restraints extracted from thermodynamic mutant cycles. Biophys J. 2002;83:2595–609. doi: 10.1016/S0006-3495(02)75270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asamoah OK, Wuskell JP, Loew LM, Bezanilla F. A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron. 2003;37:85–97. doi: 10.1016/s0896-6273(02)01126-1. [DOI] [PubMed] [Google Scholar]
- 25.Heyduk T, Heyduk E. Luminescence energy transfer with lanthanide chelates: interpretation of sensitized acceptor decay amplitudes. Anal Biochem. 2001;289:60–7. doi: 10.1006/abio.2000.4925. [DOI] [PubMed] [Google Scholar]
- 26.Shimony E, Sun T, Kolmakova-Partensky L, Miller C. Engineering a uniquely reactive thiol into a cysteine-rich peptide. Protein Eng. 1994;7:503–7. doi: 10.1093/protein/7.4.503. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein SA, Pheasant DJ, Miller C. The charybdotoxin receptor of a Shaker K+ channel: peptide and channel residues mediating molecular recognition. Neuron. 1994;12:1377–88. doi: 10.1016/0896-6273(94)90452-9. [DOI] [PubMed] [Google Scholar]
- 28.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–6. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.