Abstract
NDP kinases (NDPKs) are multifunctional proteins that regulate a variety of eukaryotic cellular activities, including cell proliferation, development, and differentiation. However, much less is known about the functional significance of NDPKs in plants. We show here that NDPK is associated with H2O2-mediated mitogen-activated protein kinase signaling in plants. H2O2 stress strongly induces the expression of the NDPK2 gene in Arabidopsis thaliana (AtNDPK2). Proteins from transgenic plants overexpressing AtNDPK2 showed high levels of autophosphorylation and NDPK activity, and they have lower levels of reactive oxygen species (ROS) than wild-type plants. Mutants lacking AtNDPK2 had higher levels of ROS than wild type. H2O2 treatment induced the phosphorylation of two endogenous proteins whose molecular weights suggested they are AtMPK3 and AtMPK6, two H2O2-activated A. thaliana mitogen-activated protein kinases. In the absence of H2O2 treatment, phosphorylation of these proteins was slightly elevated in plants overexpressing AtNDPK2 but markedly decreased in the AtNDPK2 deletion mutant. Yeast two-hybrid and in vitro protein pull-down assays revealed that AtNDPK2 specifically interacts with AtMPK3 and AtMPK6. Furthermore, AtNDPK2 also enhances the myelin basic protein phosphorylation activity of AtMPK3 in vitro. Finally, constitutive overexpression of AtNDPK2 in Arabidopsis plants conferred an enhanced tolerance to multiple environmental stresses that elicit ROS accumulation in situ. Thus, AtNDPK2 appears to play a previously uncharacterized regulatory role in H2O2-mediated MAPK signaling in plants.
All organisms produce reactive oxygen species (ROS) such as superoxide (⋅O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (⋅OH−) as by-products of normal metabolic processes. A slight alteration in the homeostatic set point of intracellular ROS levels signals the cell to modulate its cell metabolism, gene expression, and posttranslational modification of proteins (1–3), indicating that ROS are important for regulating normal cellular functions. However, when ROS levels exceed the cellular antioxidant capacity, the cellular homeostasis is altered, and this results in oxidative injuries. This imbalance is either caused by excessive intracellular ROS production or by a deficiency in antioxidant systems controlling the ROS levels (4). Excessive production of ROS can result from environmental stress. For example, in plants, cold, salt, drought, heat, excessive light, osmotic shock, UV-B irradiation, and pathogens can cause high levels of ROS to be generated and/or accumulated (3–10). These high ROS doses can damage plant tissues and, in severe cases, can lead to cell death (6–10).
The damaging effects of ROS have caused plant and animal cells to develop complex redox homeostatic mechanisms to cope with the over-production of ROS during oxidative stress (7–13). The first step in these mechanisms is the detection and signaling of ROS levels. In many eukaryotes, oxidative stress signaling involves an orchestrated sequence of intracellular signaling cascades. In particular, mitogen-activated protein kinase (MAPK) signaling pathways are reported to be actively involved in transducting oxidative signaling (14–16). MAPK-signaling pathways involve three distinct components of the protein kinase family: MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). Plants transiently elevate the expression of MAPK after exposure to various environmental stresses (17–20). Overexpression of a constitutively active deletion mutant of the MAPKKK, ANP1, activated two of the six coexpressed MAPKs in Arabidopsis protoplasts subjected to acute H2O2 stress (13). Thus, MAPKs appear to play a pivotal role in redox regulation in plants. However, the details of such MAPK-mediated oxidative stress signaling and how it is regulated remain to be elucidated.
NDP kinase (NDPK; EC 2.7.4.6) is believed to be a housekeeping enzyme that maintains the intracellular levels of all (d)NTPs used in biosynthesis except ATP. However, increasing lines of evidence suggest that NDPK also plays a significant role in signal transduction pathways (21). In animals, NDPKs play important roles in vital processes such as the control of cell proliferation, regulation of transcription, and protein phosphotransferase activity (22–24). In plants, it is associated with the phytochrome A response, UV-B signaling, heat stress, and growth (25–28). Hence, NDPK is strongly implicated in the regulation of cellular protein functions, possibly through its phosphotransferase activity (26, 29, 30). However, why NDPKs have such diverse cellular functions and how NDPKs are regulated in response to various cellular processes is still poorly understood.
Here, we describe our initial genetic and biochemical studies that show Arabidopsis thaliana NDPK (AtNDPK2) is associated with MAPK-mediated H2O2 signaling in plants. We reveal that AtNDPK2 specifically interacts with two H2O2-activated A. thaliana MAPKs, AtMPK3 and AtMPK6, and that it also enhances the myelin basic protein (MBP) phosphorylation activity of AtMPK3. We also show that overexpression of AtNDPK2 in plants down-regulates the accumulation of ROS, and that this in turn enhances the tolerance of Arabidopsis plants to multiple abiotic stresses.
Materials and Methods
Plant Materials.
Wild-type (ecotype WS), AtNDPK2 knockout (25), and AtNDPK2-overexpressing transgenic (T3) Arabidopsis thaliana plants were used in this study. Plants were transformed with the Agrobacterium binary vector pCAMBIA1300 (provided by R. Jefferson, CAMBIA, Canberra, Australia) containing AtNDPK2 cDNA (25) inserted into the XbaI and BamHI sites under the control of the CaMV35S promoter. AtNDPK2 expression in transgenic plants was confirmed by both Northern and Western analysis. The chlorophyll content in leaf samples was determined by using a standard protocol (31). Protoplasts from plants were prepared according to the protocol described (32).
Northern Blot Analysis.
Total RNA from control and transgenic plants was isolated as described (33), and 10 μg was electrophoresed on a formaldehyde-1.5% agarose gel. Then, it was transferred to a Millipore Immobilon-Ny+ transfer membrane, UV crosslinked, and hybridized with AtNDPK1, -2, and -3, (accession nos. AF017641, AF017640, and AF044265) gene-specific probe (UTR region). RNA blot hybridization and membrane washing were performed as described (34).
Western Blot Analysis.
Total proteins in Arabidopsis leaves were extracted in 30 mM Tris, pH 8.0, containing 1 mM EDTA and 1× plant protease inhibitor mixture (Sigma) and separated on a SDS/12% PAGE gel. Then, they were blotted onto a Nylon membrane (Hybond-C Extra, Amersham Pharmacia) by using a semidry electro-blotter. Anti-AtNDPK2, anti-His, and anti-GST monoclonal antibodies were used for detection by using a standard protocol (34).
Kinase and Phosphorylation Assay.
Total proteins from Arabidopsis leaves were extracted in 30 mM Tris, pH 8.0, containing 1 mM EDTA and 1× plant protease inhibitor mixture (Sigma). The protein phosphorylation activity was measured by incubating 1 μg of plant proteins at 30°C for 15 min in a solution containing 50 mM Hepes (pH 7.5), 1 mM DTT, and 10 mM MgCl2 with 0.5 μCi (1 Ci = 37 GBq) [γ-32P]ATP. Kinase activity was measured by incubating 1 μg of plant protein for 10 min at 30°C in 20 mM Hepes, pH 7.4, buffer containing 1 mM each of ATP and GDP, 0.5 μCi [γ-32P]ATP, and 3 mM MgCl2 in a total volume of 10 μl. The activity was arrested by adding SDS sample buffer containing 125 mM Tris base, 2% (vol/vol) SDS, and 10% (vol/vol) glycerol. A 4-μl aliquot of the sample was spotted onto polyethyleneimine-cellulose (PEI-cellulose) TLC plates and developed with 0.75 M KH2PO4 buffer, pH 3.6. Dried TLC plates were exposed to x-ray film to detect [γ-32P]GTP (35). For the MBP phosphorylation assay, purified His-AtMPK3 (1 μg) was incubated for 15 min at 37°C with or without GST-AtNDPK2 (0.1 μg) and myelin basic protein (1 μg) together with 0.5 μCi [γ-32P]ATP and 3 mM MgCl2. The reaction was arrested by the addition of SDS sample buffer, and the samples were subjected to SDS/PAGE on a 12% gel. Phosphorylated proteins were visualized by autoradiography at different exposure times.
Flow Cytometry and Confocal Microscopy Studies.
To measure intracellular ROS levels, Arabidopsis protoplasts were incubated for 5 min at room temperature with 2,7-dichlorohydrofluoroscein diacetate (DCFH-DA; D-399, Molecular Probes) at a final concentration of 5 μM and then analyzed microscopically by a model Axioplan2 fluorescence microscope (Zeiss) by using AXIOVISION V.3.0 software (34). The fluorescence intensity levels were measured in flow cytometer PASIII (Partec GmbH, Bioflow, Martinsried, Germany) with excitation and emission settings of 488 and 530 nm, respectively. Experiments were repeated at least three times with ≈30,000 protoplasts per assay. The intracellular ROS levels in DCFH-DA-treated protoplasts were also measured by spectrofluorometric analyses by using a UV-fluorescent spectrophotometer (SFM25, Bio-Tek, Burlington, VT) with excitation and emission settings of 488 and 530 nm, respectively.
Yeast Two-Hybrid Assays.
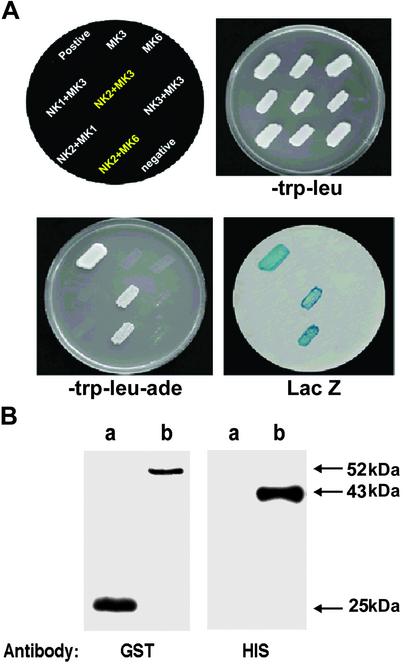
The full-length coding regions of AtMPK1, -3, and -6 were cloned into the pAS2–1 vector (carrying the TRP1 gene, CLONTECH) containing the GAL4 DNA-binding domain (BD). AtNDPK1, -2, and -3 were cloned into the pACT2 vector (carrying the LEU2 gene, CLONTECH) containing the GAL4 activation domain (AD). pTD1-1 and pVA3-1 (CLONTECH) encode the interacting proteins tumor suppressor p53 and simian virus 40 (SV40) large T-antigen (36) fused with the BD and AD, respectively, and were used as a positive control. Interaction of the encoded fusion proteins was investigated by cotransforming appropriate plasmids into the yeast reporter strain pJ69-4A (MATa trp1–90 leu2-3,112 ura3–52 his3–200 gal4 Δgal80 ΔLYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ; ref. 37). Transformed yeast cells bearing both the plasmids were selected by being plated on SD medium lacking tryptophan and leucine (SD-WL) and grown at 28°C for 4 days. Interaction of proteins encoded by recombinant pAS2–1/pACT2 was tested by growing the cells in SD medium lacking tryptophan, leucine, and adenine (SD-WLA). Adenine-positive colonies were further tested for β-galactosidase (LacZ) activation according to the manufacturer's protocol (CLONTECH). The growth of blue colonies in adenine-deficient medium indicated positive interaction. The positive yeast colonies as indicated by both reporter genes (adenine and LacZ) were independently identified and isolated. Plasmids from these clones were rescued, and the presence of the insert sequences was confirmed by restriction analysis and sequencing.
In Vitro Pull-Down Assay.
Purified GST or GST-AtNDPK2 was incubated for 2 h at 4°C with 40 μl of glutathione-Sepharose in PBST (PBS containing 1% Triton X-100). After three washes, the beads were incubated with His-AtMPK3 in 0.5 ml of PBST for 3 h on a rotary shaker at 4°C. The beads then were washed several times with PBST, boiled with SDS sample buffer, and subjected to SDS/PAGE using a 12% gel. Western analysis of the resolved proteins was carried out by using anti-His and anti-GST monoclonal antibodies.
Stress Tolerance Analysis.
Plants were evaluated for cold tolerance according to the protocol described (38). To test for salt tolerance, seeds of wild-type and T3 transgenic plants were surface sterilized, placed on 1× MS agar medium containing 50–100 mM NaCl, and grown for 3 weeks in a growth chamber under controlled conditions (12-h day/12-h night cycle, at 25°C, with a light intensity of 100 Em−2s−1). The response to salt stress was scored by counting the surviving plants. To test the effect of MV (methyl viologen), leaves from 3-week-old plants were incubated in liquid MS medium containing 1 μM MV in 0.1% Tween 20 under the controlled conditions described above. After 7 days, the sensitivity of leaves to MV was judged visually by examining the chlorophyll contents of plant survivors.
Results
AtNDPKs Respond to Oxidative Stress.
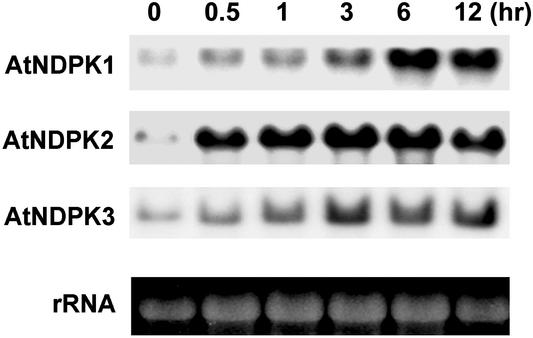
We recently developed a yeast-based genetic screening method to clone plant genes involved in cellular redox regulation (34). This method allowed us to identify a number of genes from plant that suppress Bax toxicity by inhibiting ROS generation in yeast. Several PBI (Plant Bax Inhibitor) genes were isolated, among which the PBI1 (soybean ascorbate peroxidase) gene has already been described (34). PBI2 was identified as soybean NDPK (accession no. U50150). To determine the physiological role of NDPKs in ROS-mediated signaling in plants, we used the Arabidopsis genetic system. Wild-type plants were exposed to H2O2 and the transcript levels of three NDPK isoforms, AtNDPK1, -2 and -3, were analyzed. Transcript levels, especially that of AtNDPK2, were significantly raised within 30 min of H2O2 treatment and remained high for 12 h after treatment (Fig. 1). These results suggest that AtNDPK2 plays a crucial role in the response to oxidative stress. Because light signaling, especially that mediated by phytochrome A, occurs through AtNDPK2 (25), and because several biotic and abiotic stress responses that can generate ROS are mediated in a light-dependent manner (39, 40), we investigated the functional significance of AtNDPK2 in ROS stress first of all.
Figure 1.
The AtNDPK2 transcript accumulates during oxidative stress. Total RNA was prepared from 2-week-old Arabidopsis seedlings treated with 4 mM H2O2. Total RNA from each sample (10 μg) was electrophoresed on a formaldehyde agarose gel. The number above each lane indicates the time in hours the plants were exposed to H2O2. By prestaining the gels with ethidium bromide (Bottom), it was confirmed that an equal amount of RNA was applied to each lane. The RNA blot was probed with 32P-labeled AtNDPK1, -2, and -3 cDNA.
AtNDPK2 Regulates the Cellular Redox State.
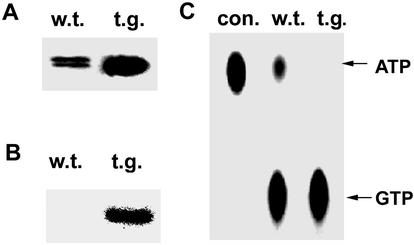
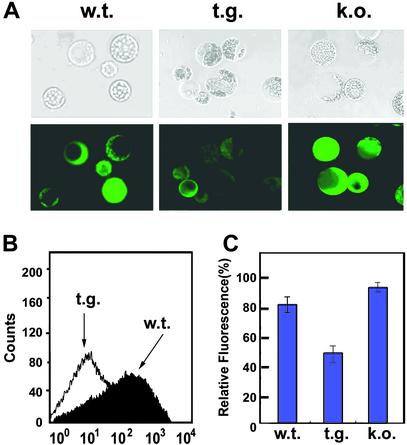
To investigate whether AtNDPK2 expression can influence the cellular redox state, transgenic Arabidopsis plants that overexpress AtNDPK2 were developed. The AtNDPK2 expression was confirmed by Western blot analysis (Fig. 2A). Compared with wild-type plants, the proteins isolated from transgenic plants were autophosphorylated at higher levels and showed greater NDPK activity (Fig. 2 B and C). ROS levels in wild-type, AtNDPK2 knockout mutant (25), and transgenic plants overexpressing AtNDPK2 were measured by analyzing the fluorescence levels in DCFH-DA-stained protoplasts (Fig. 3; ref. 41). Mutant protoplasts lacking AtNDPK2 had higher ROS levels than wild-type cells. In contrast, the AtNDPK2-overexpressing protoplasts exhibited reduced fluorescence and appeared dark against the faint background fluorescence (Fig. 3A). That AtNDPK2 inhibits ROS generation was confirmed by the flow cytometric analysis of protoplasts (Fig. 3B). These experiments reveal that, whereas the wild-type protoplasts showed detectable levels of DCF fluorescence, the protoplasts overexpressing AtNDPK2 had very low fluorescence. When the protoplast fluorescence was measured by fluorescent spectrophotometry, the relative DCF fluorescence intensity was lower in the AtNDPK2 overexpressing cells (50%) than in the wild-type (80%) or knockout mutant (95%) protoplasts (Fig. 3C). These observations further support the notion that AtNDPK2 is involved in ROS regulation in planta.
Figure 2.
Overexpression of AtNDPK2 in Arabidopsis plants enhances NDPK and autophosphorylation activity. (A) Western blot analysis. Crude protein from plants (1 μg) was resolved by SDS/12% PAGE and transferred to nitrocellulose. Membrane strips were immunoblotted with AtNDPK2-specific antibody. (B) Autophosphorylation assay. The samples used in A were incubated at 30°C for 15 min with 0.5 μCi [γ-32P]ATP. The reaction mixtures then were subjected to electrophoresis on a 12% acrylamide gel and autoradiographed. (C) NDPK assay. The protein samples used in A were incubated with GDP and [γ-32P]ATP, resolved on PEI cellulose TLC plates, and exposed to x-ray film. Con, control with only GDP and [γ-32P]ATP; w.t., wild type; t.g., AtNDPK2-overexpressing transgenic plant.
Figure 3.
AtNDPK2 regulates the cellular redox state. Intracellular ROS in Arabidopsis wild-type (w.t.), AtNDPK2-overexpressing transgenic (t.g.), and AtNDPK2-knockout mutant (k.o.) protoplasts were measured. (A) Microscopic analysis. Phase-contrast display (Upper) and the corresponding fluorescence data after incubation with DCFH-DA (Lower) are depicted. (B) Flow cytometric analysis. The intracellular ROS contents of wild-type and AtNDPK2-transgenic cells were measured by flow cytometry. (C) Spectrofluorometric analysis. The intracellular ROS abundance in the protoplasts used in A was measured by fluorescence spectrophotometry. Results represent the mean of three separate experiments.
AtNDPK2 Enhances the Phosphorylation of H2O2-Activated Endogenous Proteins.
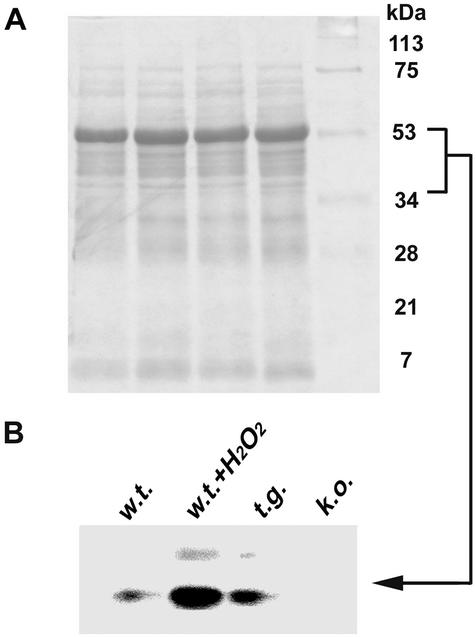
Plant cells produce ROS in response to a broad range of biological and physiological environmental stimuli. AtNDPK2 regulates the accumulation of ROS inside the cells (Fig. 3), most likely by phosphorylating specific proteins. To determine the targets of AtNDPK2 activity, we investigated the protein-phosphorylation profiles of untreated and H2O2-treated wild-type cells by performing the protein phosphorylation assay. Two abundantly phosphorylated proteins were detected in H2O2-treated wild-type cells (Fig. 4). The molecular sizes of these proteins suggest that they may be AtMPK3 and AtMPK6, the two MAPKs in Arabidopsis known to become activated by H2O2 treatment (13). In untreated AtNDPK2 knockout cells, the phosphorylation of these two proteins was greatly decreased. In contrast, in untreated AtNDPK2-overexpressing transgenic cells, their phosphorylation was slightly increased above the levels in wild-type plants (Fig. 4).
Figure 4.
AtNDPK2 is involved in H2O2-stimulated phosphorylation of two putative oxidative stress-activated proteins. Crude proteins (1 μg) from untreated wild-type cells (w.t.), wild-type cells treated with 0.2 mM H2O2 for 15 min (w.t. + H2O2), untreated AtNDPK2-overexpressing transgenic cells (t.g.), and AtNDPK2-knockout mutant (k.o.) were incubated at 30°C for 15 min with 0.5 μCi [γ-32P]ATP. The reaction mixtures then were subjected to SDS/15% PAGE and autoradiographed. (A) Total protein stained with Coomassie brilliant blue. (B) Phosphorylase activity.
AtNDPK2 Specifically Interacts with AtMPK3 and AtMPK6.
To determine whether the regulation of the cellular redox state is directly linked to AtNDPK2 and H2O2–activated MAPK signaling in plants, we next investigated whether AtNDPK2 can interact directly with AtMPK3 and AtMPK6 by performing yeast two-hybrid assays. We found that AtNDPK2 interacts with AtMPK3 and AtMPK6 (Fig. 5A) but not with AtMPK1, another Arabidopsis MAPK (42). We also determined whether this interaction is specific to AtNDPK2 or whether other members of the AtNDPK gene family could perform the same function. Members of the AtNDPK family have different localizations, and each member most likely has a distinct role. AtNDPK1 does not have an identifiable targeting sequence and is thus presumably localized to the cytosol; AtNDPK3 localizes to the mitochondria membrane (43). AtNDPK2 has an N-terminal extension and localizes mainly in nucleus and cytoplasm (25, 27). Yeast two-hybrid assays reveal that AtNDPK1 and AtNDPK3 did not interact with AtMPK3 and AtMPK6 (Fig. 5 and data not shown). Taken together, the results indicate that AtNDPK2 binds specifically to AtMPK3 and AtMPK6.
Figure 5.
AtNDPK2 specifically interacts with AtMPK3 and AtMPK6. (A) Yeast two-hybrid assay. Full-length Arabidopsis NDPK1, -2, and -3 (NK1, -2, and -3) cDNAs were cloned into pACT2 as in-frame fusions with the AD of GAL4. Full-length AtMPK1, -3, and -6 (MK1, -3, and -6) cDNAs were cloned into pAS2-1 as in-frame fusions with the BD of GAL4. Yeast cells (pJ69-4A) were transformed with the plasmids (depicted at Upper Left) and incubated at 30°C for 3 days to select for interactions. trp-leu, transformants selected on SD media lacking tryptophan and leucine; trp-leu-ade, transformants selected on SD media lacking leucine, tryptophan, and adenine; LacZ, β-galactosidase assay for the transformants. The interacting proteins, tumor suppressor p53, and SV40 large T-antigen, fused with the BD and the AD of GAL4, respectively, were used as a positive control. Cells transformed with pACT2 and pAS2-1 were used as a negative control. (B) In vitro protein pull-down assay. Approximately 1 μg of GST (lane a) or recombinant GST-tagged AtNDPK2 (lane b) was incubated with 1 μg of recombinant His-tagged AtMPK3 and pulled down with GST resin after preblocking with 1% BSA. After washing, the bound protein was fractionated by SDS/12% PAGE and subjected to Western blot analysis with anti-GST and anti-His monoclonal antibodies.
Direct interaction of AtNDPK2 with AtMPK3 was confirmed by performing the GST pull-down assay with the Escherichia coli-expressed recombinant proteins His-AtMPK3 and GST-AtNDPK2. After incubating the proteins together, the mixture was fractionated by SDS/PAGE and subjected to Western blot analysis by using monoclonal antibodies specific for GST or His (Fig. 5B). His-AtMPK3 was pulled down with GST-AtNDPK2 but not with GST, indicating that AtNDPK2 and AtMPK3 specifically and directly bind to each other in vitro.
AtNDPK2 Enhances the MAPK Activity of AtMPK3.
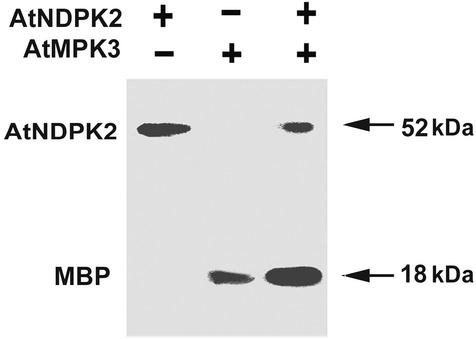
We demonstrated that AtNDPK2 also modulates the kinase activity of AtMPK3 by performing an in vitro MAPK activity assay by using MBP, which is a MAPK-specific substrate. The recombinant proteins GST-AtNDPK2 and/or His-AtMPK3 were incubated with [γ-32P]ATP and MBP. AtNDPK2 exhibited very good autophosphorylation activity but failed to phosphorylate MBP (Fig. 6). Increasing the amount of AtNDPK2 (1 μg) and extending the film exposure times failed to reveal any additional MBP phosphorylation (data not shown), suggesting that MBP is a poor substrate for AtNDPK2. In contrast, recombinant AtMPK3 readily phosphorylated MBP. When the two proteins were incubated together, AtMAPK3-mediated MBP phosphorylation was elevated (Fig. 6), indicating that AtNDPK2 significantly enhances the phosphorylation ability of AtMPK3. Interestingly, the autophosphorylation activity of AtNDPK2 was greatly reduced or sometimes disappeared when it was incubated together with AtMPK3 and MBP (Fig. 6 and data not shown). Furthermore, we could not detect phosphorylated AtMPK3 either in the presence or absence of AtNDPK2 and MBP. It is possible that the phosphotransferase activity of AtMPK3 is more efficient than its autophosphorylation activity, and, as a result, the phosphate bound to the enzyme is immediately transferred to its substrate, MBP. These results collectively suggest that AtNDPK2 autophosphorylates itself but is unable to phosphorylate MBP, and that it enhances the ability of AtMPK3 to phosphorylate MBP.
Figure 6.
AtNDPK2 enhances the MBP phosphorylation activity of AtMPK3. Recombinant GST-AtNDPK2 (0.1 μg) alone or His-AtMPK3 (1 μg) with (+) or without (−) GST-AtNDPK2 (0.1 μg) was incubated with MBP (final concentration of 1.0 μg/μl) and 0.5 μCi [γ-32P]ATP. The proteins were subjected to SDS/12% PAGE and autoradiographed to detect the phosphorylated proteins.
Plants Overexpressing AtNDPK2 Are Tolerant to Multiple Stresses.
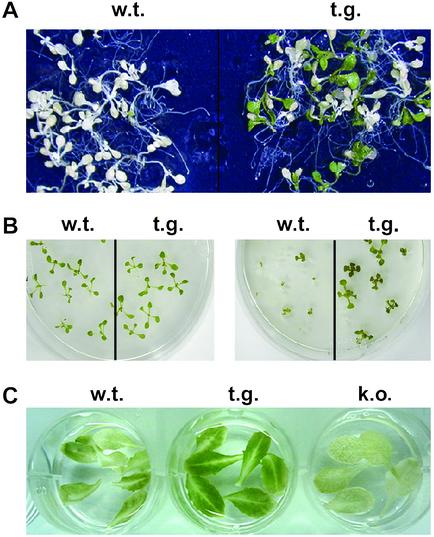
Recent studies indicate that H2O2 can activate a specific Arabidopsis MAPKKK, ANP1, which initiates the phosphorylation cascade that involves AtMPK3 and AtMPK6 activation (13). The activated MAPK cascade in turn activates the stress-response genes that protect plants from diverse environmental stresses (17, 18, 20). Because AtNDPK2 is associated with the cellular redox state (Fig. 1) and enhances the MAPK activity of AtMPK3 (Fig. 6), we investigated whether overexpression of AtNDPK2 influences the response of Arabidopsis plants to cold, salt, and ROS stress (Fig. 7). The cold tolerance assay was performed by freezing the wild-type and transgenic Arabidopsis plants overexpressing AtNDPK2 at −7°C (38), after which the plants were returned to normal growth conditions, and surviving plants were counted 1 week later. The transgenic plants were 50% more tolerant to cold stress than the wild type (Fig. 7A). The surviving plants of both types retained their chlorophyll pigmentation. Further, 50 mM NaCl severely affected the germination and growth rate of the wild-type plants, and >80% of transgenic plants expressing AtNDPK2 survived (Fig. 7B). Phenotypically, the transgenic plants did not differ from the wild-type plants under normal growth conditions (Fig. 7B). The survival rates of transgenic plants to cold and salt stress correlate positively with the ectopic expression level of AtNDPK2 (data not shown).
Figure 7.
AtNDPK2-overexpressing transgenic plants are protected from multiple stress. The environmental stress tolerance of wild-type (w.t.), AtNDPK2-overexpressing transgenic (t.g.), and AtNDPK2-knockout mutant (k.o.) plants was investigated. (A) Tolerance to cold stress. Arabidopsis plants were frozen at −7°C for 1 h, returned to the original growth conditions (38), and photographed 1 week later. The surviving plants all showed green pigmentation. (B) Tolerance to salt stress. Arabidopsis seedlings were raised for 2 weeks on MS medium (Left) or for 3 weeks in MS medium containing 50 mM NaCl (Right) to assess their survival under salt stress. (C) Tolerance to MV. Fully expanded leaves from 3-week-old plants were transferred to MS liquid medium containing 1.0 μM MV for 7 days, and the percentage of plants that survived was recorded. Dying plants were albino and displayed a loss of chlorophyll content.
As ROS enhance the expression of AtNDPK2 (Fig. 1), we speculated that overexpressing AtNDPK2 would protect the plant from ROS stress. To test this hypothesis, AtNDPK2-transgenic and -knockout mutants were subjected to oxidative stress induced by MV, an herbicide that elevates cellular ROS levels by inhibiting photosynthesis and photorespiration. MV is widely used as source of superoxide radicals in studies related to photosynthesis (44). The AtNDPK2-overexpressing transgenic plants were more tolerant to MV-mediated oxidative stress than were the wild type, whereas the mutants lacking AtNDPK2 were more sensitive (Fig. 7C). On the phenotypic level, the leaves from the transgenic plants appeared greener than those of wild type and the AtNDPK2-knockout mutants (Fig. 7C). This observation was confirmed by measuring the chlorophyll content of the leaves, which showed that the transgenic plants had 23% and 59.5% more chlorophyll than did the wild type and knockout mutant, respectively (data are the mean of four independent analysis). These results suggest that overexpression of AtNDPK2 enhances the tolerance of Arabidopsis plants to multiple stresses.
Discussion
In this paper, we describe biochemical and genetic studies that reveal AtNDPK2 participates in cellular redox regulation. We showed that (i) H2O2 induces the transient expression of AtNDPK2, (ii) AtNDPK2 specifically interacts with two H2O2-activated MAPKs, AtMPK3 and AtMPK6, as well as enhancing the MBP phosphorylation ability of AtMPK3, and (iii) overexpression of AtNDPK2 in plants leads to decreased constitutive ROS levels and enhanced tolerance to multiple environmental stresses that elicit ROS accumulation in situ. Although how these proteins interact and function together remains to be determined in planta, our observations suggest that AtNDPK2 is a component of a pathway specific to H2O2-activated MAPK signaling in plants. We suggest that the stress-tolerance function of AtNDPK2 resides in its ability to down-regulate cellular redox states caused by environmental stress, via specific activation of the H2O2-activated MAPKs, AtMPK3 and AtMPK6. Because AtNDPK2 was identified as a phytochrome-mediated light signal mediator (25), this model also suggests that light signals may modulate stress responses, in addition to protecting plants from stress.
Generation of ROS in plants has been implicated in abiotic and biotic stress responses, in which the level of ROS is an important cellular regulator for stress response as well as oxidative cell death. Therefore, it is crucial that plants maintain an adequate cellular redox state to make proper stress responses and overcome stress. The environmental stress response is accelerated under light, although there is no direct evidence that light signals are mediated by ROS in plants. For instance, phytochrome-mediated light signaling modulates cold/drought-induced gene expression (39) and salicylic acid (SA)-induced pathogenesis-related gene expression, as well as the hypersensitive response to pathogens (40, 45). In contrast, antioxidant-deficient transgenic plants induce lesions under strong light (8, 46), suggesting that light may be required for the amplification of an ROS response of sufficient amplitude to induce stress-mediated cell death. This possibility implies that plants need to activate ROS scavenger enzymes for normal growth and development under strong light.
Overexpression of AtNDPK2 clearly resulted in tolerance against several environmental stresses such as cold, salt, and H2O2 (Fig. 7), supporting our proposal that AtNDPK2 functions as a positive regulator in the down-regulation of the cellular redox state. However, because the atndpk2-null mutant was only slightly sensitive to cold and NaCl stress compared with the wild type (data not shown), we cannot completely rule out the possibility that other AtNDPK isoforms also may make a minor contribution to the regulation of the cellular redox state.
We found that AtNDPK2 is not rephosphorylated after its phosphate group has been transferred to its substrate protein (Fig. 6). This observation is consistent with an earlier observation (47) that Nm23, a human NDPK, has protein phosphotransferase activity rather than protein kinase activity. There may be two possible reasons why AtNDPK2 does not get rephosphorylated. First, AtNDPK2 is released from the interaction with its substrate in such a state that it cannot get rephosphorylated. Second, AtNDPK2 remains bound to its substrate protein AtMPK3 and is, therefore, not available for a new cycle of ATP binding and phosphate transfer. To elucidate further the mechanism by which AtNDPK2 regulates AtMPK3, it would be most helpful to study the structure and functions of these two proteins and focus on identification of the previously uncharacterized components that are associated with AtNDPK2-MAPK-signaling pathways.
The multiple stress tolerance of transgenic plants overexpressing AtNDPK2 is similar to that of transgenic plants overexpressing the constitutively active deletion mutant of ANP1 (13, 25). ANP1 initiates the phosphorylation cascade that involves AtMPK3 and AtMPK6 in a ROS- and light-dependent manner (13, 25). Dual localizations of AtNDPK2 to the nucleus and cytoplasm and its involvement in phytochrome A signaling and ROS-dependent up-regulation (Fig. 1) suggest an important role for NDPK2 in the environmental stress associated with ROS generation (25, 27). These results strongly indicate that AtNDPK2 may be an important upstream signaling component of the AtMPK3 and AtMPK6-mediated signaling cascade associated with stress tolerance in plants. How such multiple stress tolerance can arise from AtNDPK2 overexpression is suggested by our cDNA microarray studies (H.M., Gyeonghae Yang, Gyutae Kim, C. O. Lim, and D.-J. Yun, unpublished results), which showed that AtNDPK2 overexpression is associated with increased expression of a number of antioxidant genes, including peroxidase, glutathione reductase, glutathione transferase, thioredoxin reductase, peroxiredoxin, and protective genes encoding several heat-shock proteins. Thus, we suggest that AtNDPK2 mediates multiple stress tolerance by signaling the transient expression of genes involved in antioxidant and protective functions, possibly through activation of MAPK cascade.
Acknowledgments
This research was partially supported by Grant PF003103-00 from the Plant Diversity Research Center of the 21st Century Frontier Research Program, Ministry of Science and Technology (to D.-J.Y.), by Project Grant 297076-5 from the Ministry of Agriculture and Forestry, Korea (to D.-J.Y.), a grant from the Takara Biomedical Company, Korea (to C.O.L.), and a grant from the Biogreen 21 Program, Rural Development Administration, Korea (to S.-S.K.). H.M. and D.S. are supported by scholarships from the BK21 Program, Ministry of Education, Korea.
Abbreviations
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinase
- MAPKK
MAPK kinase
- NDPK
NDP kinase
- AtNDPK
Arabidopsis thaliana NDPK
- AtMPK
A. thaliana MAPK
- MBP
myelin basic protein
- AD
activation domain
- MV
methyl viologen
References
- 1.Finkel T, Holbrook N J. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Kannan K, Jain S K. Pathophysiology. 2000;7:153–163. doi: 10.1016/s0928-4680(00)00053-5. [DOI] [PubMed] [Google Scholar]
- 3.Hallwell B, Gutteridge J M C. Free Radicals in Biology and Medicine. 3rd Ed. New York: Oxford Univ. Press; 2002. [Google Scholar]
- 4.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 5.Bolwell G P, Wojtaszek P. Physiol Mol Plant Pathol. 1997;51:347–366. [Google Scholar]
- 6.Noctor G, Foyer C H. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 7.Grant J J, Loake G J. Plant Physiol. 2000;124:21–29. doi: 10.1104/pp.124.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamnongpol S, Willekens H, Langebartels C, Vanmontagu M, Inze D, Vancamp W. Plant J. 1996;10:491–503. [Google Scholar]
- 9.Takahashi H, Chen Z X, Du H, Liu Y D, Klessig D F. Plant J. 1997;11:993–1005. doi: 10.1046/j.1365-313x.1997.11050993.x. [DOI] [PubMed] [Google Scholar]
- 10.Jabs T. Biochem Pharmacol. 1999;57:231–245. doi: 10.1016/s0006-2952(98)00227-5. [DOI] [PubMed] [Google Scholar]
- 11.Sandermann H, Ernst D, Heller W, Langebartels C. Trends Plant Sci. 1998;3:47–50. [Google Scholar]
- 12.Schoffl F, Prandl R, Reindl A. Plant Physiol. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovtun Y, Chiu W L, Tena G, Sheen J. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tournier C, Thomas G, Pierre J, Jacquemin C, Pierre M, Saunier B. Eur J Biochem. 1997;244:587–595. doi: 10.1111/j.1432-1033.1997.00587.x. [DOI] [PubMed] [Google Scholar]
- 15.Wrzaczek M, Hirt H. Biol Cell. 2001;93:81–87. doi: 10.1016/s0248-4900(01)01121-2. [DOI] [PubMed] [Google Scholar]
- 16.Klotz L O, Pellieux C, Briviba K, Pierlot C, Aubry J M, Sies H. Eur J Biochem. 1999;260:917–922. doi: 10.1046/j.1432-1327.1999.00255.x. [DOI] [PubMed] [Google Scholar]
- 17.Machida Y, Nishihama R, Kitakura S. Crit Rev Plant Sci. 1997;16:481–496. [Google Scholar]
- 18.Mizoguchi T, Ichimura K, Shinozaki K. Trends Biotechnol. 1997;15:15–19. doi: 10.1016/S0167-7799(96)10074-3. [DOI] [PubMed] [Google Scholar]
- 19.Desikan R, Hancock J T, Ichimura K, Shinozaki K, Neill S J. Plant Physiol. 2001;126:1579–1587. doi: 10.1104/pp.126.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant J J, Yun B W, Loake G J. Plant J. 2000;24:569–582. doi: 10.1046/j.1365-313x.2000.00902.x. [DOI] [PubMed] [Google Scholar]
- 21.Otero A S. J Bioenerg Biomembr. 2000;32:269–275. doi: 10.1023/a:1005589029959. [DOI] [PubMed] [Google Scholar]
- 22.Cipollini G, Berti A, Fiore L, Rainaldi G, Basolo F, Bevilacqua G, Caligo M A. Int J Cancer. 1997;73:297–302. doi: 10.1002/(sici)1097-0215(19971009)73:2<297::aid-ijc22>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 23.Engel M, Seifert M, Theisinger B, Seyfert U, Welter C. J Biol Chem. 1998;273:20058–20065. doi: 10.1074/jbc.273.32.20058. [DOI] [PubMed] [Google Scholar]
- 24.Wagner P D, Vu N D. Biochem J. 2000;346:623–630. [PMC free article] [PubMed] [Google Scholar]
- 25.Choi G, Yi H, Lee J, Kwon Y K, Soh M S, Shin B, Luka Z, Hahn T R, Song P S. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- 26.Galvis M L, Marttila S, Hakansson G, Forsberg J, Knorpp C. Plant Physiol. 2001;126:69–77. doi: 10.1104/pp.126.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann S, Baumann A, Jaekel K, Marbach I, Engelberg D, Frohnmeyer H. J Biol Chem. 1999;274:17017–17024. doi: 10.1074/jbc.274.24.17017. [DOI] [PubMed] [Google Scholar]
- 28.Pan L, Kawai M, Yano A, Uchimiya H. Plant Physiol. 2000;122:447–452. doi: 10.1104/pp.122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barthel T K, Walker G C. J Biol Chem. 1999;274:36670–36678. doi: 10.1074/jbc.274.51.36670. [DOI] [PubMed] [Google Scholar]
- 30.Mesnildrey S, Agou F, Vero M. FEBS Lett. 1997;418:53–57. doi: 10.1016/s0014-5793(97)01292-1. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Lightner J, Warwick N, Browse J. Plant Physiol. 1997;113:347–356. doi: 10.1104/pp.113.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S G, Jin J B, Piao H L, Pih K T, Jang H J, Lim J H, Hwang I. Plant Mol Biol. 1998;38:437–447. doi: 10.1023/a:1006099718761. [DOI] [PubMed] [Google Scholar]
- 33.Hong J C, Nagao R T, Key J L. J Biol Chem. 1987;262:8367–8376. [PubMed] [Google Scholar]
- 34.Moon H, Baek D, Lee B, Prasad D T, Lee S Y, Cho M J, Lim C O, Choi M S, Bahk J, Kim M O, et al. Biochem Biophys Res Commun. 2002;290:457–462. doi: 10.1006/bbrc.2001.6208. [DOI] [PubMed] [Google Scholar]
- 35.Song E J, Kim Y S, Chung J Y, Kim E, Chae S K, Lee K J. Biochemistry. 2000;39:10090–10097. doi: 10.1021/bi000267a. [DOI] [PubMed] [Google Scholar]
- 36.Iwabuchi K, Li B, Bartel P, Fields S. Oncogene. 1993;8:1693–1696. [PubMed] [Google Scholar]
- 37.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J C, Lee S H, Cheong Y H, Yoo C M, Lee S I, Chun H J, Yun D J, Hong J C, Lee S Y, Lim C O, Cho M J. Plant J. 2001;25:247–259. doi: 10.1046/j.1365-313x.2001.00947.x. [DOI] [PubMed] [Google Scholar]
- 39.Kim H J, Kim Y K, Park J Y, Kim J. Plant J. 2002;29:693–704. doi: 10.1046/j.1365-313x.2002.01249.x. [DOI] [PubMed] [Google Scholar]
- 40.Genoud T, Millar A J, Nishizawa N, Kay S A, Schafer E, Nagatani A, Chua N H. Plant Cell. 1998;10:889–904. doi: 10.1105/tpc.10.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H, Bannenberg G L, Moldeus P, Shertzer H G. Arch Toxicol. 1994;68:582–587. doi: 10.1007/s002040050118. [DOI] [PubMed] [Google Scholar]
- 42.Mizoguchi T, Gotou Y, Nishida E, Yamaguchi-Shinozaki K, Hayashida N, Iwasaki T, Kamada H, Shinozaki K. Plant J. 1994;5:111–122. doi: 10.1046/j.1365-313x.1994.5010111.x. [DOI] [PubMed] [Google Scholar]
- 43.Sweetlove L J, Mowday B, Hebestreit H F, Leaver C J, Millar A H. FEBS Lett. 2001;508:272–276. doi: 10.1016/s0014-5793(01)03069-1. [DOI] [PubMed] [Google Scholar]
- 44.Kurepa J, Smalle J, Montagu M, Inze D. Plant J. 1998;14:759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- 45.Genoud T, Buchala A J, Chua N H, Metraux J P. Plant J. 2002;31:87–95. doi: 10.1046/j.1365-313x.2002.01338.x. [DOI] [PubMed] [Google Scholar]
- 46.Mach J M, Castillo A R, Hoogstraten R, Greenberg J T. Proc Natl Acad Sci USA. 2001;16:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engel M, Veron M, Theisinger B, Lacombe M L, Seib T, Dooley S, Welter C. Eur J Biochem. 1995;234:200–207. doi: 10.1111/j.1432-1033.1995.200_c.x. [DOI] [PubMed] [Google Scholar]